Abstract
We report an electrochemical biosensor combined with recombinase polymerase amplification (RPA) for rapid and sensitive detection of severe acute respiratory syndrome coronavirus 2. The electrochemical biosensor based on a multi-microelectrode array allows the detection of multiple target genes by differential pulse voltammetry. The RPA reaction involves hybridization of the RPA amplicon with thiol-modified primers immobilized on the working electrodes, which leads to a reduction of current density as amplicons accumulate. The assay results in shorter “sample-to-answer” times than conventional PCR without expensive thermo-cycling equipment. The limits of detection are about 0.972 fg/μL (RdRP gene) and 3.925 fg/μL (N gene), which are slightly lower than or comparable to that of RPA assay results obtained by gel electrophoresis without post-amplification purification. The combination of electrochemical biosensors and the RPA assay is a rapid, sensitive, and convenient platform that can be potentially used as a point-of-care test for the diagnosis of COVID-19.
Keywords: Coronavirus, Recombinase polymerase amplification, Electrochemical detection method, COVID-19, SARS-CoV-2
1. Introduction
Coronavirus disease 2019 (COVID-19) is a respiratory infectious disease and, based on the pairwise protein sequence analysis, it is part of the species of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Gorbalenya et al., 2020; Kilic et al., 2020; Organization, 2020; Zhou et al., 2020). The outbreak of COVID-19 started in December 2019, with the first case reported in Wuhan City, China, this virus outbreak is the result of an animal-to-human transmission event (Andersen et al., 2020; Broughton et al., 2020; Gorbalenya et al., 2020; Huajun et al., 2020; Kilic et al., 2020; Tan et al., 2020; C. Wang et al., 2020). COVID-19 is clinically mild but more contagious and spreads much faster than MERS or SARS (Ji et al., 2020). Also, there is increasing evidence that many patients with COVID-19 are asymptomatic, and these patients are likely to cause a new round of outbreaks (Broughton et al., 2020; Carter et al., 2020; Huang et al., 2020; Silveira et al., 2016; Won et al., 2020; Xiang et al., 2020; Yuan et al., 2020). Since the distribution of antiviral treatment or vaccines is still in the early stages, early diagnosis is important to reduce virulence (Lee and Lee, 2020). Hence, laboratory diagnosis of SARS-CoV-2 holds the key for the timely management and isolation of confirmed cases to prevent further transmission (Miripour et al., 2020).
Coronaviruses have several molecular targets within their positive-sense, single-stranded RNA genome that can be used for PCR assays, including the ORF1b or ORF8 regions and the nucleocapsid (N), spike (S) protein, RNA-dependent RNA polymerase (RdRP), and envelope (E) genes (Broughton et al., 2020; Chan et al., 2020; Chu et al., 2020; Corman et al., 2012; Huajun et al., 2020). Since these tests use two or more primers and probe sets to detect multiple regions in the virus, fluorescence resonance energy transfer (FRET) quenching is employed to monitor the increase in the number of amplicons indirectly during the reaction in real-time (Liang et al., 2020; Qu et al., 2020). However, this testing scheme requires complex analytical instruments, expensive reagents, and considerable expertise (Kilic et al., 2020).
Current serological tests, whether realized using laboratory-based enzyme-linked immunosorbent assay (ELISA) platforms or lateral flow serological assays designed for use as point-of-care tests (POCTs), have rapidly become available but with low accuracy, because antibodies might not be detected in the early stages of infection (Carter et al., 2020; Chan et al., 2020; Espejo et al., 2020; Ji et al., 2020; Tan et al., 2020; Vashist, 2020; Xiang et al., 2020). Currently, only molecular quantitative reverse transcription-polymerase chain reaction (PCR) (RT-qPCR) testing of respiratory tract samples is recommended for the identification and diagnosis of COVID-19 cases (Chan et al., 2020; Corman et al., 2012; Huajun et al., 2020; Joung et al., 2020; Kilic et al., 2020; Nagura-Ikeda et al., 2020; Tahamtan and Ardebili, 2020; Tang et al., 2020; Vashist, 2020; Yishan Wang et al., 2020; Yang et al., 2020; Yuan et al., 2020; Zhang et al., 2020). Isothermal amplification techniques, such as recombinase polymerase amplification (RPA) and loop-mediated isothermal amplification (LAMP), are alternative methods that feature high sensitivity, specificity, and rapidity under isothermal conditions (Esbin et al., 2020; Joung et al., 2020; Kashir and Yaqinuddin, 2020; Kim et al., 2019b; Nassir et al., 2020; Won et al., 2020; Zhang et al., 2020). Because these methods significantly simplify the protocol without requiring a thermocycler, isothermal amplification has been combined with various forms of biosensors that provide simple, fast, disposable, and low-cost diagnosis.
In this study, we report an electrochemical biosensor coupled with RPA for the rapid and sensitive detection of SARS-CoV-2. An electrochemical biosensor based on microelectrode array microchips, comprising a reference electrode, counter electrode, and five individual working electrodes, allows for the detection of multiple target genes by differential pulse voltammetry (DPV). The isothermal RPA reaction involves hybridization of RPA amplicons with thiol-modified primers immobilized on the working electrodes, reducing the current density as amplicons accumulate (Fig. 1 ).
Fig. 1.
Electrochemical biosensor combined with isothermal amplification. Schematic of an electrochemical biosensor combined with recombinase polymerase amplification (RPA). The RPA reaction occurs on the working electrodes, and amplicon detection is quantified by differential pulse voltammetry (DPV).
2. Materials and methods
2.1. Reagents and equipment
The TwistAmp® Basic kit for RPA was purchased from TwistDx Limited (UK). The primers were synthesized and purified by Bionics Inc. (Korea). The plasmids containing a sequence of SARS-CoV-2 N and RdRP genes were purchased from GenScript (USA) and used as a template for the RPA assay. Polydimethylsiloxane (PDMS) was obtained from Dow Corning (USA). K3[Fe(CN)6] powder and KCl were obtained from Sigma-Aldrich (Korea). All the chemicals used were of analytical grade and used as received without further purification. Electrochemical measurements were carried out using an electrochemical analyzer (CHI 830B from CH Instruments, USA). X-ray photoelectron spectroscopy (XPS, ESCALAB 250, USA) with an AlKα X-RAY source and a monochromator under ultrahigh vacuum (1 × 10−9 Torr) was carried out to confirm the presence of the AgCl layer.
2.2. Fabrication of electrochemical device with coplanar electrode configuration
The technique used to produce microelectrodes was adapted from our previous work (Kim et al., 2019; Kim et al., 2020; Schuck et al., 2020). A 5 cm × 5 cm bare glass substrate was used for the coplanar microelectrode array. The substrate was cleaned by immersing in 50 mL of acetone and sonicating for 20 min. Acetone residues and remaining foreign substances were cleaned with isopropanol in the same manner as the acetone step, and the substrate was placed in an oven (75 °C) to dry for 10 min. The residue-free bare glass was spin-coated with a positive photoresist (PR) and pre-baked for 10 min at 95 °C. Subsequently, the photomask was aligned over the PR and patterned by a photolithography method using a mask aligner (MA-6, Karl-Suss). Ag was deposited over the substrate (glass/patterned PR layer) using a thermal evaporator system (Evaporation System SHE-6T-350D). The fabricated glass/patterned PR/Au thin film layer was soaked in acetone and sonicated until the PR/Au thin film layer was lifted-off. Using the same fabrication method as for the Ag electrodes (RE), the counter electrode (CE) and working electrode (WE) were fabricated using Au. For the formation of the AgCl layer, simple immersion was performed under FeCl3 as a bleach solution for 1 min at 30 s intervals, which showed the highest current density of the redox reaction in cyclic voltammetry (CV) measurements.
2.3. Recombinase polymerase amplification assay preparation
The N gene and RdRP gene of SARS-CoV-2 were used as target templates (Cheong et al., 2020; Kashir and Yaqinuddin, 2020; Kilic et al., 2020; Won et al., 2020). The primers were ordered from Bionics (Suwon, Korea) with requested thiol modification at the end of the forward primer. The NTC, including all reagents except template DNA, was used as a negative control. For the RPA reaction, the TwistDx Limited (Maidenhead, UK) protocol was used (Huang et al., 2020; Kilic et al., 2020; Magro et al., 2017; Yeh et al., 2017). Rehydration buffer (29.5 μL), template (1 μL), 10 μM thiol-modified forward primer and reverse primer (2.4 μL each), nuclease-free water (11.2 μL), and 280 mM of MgOAc solution (2.5 μL) were added in a microtube or slab with a cylindrical hole to start the RPA reaction. The prepared RPA solution (50 μL of total volume) was stored at human body temperature for 20 min.
2.4. Measurement and characterization of the electrochemical device
Cyclic voltammetry (CV) and differential pulse voltammetry (DPV) for electrochemical measurements were carried out using CHI 830B instrument (CH Instruments, USA). Potassium ferricyanide (K3[Fe(CN)6]) solution (5 mM) containing KCl (0.1 M) was used as the supporting electrolyte for both CV and DPV measurements (Akanda et al., 2016; Vogt et al., 2016). CV measurements were used to investigate the stability and reproducibility of the electrochemical device. The scanning potential range was set from −0.2 V to 0.4 V at a scan rate of 100 mV/s. The DPV measurement was made to investigate the movement of electrons of K3[Fe(CN)6] on the working electrode. The DPV study was performed in a scan range of −0.2 V to 0.4 V, with a pulse amplitude of 0.05 V, a pulse width of 0.05 s, and a pulse period of 0.2 s. All the electrical measurements were performed at room temperature.
3. Results and discussion
3.1. Characterization of the electrochemical biosensor
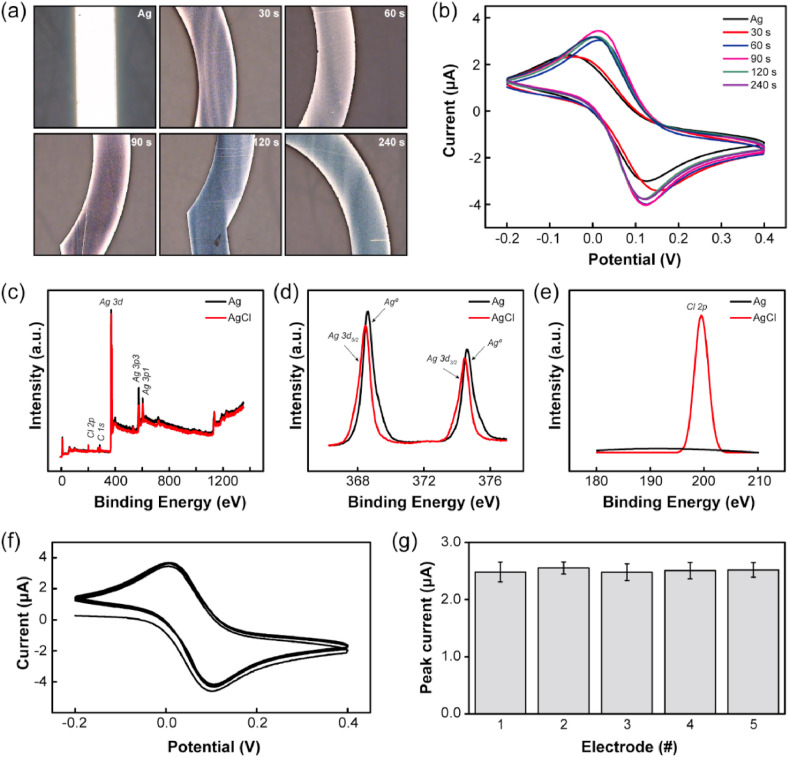
An electrochemical biosensor with a microelectrode array was fabricated on a glass substrate using microfabrication (Figure S1 and Method section for more details). The multi-electrode array structure guarantees uniformity, increasing the reproducibility of our device since they were all fabricated under the same conditions. Also, each electrode can be used for different targets or to confirm the presence of a single target at least three times, then avoiding an incorrect diagnosis. As a reference electrode, Ag/AgCl was used because it is simple, stable, and has less potential hysteresis against temperature cycles (Kurkina et al., 2011; Punter-Villagrasa et al., 2017; Yoon et al., 2020). For the formation of the AgCl layer, simple immersion in a bleach solution was performed for a short time (up to 4 min in 30 s intervals). Optical images before and after chlorination showed that the part of the electrode in contact with the solution became dark purple because of the formation of silver chloride particles (Fig. 2 a). The highest current density of the redox reaction was shown by cyclic voltammetry (CV) measurements in the 90 s sampling interval (Fig. 2b). The formation of the AgCl layer was confirmed by X-ray photoelectron spectroscopy (XPS) (Fig. 2c–e). The peaks at 367.5 and 373.5 eV were assigned to the binding energies of Ag 3d 5/2 and Ag 3d 3/2 of Ag+ in AgCl, respectively, whereas the peaks at 368.8 and 374.6 eV are attributed to metallic Ag0. For Cl 2p, a peak was observed at a binding energy of approximately 200 eV.
Fig. 2.
Characterization of the Ag/AgCl electrode. (a–b) Optical images (a) and cyclic voltammetry measurements (b) obtained from an Ag electrode after AgCl formation by immersing in a bleach solution for 30 s intervals. (c–e) X-ray photoelectron spectroscopy (XPS) spectra of Ag and AgCl electrodes: (c) survey spectrum, (d) Ag 3d 5/2 and Ag 3d 3/2 and (e) Cl 2p regions. (f–g) stability (f) and reproducibility (g) of the electrochemical biosensors by CV measurements.
For the stability and reproducibility of the electrochemical biosensors, multi-array working electrodes were fabricated in the same conditions with Au. The gold electrode has excellent binding properties with thiol modified primer and it is a biocompatibility material. In addition, since the oxidation-reduction reaction does not occur easily, it has stable properties during the electrochemical reaction (Aravamudhan et al., 2007; Walter et al., 2002). The electrochemical biosensors were investigated via CV measurements under a potential range of −0.2 to 0.4 V at a scan rate of 0.1 V/s in 5 mM of potassium ferricyanide (K3[Fe(CN)6]) solution containing 0.1 M KCl as a supporting electrolyte. The supporting electrolyte was sufficiently saturated within three to five consecutive cycles (Fig. 2f). To evaluate the reproducibility, five individual working electrodes were tested. The relative standard deviation (RSD) of the peak currents of these five electrodes was less than 0.07 μA after three consecutive cycles, which is negligible for quantitative detection (Fig. 2g).
3.2. Verification of RPA in combination with electrochemical detection
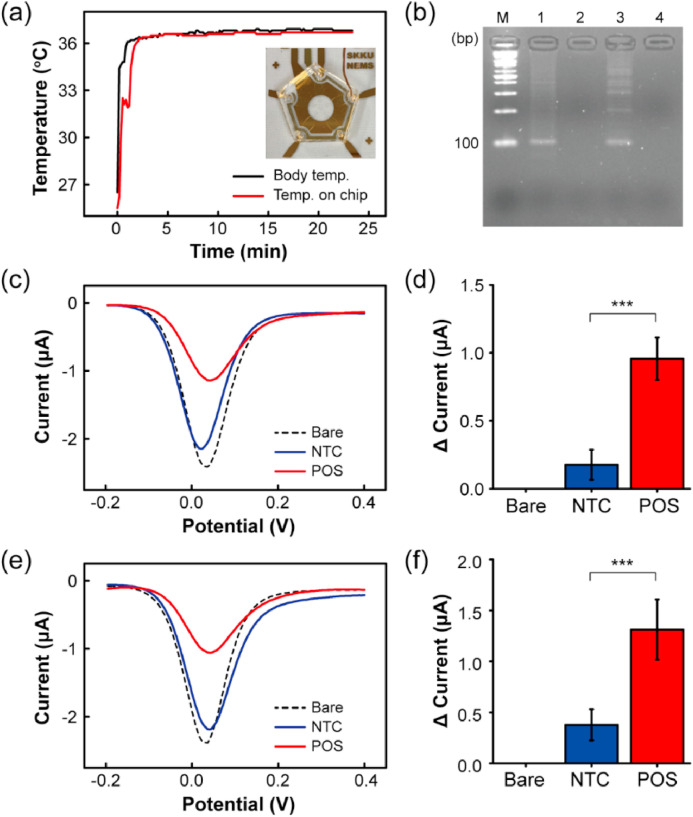
The RPA reactions were carried out according to the manufacturer's instructions (see the Methods section for details) using sequences published previously for targeting the nucleocapsid (N) gene and RNA-dependent RNA polymerase (RdRP) gene of SARS-CoV-2 (Binnicker, 2020; Chan et al., 2020; Kashir and Yaqinuddin, 2020). The sequence information of the RPA primers is described in Table S1. The primers were thiol-modified to increase adsorption on the electrode (Jayakumar et al., 2018; White et al., 2015). The feasibility of the assay was confirmed by gel electrophoresis following DNA purification (Figure S2). A no-template control (NTC), including all reagents except for the template DNA, was used in all the reactions. Gel electrophoresis confirmed the presence of the amplified target DNA (~100 bp). To validate the feasibility of our electrochemical biosensor under human body temperature, on-chip RPA was carried out for 20 min. Reactions for targeting the N gene and RdRP gene were performed in different electrodes, and a polydimethylsiloxane (PDMS) slab with a cylindrical hole of 25 mm diameter was prepared as a reaction chamber. During the RPA reaction, the surface temperature of one electrode was measured using a thermocouple. In less than 3 min, the reaction chamber reached a temperature of 36.4 ± 0.2 °C, close to the human body temperature (Fig. 3 a). A specific target amplicon was observed by agarose gel electrophoresis (Fig. 3b). For the DPV measurement, unhybridized DNA fragments were removed with PBS so that the amplicon hybridized with thiol-modified primers remained on the working electrode. DPV was recorded in a potential range of −0.2 to 0.4 V, pulse amplitude of 0.05 V, pulse width of 0.05, and pulse period of 0.2 s in 5 mM potassium ferricyanide (K3[Fe(CN)6]) solution containing 0.1 M KCl. The peak current decreased compared to that measured at the working electrode without the template DNA for targeting the N gene and RdRP gene because the amplicons hindered the movement of K3[Fe(CN)6] electrons (Fig. 3c and e). A slight decrease in the peak current of the NTC appeared because of the adsorption of primers on the working electrode. A significant decrease in the peak current was observed for the positive samples and was significantly different from the NTC (***: p-value < 0.001) (Fig. 3d and f).
Fig. 3.
Feasibility verification of RPA in combination with electrochemical detection. The surface temperature of the Ag/AgCl electrode (a) and agarose gel electrophoresis analysis (b) for confirmation of the RPA assay. DPV signals obtained by on-chip RPA performed with specific target N gene (c) and RdRP gene (e). Change in the peak current for N gene (d) and RdRP gene (f) compared with an NTC and a blank control (***: p-value < 0.001.
3.3. Sensitivity of RPA in combination with electrochemical detection
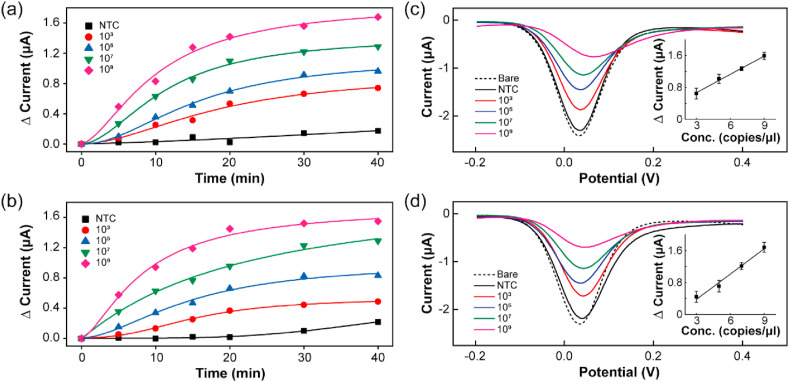
The sensitivity of the RPA-coupled electrochemical biosensor was investigated by detecting DPV signals at different template concentrations ranging from 109 to 103 copies. The assay was carried out for 40 min at human body temperature, and the signals were collected at 5 min intervals. The logistic fittings of the peak current change (Δcurrent) depending on the time for targeting the N gene and RdRP gene are shown in Fig. 4 a and Fig. 4b, respectively. The DPV signals depending on the time for each concentration of the N gene and RdRP gene are shown in Figure S3 and Figure S4, respectively. The absolute current intensity decreased with increasing template concentration and time due to increased adsorption of the amplicons on the electrode. Thus, the Δcurrent values were proportional to concentrations in the range of 109 to 103 copies within 20 min. After 20 min, the Δcurrent was saturated with complete adsorption of the amplicons on the electrode. The DPV signals and Δcurrent values for the N gene and RdRP gene at 20 min depending on the template concentration are shown in Fig. 4c and d, respectively. Inset shows the linearity of the peak current to the target gene concentration. The relative standard deviation (RSD) of the peak currents of these error bar was less than 0.14 μA. The standard deviation (SD) of NTC and the slope (S) were used to calculate the LoD applying the formula: 3 × (SD/S) (Hong et al., 2018). The standard curves in the inset show a linear relationship (R2 > 0.99) for both genes (RdRp, N) with a detection limits of 0.972 fg/μL (RdRP gene) and 3.925 fg/μL (N gene), which are slightly lower than or comparable to that of the RPA assay results obtained by gel electrophoresis without post-amplification purification (Figure S5). The specificity of our device could be improved by employing carbon nanomaterials (such as graphene oxide and graphene nanowalls) over the working electrode for detection using electrochemical methods as demonstrated by other studies (Akhavan et al, 2012, 2014; Yi Wang et al., 2020). However, the performance of our RPA-coupled electrochemical device has already potential to provide a fast and accurate detection for the SARS-CoV-2 genome and the multiplexed structure could be an advantage to detect different strains of the virus, allowing a more varied and safe diagnosis.
Fig. 4.
Reaction sensitivity of RPA coupled with electrochemical detection. The assay concentration ranged from 103 to 109 copies for 40 min under human body temperature. The variation in the peak current for each DPV curve measured during 40 min depending on different target concentrations: N gene (a) and RdRP gene (b) as well as for an NTC. DPV signals obtained at 20 min depending on template concentration: N gene (c) and RdRP gene (d). Inset shows the change in the peak current with respect to the target gene concentration.
4. Conclusions
In summary, the RPA-coupled electrochemical detection method was proposed for SARS-CoV-2 genome detection. This protocol allows the detection of target genes in less than 20 min at the human body temperature. For on-chip RPA, a PDMS slab with a cylindrical hole 25 mm in diameter was prepared for a reaction chamber, and electrodes were coated with thiol-modified primers. During RPA, the amplicon demonstrated a lower DPV signal because of the electrostatic repulsion between the negatively charged amplicon and the electrode. The limit of detection was slightly lower than or comparable to that of the RPA assay results obtained by gel electrophoresis without post-amplification purification. The data suggest that the combination of RPA with an electrochemical biosensor can be utilized as a rapid, sensitive, and convenient DNA detection platform under human body temperature instead of using an external temperature controller.
CRediT authorship contribution statement
Hyo Eun Kim: Conceptualization, Methodology, Investigation, Software, Formal analysis, Writing – original draft, Writing – review & editing. Ariadna Schuck: Methodology, Software, Formal analysis, Writing – review & editing. See Hi Lee: Formal analysis, Investigation. Yunjong Lee: Samples supply, Conceptualization. Minhee Kang: Samples supply, Conceptualization, Formal analysis, Writing – review & editing, Supervision. Yong-Sang Kim: Conceptualization, Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported in part by a National Research Foundation of Korea grant funded by the Korean government (No. NRF-2018R1D1A1B05049787).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113168.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Akanda M.R., Sohail M., Aziz M.A., Kawde A.N. Electroanalysis. 2016;28:408–424. [Google Scholar]
- Akhavan O., Ghaderi E., Rahighi R. ACS Nano. 2012;6:2904–2916. doi: 10.1021/nn300261t. [DOI] [PubMed] [Google Scholar]
- Akhavan O., Ghaderi E., Rahighi R., Abdolahad M. Carbon N. Y. 2014;79:654–663. [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravamudhan S., Kumar A., Mohapatra S., Bhansali S. Biosens. Bioelectron. 2007;22:2289–2294. doi: 10.1016/j.bios.2006.11.027. [DOI] [PubMed] [Google Scholar]
- Binnicker M.J. Clin. Chem. 2020;66:664–666. doi: 10.1093/clinchem/hvaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Yip C.C.-Y., To K.K.-W., Tang T.H.-C., Wong S.C.-Y., Leung K.-H., Fung A.Y.-F., Ng A.C.-K., Zou Z., Tsoi H.-W., Choi G.K.-Y., Tam A.R., Cheng V.C.-C., Chan K.-H., Tsang O.T.-Y., Yuen K.-Y. J. Clin. Microbiol. 2020;58:1–10. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong J., Yu H., Lee C.Y., Lee J., Choi H., Lee J.-H., Lee H., Cheon J. Nat. Biomed. Eng. 2020;4:1159–1167. doi: 10.1038/s41551-020-00654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Clin. Chem. 2020;555:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M., van Boheemen S., Gopal R., Ballhause M., Bestebroer T.M., Muth D., Müller M.A., Drexler J.F., Zambon M., Osterhaus A.D., Fouchier R.M., Drosten C. Euro Surveill. 2012;17:1–6. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Rna. 2020;26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo A.P., Akgun Y., Al Mana A.F., Tjendra Y., Millan N.C., Gomez-Fernandez C., Cray C. Am. J. Clin. Pathol. 2020;154:293–304. doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.A., Kim Y.J., Kim S.J., Yang S. Biosens. Bioelectron. 2018;107:103–110. doi: 10.1016/j.bios.2018.01.067. [DOI] [PubMed] [Google Scholar]
- Huajun B., Cai X., Zhang X. 2020. OSF. [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakumar K., Camarada M.B., Dharuman V., Rajesh R., Venkatesan R., Ju H., Maniraj M., Rai A., Barman S.R., Wen Y. ACS Appl. Mater. Interfaces. 2018;10:21541–21555. doi: 10.1021/acsami.8b03236. [DOI] [PubMed] [Google Scholar]
- Ji T., Liu Z., Wang G., Guo X., Akbar khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Segel M., Bruneau R., Huang M.-L.W., Kim N.-G., Yu X., Li J., Walker B.D., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. medRxiv Prepr. Serv. Heal. Sci. 2020 [Google Scholar]
- Kashir J., Yaqinuddin A. Med. Hypotheses. 2020;141:109786. doi: 10.1016/j.mehy.2020.109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic T., Weissleder R., Lee H. iScience. 2020;23:101406. doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.E., Schuck A., Lee J.H., Kim Y.-S. Sensor. Actuator. B Chem. 2019;291 [Google Scholar]
- Kim H.E., Schuck A., Oh J., Jung K.-M., Kim Y.-S. Solid State Electron. 2020;167:107750. [Google Scholar]
- Kim J.H., Kang M., Park E., Chung D.R., Kim J., Hwang E.S. BioChip J. 2019;13:341–351. doi: 10.1007/s13206-019-3404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkina T., Vlandas A., Ahmad A., Kern K., Balasubramanian K. Angew. Chem. Int. Ed. 2011;50:3710–3714. doi: 10.1002/anie.201006806. [DOI] [PubMed] [Google Scholar]
- Lee D., Lee J. Transp. Res. Interdiscip. Perspect. 2020;5:100111. doi: 10.1016/j.trip.2020.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K.-H., Chang T.-J., Wang M.-L., Tsai P.-H., Lin T.-H., Wang C.-T., Yang D.-M. J. Chinese Med. Assoc. Publish Ah. 2020:701–703. doi: 10.1097/JCMA.0000000000000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro L., Jacquelin B., Escadafal C., Garneret P., Kwasiborski A., Manuguerra J.-C., Monti F., Sakuntabhai A., Vanhomwegen J., Lafaye P., Tabeling P. Sci. Rep. 2017;7:1347. doi: 10.1038/s41598-017-00758-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miripour Z.S., Sarrami-Forooshani R., Sanati H., Makarem J., Taheri M.S., Shojaeian F., Eskafi A.H., Abbasvandi F., Namdar N., Ghafari H., Aghaee P., Zandi A., Faramarzpour M., Hoseinyazdi M., Tayebi M., Abdolahad M. Biosens. Bioelectron. 2020;165:112435. doi: 10.1016/j.bios.2020.112435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagura-Ikeda M., Imai K., Tabata S., Miyoshi K., Murahara N., Mizuno T., Horiuchi M., Kato K., Imoto Y., Iwata M., Mimura S., Ito T., Tamura K., Kato Y. J. Clin. Microbiol. 2020;58:1–9. doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassir A.A., Baptiste M.J., Mwikarago I., Habimana M.R., Ndinkabandi J., Murangwa A., Nyatanyi T., Muvunyi C.M., Nsanzimana S., Leon M., Musanabaganwa C. medRxiv. 2020 2020.09.17.20196402. [Google Scholar]
- Organization, W.H . 2020. Coronavirus Disease ( COVID-19) ([WWW Document]) [Google Scholar]
- Punter-Villagrasa J., Colomer-Farrarons J., del Campo F.J., Miribel P. Springer International Publishing; Cham: 2017. Amperometric and Impedance Monitoring Systems for Biomedical Applications, Bioanalysis. [Google Scholar]
- Qu S., Sun F., Qiao Z., Li J., Shang L. Small. 2020;16:1907633. doi: 10.1002/smll.201907633. [DOI] [PubMed] [Google Scholar]
- Schuck A., Kim H.E., Jung K.-M., Hasenkamp W., Kim Y.-S. Biosens. Bioelectron. 2020;157:112167. doi: 10.1016/j.bios.2020.112167. [DOI] [PubMed] [Google Scholar]
- Silveira C., Monteiro T., Almeida M. Biosensors. 2016;6:51. doi: 10.3390/bios6040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.C., Tiu C., Hu Z., Chen V.C.W., Young B.E., Sia W.R., Tan Y.J., Foo R., Yi Y., Lye D.C., Anderson D.E., Wang L.F. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Tang Y., Schmitz J.E., Persing D.H., Stratton C.W. J. Clin. Microbiol. 2020;58:1–9. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S.K. 2020. Diagnostics 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt S., Su Q., Gutiérrez-Sánchez C., Nöll G. Anal. Chem. 2016;88:4383–4390. doi: 10.1021/acs.analchem.5b04814. [DOI] [PubMed] [Google Scholar]
- Walter E.C., Favier F., Penner R.M. Anal. Chem. 2002;74:1546–1553. doi: 10.1021/ac0110449. [DOI] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Yishan, Kang H., Liu X., Tong Z. J. Med. Virol. 2020;92:538–539. doi: 10.1002/jmv.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Yi, Jiao W., Wang Yu, Wang Ya-cui, Shen C., Qi H., Shen A.-D. Microchim. Acta. 2020;187:667. doi: 10.1007/s00604-020-04637-5. [DOI] [PubMed] [Google Scholar]
- White S.P., Dorfman K.D., Frisbie C.D. Anal. Chem. 2015;87:1861–1866. doi: 10.1021/ac503914x. [DOI] [PubMed] [Google Scholar]
- Won J., Lee S., Park M., Kim T.Y., Park M.G., Choi B.Y., Kim D., Chang H., Kim V.N., Lee C.J. Exp. Neurobiol. 2020;29:107–119. doi: 10.5607/en20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J., Zhou Q., Ye H., Ma Y., Li H., Wei X., Cai P., Ma W.L. Clin. Infect. Dis. 2020:1–5. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Wang Y.C., Shen C.F., Cheng C.M. Diagnostics. 2020;10:9–11. [Google Scholar]
- Yeh, E.-C., Fu, C.-C., Hu, L., Thakur, R., Feng, J., Lee, L.P., 2017. 1–12...
- Yoon S., Kang D., Sohn S., Park J., Lee M., Choi S. J. Nucl. Fuel Cycle Waste Technol. 2020;18:143–155. [Google Scholar]
- Yuan X., Yang C., He Q., Chen J., Yu D., Li J., Zhai S., Qin Z., Du K., Chu Z., Qin P. ACS Infect. Dis. 2020 doi: 10.1021/acsinfecdis.0c00365. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Odiwuor, N., Xiong, J., Sun, L., Nyaruaba, R.O., Wei, H., Tanner, N., 2020. 2...
- Zhou P., Yang X.Lou, Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.Di, Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.