Abstract
Mutations in nuclear genes encoding for mitochondrial proteins very long-chain acyl-CoA dehydrogenase (VLCAD) and trifunctional protein (TFP) cause rare autosomal recessive disorders. Studies in fibroblasts derived from patients with mutations in VLCAD and TFP exhibit mitochondrial defects. To gain insights on pathological changes that account for the mitochondrial deficits we performed quantitative proteomic, biochemical, and morphometric analyses in fibroblasts derived from subjects with three different VLCAD and three different TFP mutations. Proteomic data that was corroborated by antibody-based detection, indicated reduced levels of VLCAD and TFP protein in cells with VLCAD and TFP mutations respectively, which in part accounted for the diminished fatty acid oxidation capacity. Decreased mitochondrial respiratory capacity in cells with VLCAD and TFP mutations was quantified after glucose removal and cells with TFP mutations had lower levels of glycogen. Despite these energetic deficiencies, the cells with VLCAD and TFP mutations did not exhibit changes in mitochondria morphology, distribution, fusion and fission, quantified by either confocal or transmission electron microscopy and corroborated by proteomic and antibody-based protein analysis. Fibroblasts with VLCAD and to a lesser extend cells with TFP mutations had increased levels of mitochondrial respiratory chain proteins and proteins that facilitate the assembly of respiratory complexes. With the exception of reduced levels of catalase and glutathione S-transferase theta-1 in cells with TFP mutations, the levels of 45 proteins across all major intracellular antioxidant networks were similar between cells with VLCAD and TFP mutations and non-disease controls. Collectively the data indicate that despite the metabolic deficits, cells with VLCAD and TFP mutations maintain their proteomic integrity to preserve cellular and mitochondria architecture, support energy production and protect against oxidative stress.
Keywords: Long-chain fatty acids (LCFA), Beta-oxidation, Mitochondria, Proteomics, Very long-chain acyl-CoA dehydrogenase (VLCAD), Trifunctional protein (TFP)
1. Introduction
Fatty acid β-oxidation is the primary pathway for the metabolism and conversion of long-chain fatty acids (LCFA) to energy [[1], [2], [3]]. Approximately 25 proteins that are broadly subdivided into three functional clusters participate in this metabolic pathway. The first cluster includes transporters and enzymes that import and convert fatty acids to acyl CoA species. The second group includes proteins that participate in the carnitine cycle, which transforms fatty acids to acylcarnitines and actively transport acylcarnitines into mitochondria. Finally, proteins embedded within the inner mitochondrial membrane execute the β-oxidation of fatty acids in a series of four cyclical reactions to generate acetyl-CoA. The β-oxidation of fatty acids also generates NADH and FADH2 to support oxidative phosphorylation and ATP production. The first step in the oxidation of fatty acids with 14 or more carbons is executed by VLCAD, a homodimer encoded by the ACADVL gene. The next 3 enzymatic steps are executed by TFP, which is composed of four α and four β subunits (α4β4) that are encoded by two different genes HADHA and HADHB, respectively. The HADHA encoded α-subunits contain the activities of enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase (LCHAD) and the HADHB encoded β-subunits the 3-ketoacyl-CoA thiolase activity.
Defects in nuclear genes encoding for VLCAD and TFP cause rare inherited autosomal recessive diseases that present primarily with hypoglycemia, cardiomyopathy, intermittent muscle breakdown (rhabdomyolysis), and liver failure [[1], [2], [3], [4], [5], [6], [7], [8], [9]]. Clinical management includes reduction of dietary intake of LCFA, fasting avoidance, high carbohydrate diet supplemented with medium chain triglycerides and in some cases carnitine [10,11]. Despite the advances in detection and clinical management, patients with these disorders still experience life-long symptoms since the molecular and biochemical mechanisms that drive disease phenotypes remain insufficiently characterized.
Skin-derived fibroblasts from subjects with mutations in either VLCAD or TFP have been used for routine disease diagnosis, investigations relating to disease mechanisms and screening of potential therapeutics [5,6,[12], [13], [14], [15], [16], [17], [18], [19]]. Studies have also explored the metabolic deficits that result from VLCAD or TFP mutations and the potential impact on mitochondrial bioenergetics [[20], [21], [22], [23]]. Case reports [24,25], and a comprehensive study [26], explored the impact of TFP and VLCAD mutations on mitochondria morphology using muscle and liver biopsies. Swollen mitochondrial with distorted cristae primarily in degenerating muscle fibers, in some but not all subjects with the common c.1528G > C mutation in the α-subunit of TFP were observed [26]. A recent study expanded these observations to show a reduction in mitochondrial networks and fusion/fission defects in fibroblasts from 8 individuals with the c.1528G > C mutation TFP and 1 individual with c.881T > C/c.1438C > T VLCAD mutation [22]. To further improve our basic understanding of how mutations in these proteins impact mitochondria morphology and function we performed label-free proteomic quantification, biochemical assays and morphometric evaluation using both confocal and transmission electron microscopy in fibroblasts with 3 different VLCAD or 3 different TFP mutations. The proteomic profiling coupled with the biochemical and morphometric analyses indicate that proteomic changes in cells with VLCAD and TFP mutations may lessen the metabolic deficits by augmenting the levels of proteins that support mitochondrial structure and function as well as cellular structural integrity.
2. Materials and methods
2.1. Fibroblasts
All fibroblasts utilized in this study are human primary cell lines. The cells lines with mutations in VLCAD and TFP proteins were purchased from the Coriell Institute for Medical Research (Camden, New Jersey, USA) or collected by the Vockley laboratory with written informed consent obtained from patients and/or parents under protocols approved by the Institutional Review Board at the University of Pittsburgh. The non-disease cell lines were purchased from ATCC (PCS-201-012-control 1), Sigma-Aldrich (#106-05A-control 2) and Cell Science (#2320-control 3), control 4 was a gift from the laboratory of Jordan S. Orange, MD, PhD Columbia Vagelos College of Physicians and Surgeons. The study was carried out with the approval of the Children's Hospital of Philadelphia institutional biosafety committee (IBC# 198-03-001) in accordance with the approved guidelines and regulations. Cells were grown in Dulbecco's Modified Eagle's Medium DMEM (Invitrogen) containing 4.5 g/L glucose, 15% fetal bovine serum (Sigma-Aldrich) in 5% CO2 at 37 °C. The fibroblasts with VLCAD and TFP mutations shared a similar spindle-shaped morphology to non-disease controls and they adhered to plastic. Routine testing for cell duplication and viability did not reveal any significant changes between cells with mutations and controls.
2.2. Fatty acid oxidation (FAO) flux
FAO flux quantifies the rate of [9,10-3H] Palmitate (PerkinElmer, Waltham, MA) oxidation. [9,10-3H] Palmitate is emulsified in Krebs/BSA (4.5 mg/ml) solution to a final concentration of 23 μM overnight at 37 °C under continuous agitation. The fibroblasts were seeded in 24-well plates (1 × 105 cell/well) and were incubated for 4 h with 200 μl of the [9,10-3H] palmitate emulsified Krebs/BSA solution. The 3H2O released in the cell media was quantified at the end of the 4-h incubation. Etomoxir (2[6(4-chlorophenoxy) hexyl]oxirane-2-carboxylate, 10 μM) an irreversible inhibitor of carnitine palmitoyltransferase-1 (CPT-1α) was used to document mitochondrial-dependent β-oxidation of palmitate. The assay was performed in three different cell preparations for each cell type using cells of similar passage (p4-p10). For each independent determination, non-disease and fibroblasts with mutations were analyzed in duplicate. For this assay we used control, non-disease fibroblast lines 1 and 2. The flux is expressed in pmol/h/mg and the data is reported as the percent of the control 1 fibroblasts.
2.3. Proteomic analysis, mass spectrometry data acquisition, spectral library generation and quantification of protein relative levels
Fibroblasts (non-disease control lines 1, 2, 3 and cells with mutations in VLCAD and TFP Supplementary Fig. S1) were grown in T75 flasks at 90% confluence (about 1 × 106 cells) harvested by gentle scraping and cell pellets were collected. The pellets were washed 3 times with cold PBS. Cell pellets were lysed by addition of SDC buffer containing TCEP and 2-chloroacetamide (PreOmics). Benzonase was added and samples were triturated and heated at 95 °C for 10 min. Debris were cleared by centrifugation; protein concentration was quantified by the Bradford assay and 50 μg of protein was acetone/trichloroacetic acid precipitated. The resulting pellet was solubilized and digested with the iST kit (PreOmics GmbH, Martinsried, Germany). Briefly, solubilization, reduction, and alkylation were performed in sodium deoxycholate (SDC) buffer containing TCEP and 2-chloroacetamide. Proteins were enzymatically hydrolyzed for 1.5 h at 37 °C by addition of LysC and trypsin. Peptides were de-salted, dried by vacuum centrifugation and were reconstituted in 0.1% TFA containing indexed Retention Time (iRT) peptides (Biognosys).
Samples for the acquisition of the whole proteome were analyzed on a QExactive HF mass spectrometer (Thermofisher Scientific San Jose, CA) as described in detail previously [27]. Briefly, tryptic digests of cell protein pellets were separated by reverse-phase (RP)-HPLC and spectra were collected in data-independent acquisition (DIA) and data-dependent acquisition (DDA). For DDA, the mass spectrometer was set to repetitively scan m/z from 300 to 1400 (R = 240,000) followed by data-dependent MS/MS scans on the twenty most abundant ions, minimum AGC 1e4, dynamic exclusion with a repeat count of 1, repeat duration of 30s, (R = 15000) FTMS full scan AGC target value was 3e6, while MSn AGC was 1e5, respectively. MSn injection time was 160 ms; microscans were set at one. Rejection of unassigned and 1+, 6–8 charge states were set. The raw files for Data Independent Acquisition (DIA) were collected using the following settings: one full MS scan at 120,000 resolution and a scan range of 300–1650 m/z with an AGC target of 3e6 and a maximum inject time of 60 ms. This was followed by 22 (DIA) isolation windows with varying sizes at 30,000 resolution, an AGC target of 3e6, injection times set to auto, loop count and msx count of 1. The default charge state was 4, the first mass was fixed at 200 m/z and the normalized collision energy (NCE) for each window was stepped at 25.5, 27 and 30. MS/MS raw files for the DDA were searched against a reference human protein sequence database including reviewed isoforms from the Uniprot database using MaxQuant version 1.6.1.0. Trypsin was specified as an enzyme with two possible missed cleavages. Carbamidomethyl of cysteine was specified as fixed modification and protein N-terminal acetylation and oxidation of methionine were considered variable modifications. The MS/MS tolerance FTMS was set to 20 ppm. The false discovery rate limit of 1% was set for peptide and protein identification and the remaining search parameters were set to the default values.
The MaxQuant output was used to generate the project-specific spectral library for DIA analysis. This library was supplemented with additional library generated using direct DIA approach in Spectronaut. The raw files for DIA analysis were processed with Spectronaut version 13. The default setting in Spectronaut was used for peptide and protein quantification at MS2 level. Perseus (1.6.6.0) was used for proteomics data processing and statistical analysis. The MS2 intensity values generated by Spectronaut were used to analyze the proteome data. The data were log2 transformed and normalized by subtracting the median for each sample. We filtered the data to have three values in each group. Student's t-test was employed to identify differentially expressed proteins and volcano plots were generated to visualize the affected proteins while comparing different groups of samples. Lists of differentially abundant proteins (with P.Value < 0.05 and |log2FC|>1) were selected and used for downstream bioinformatics analysis. For mitochondrial protein annotation, we utilized the newly released Mitocarta 3.0 (https://www.broadinstitute.org/files/shared/metabolism/mitocarta/human.mitocarta3.0.html).
2.4. Quantitative Western blot analysis
Fibroblasts grown in T75 flasks at 90–95% confluence were harvested by trypsinization, pelleted by centrifugation and stored at -80 °C. The BCA Protein Assay (Thermo scientific) quantified protein concentration. Proteins were separated on NUPAGE gels (Invitrogen) and transferred to nitrocellulose membranes. After 1 h of blocking, membranes were incubated with primary antibodies. The primary antibodies used as follows: VLCAD (GeneTex, Rb pAb GTX114232) 1:1000, MCAD (Abcam, Rb mAb ab92461) 1:1000, LCAD (Abcam, Rb mAb ab196655) 1:1000, TFPa (Abcam, Rb pAb ab54477) 1:500, TFPb (Bethyl, Rb A305-020A) 1:3000, CPT1α (Abcam, Ms mAb ab128568) 1:1000, MFN1 (Abcam, Ms mAb ab57602) 1:400, MFN2 (Abcam, Ms mAb ab56889) 1:400, DRP1 (Abcam, Rb mAb ab184247) 1:100 and GAPDH (Cell Signaling Technology, Rb mAb 2118S) 1:30000 dilutions overnight at 4 °C. The membranes were then incubated with fluorescent conjugated secondary antibodies for 1 h. The secondary antibodies used and their concentration as follows: Antibody DyLight 800 conjugated Anti-Rabbit IgG made in goat (611-145-002), Antibody DyLight 680 conjugated Anti-Rabbit IgG made in goat (611-144-003), Antibody DyLight 800 conjugated Anti-Mouse IgG made in goat (610-145-002), and Antibody DyLight 680 conjugated Anti-Mouse IgG made in donkey (610-744-124). Band signal intensities were obtained using Odyssey imaging system.
2.5. Immunofluorescence
The cells are fixed with 4% PFA for 10 min at room temperature. The fixed cells were incubated in a solution of PBS/BSA 3% saponin 0.03% for 1 h at room temperature. The same solution was used to dilute the antibodies. The phalloidin dye (Alexa Fluor 555 Phalloidin, 8953S Cell Signaling Technology) was used at a dilution of 1:200. Antibodies against CPT1α (mouse monoclonal Ab, ab128568 Abcam) and MFN1(mouse monoclonal Ab, ab57602 Abcam) were diluted 1:500 and incubated overnight at 4 °C. Secondary antibodies conjugated to the Alexa Fluorophore 488 (donkey anti-mouse IgG, A21202 Invitrogen) diluted at 1:800 were incubated 1 h at room temperature. For Nile red (9-diethylamino-5H-benzo[alpha]phenoxazine-5-one) staining, cells were fixed with 4% PFA for 10 min at room temperature and then incubated with 3 nM Nile red solution for 15 min. Cells were washed extensively and treated with Vectashield containing DAPI mounting medium. The Mitotracker green staining was performed in live cells. The cells were incubated with 500 nM Mitotracker solution (MitoTracker Green FM, M7514 Invitrogen) diluted in DMEM for 45 min in 5% CO2 at 37 °C. The slides were washed extensively to remove the precipitated dye and fixed with 4% PFA for 10 min at room temperature. Fluorescence images were collected using the 63× oil objective of a Leica microsystems confocal microscope under the same exposure conditions.
2.6. Mitochondrial respiration
Mitochondrial respiration was quantified using the Oroboros O2K high-resolution respirometry system. Fibroblasts were seeded in a 75-cm plate in 25 mM glucose DMEM media at a density of 1 × 106 cells per well. Fibroblasts were incubated for 1 h with DMEM without glucose, FBS and PenStrep, harvested, counted and re-suspended in 200 μl of the same media. After recording basal oxygen consumption, 2.5 μM oligomycin was added and after recording that ATPase inhibited respiration, 7.5 μM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) was added to dissipate the proton gradient and measure maximal respiration rate. In the end, 1 μM of rotenone and antimycin were added to quantify non-mitochondrial oxygen consumption.
2.7. Glycogen assay
The fibroblasts were grown in T75 flasks and at 90% confluence (about 1 × 106 cells), harvested by trypsinization and pellets collected after 5-min centrifugation at 1200×g. The cell pellets were washed with cold PBS twice. The cells were suspended in 200 μl ddH2O homogenized by pipetting on ice and cell lysates were boiled for 10 min. The boiled samples were centrifuged at 4 °C for 10 min, the supernatant was collected in new tubes, and glucose generated from the hydrolysis of glycogen was quantified by a colorimetric assay.
2.8. Transmission electron microscopy
For these studies, we utilized the non-disease control 1 and 2, VLCAD mutant 1 and 3, and TFP mutant 1 and 3 fibroblasts. The cells were grown in T75 flasks at 90% confluence (about 1 × 106 cells). Cells were harvested by trypsinization, fixed with 2.5% glutaraldehyde, 2.0% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, overnight at 4 °C. After several washes, the samples were fixed with a 2.0% osmium tetroxide for 1 h at room temperature. Samples were rinsed with ddH2O and treated with 2% uranyl acetate. After dehydration through a graded ethanol series, the cells were embedded in EMbed-812 (Electron Microscopy Sciences, Fort Washington, PA). Thin sections were stained with uranyl acetate and lead citrate and examined with a JEOL 1010 electron microscope fitted with a Hamamatsu digital camera and AMT Advantage image capture software.
3. Results
3.1. Evaluation of the LCFA oxidation pathway in non-disease and mutant primary human fibroblasts
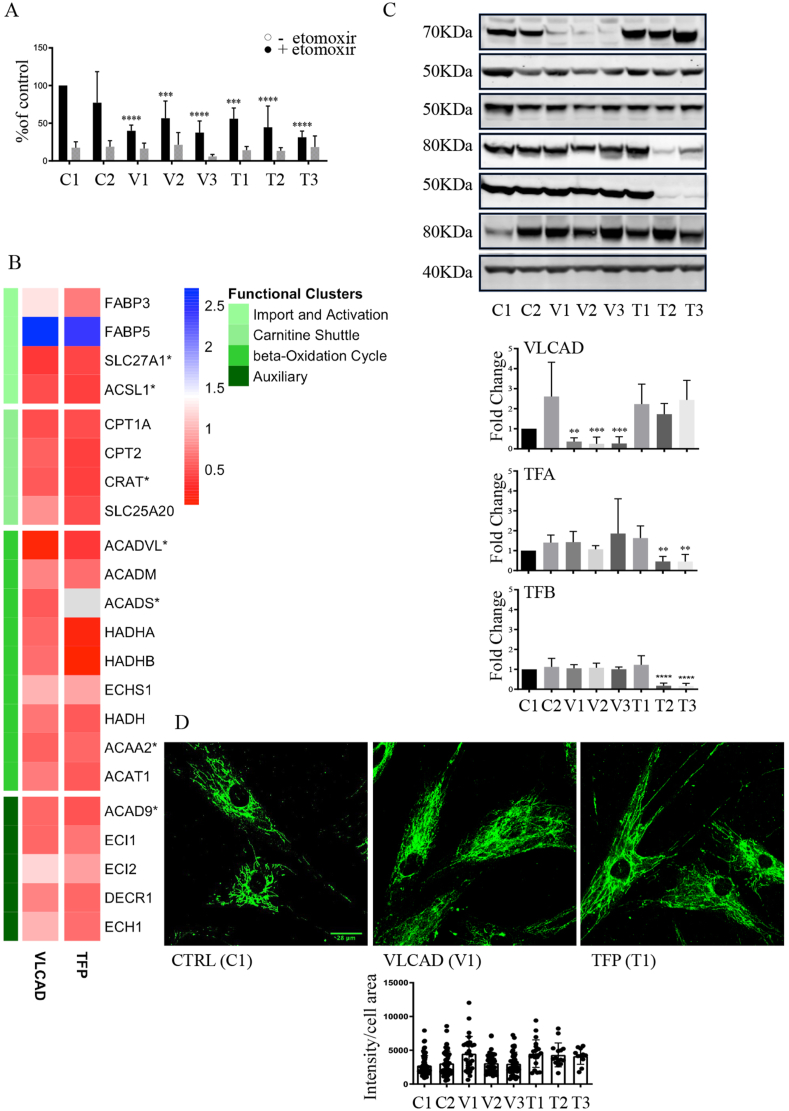
Early passage (4-10) human skin-derived fibroblasts derived from non-disease controls 1 and 2, as well as subjects with three different mutations in VLCAD, and three different mutations in TFP (one with the mutation in the alpha and two in the beta subunits) were utilized in this study (Supplementary Fig. S1). Using the [9, 10-3H] palmitate flux assay we confirmed that cells with mutations in VLCAD and TFP had significantly reduced capacity to oxidize palmitate as compared to non-disease control fibroblasts (Fig. 1A). Pre-incubation of fibroblasts with etomoxir, an inhibitor of the mitochondrial transporter carnitine O-palmitoyltransferase 1α (CPT1α), reduced palmitate flux in all cells indicating that the majority of measured activity was derived by mitochondrial fatty acid oxidation (Fig. 1A).
Fig. 1.
Evaluation of the LCFA pathway in non-disease and mutant primary human fibroblasts.
A. Oxidation of [9,10-3H] Palmitate in non-disease control and mutant primary fibroblasts quantified as pmol of palmitate oxidized per mg of protein per hour in the absence or presence of 10 μM etomoxir. Control non-disease fibroblasts line 1 and 2 were used (material and methods). The data is expressed as percent of control 1 and depicted as mean ± SD, n= 3 (three different cell cultures), ***P < 0.001, ****P < 0.0001 compared to non-disease control by one-way ANOVA.
B. Heat map depicting the changes in the relative levels (log 2 of fold change) of proteins participating in LCFA oxidation quantified by the proteomic analysis. The proteomic analyses quantified and compared changes in the relative levels of proteins in non-disease control lines 1, 2 and 3 to cells with either VLCAD or TFP mutations. The proteins are clustered based on biological function (1) transport of LCFA into cells and activation to LCFA acyl-CoA, (2) generation of LCAF-acylcarnitines and transport into mitochondria, (3) β-oxidation, and (4) auxiliary proteins. *P < 0.05 protein levels that were different between VLCAD and controls.
C. Representative western blots, original blots are shown in supplementary Fig S8-9. And densitometric quantification of relative protein levels from western blots. Data are depicted as mean ± SD, n = 3, **P < 0.01, ***P < 0.001 and ****P < 0.0001 by one-way ANOVA. Intracellular transport, activation, mitochondrial transport, β-oxidation, carnitine shuttle, and auxiliary proteins.
D. Representative confocal fluorescence images of cells stained with antibodies against CPT1α to visualize mitochondria. For the quantification of anti-CPT1α fluorescence intensities, 3–6 independent experiments were analyzed, and quantification was acquired from 16 images for TFP 2 and TFP 3 and 38–44 images for the rest of the cell lines. Analyzed using the image processing application Image J.
The palmitate flux is primarily dependent on the levels and activity of approximately 25 proteins, some of which are relatively stable [1,2]. The levels of 22 proteins that participate in LCFA oxidation were quantified in the proteomic study (Fig. 1B and supplementary data file 1). The cells with VLCAD mutations showed a significant decrease in VLCAD protein levels. The average level of VLCAD protein in the mutant lines were 11% of the non-disease controls quantified by mass spectrometry and 10% by Western blot analysis (Fig. 1C, original blots in supplementary Fig. S8). In addition to VLCAD, a significant decrease in the levels of long-chain fatty acid transport protein 1, long-chain-fatty-acid-CoA ligase 1, carnitine O-acetyltransferase, short-chain specific acyl-CoA dehydrogenase, and 3-ketoacyl-CoA thiolase, were quantified in cells with VLCAD mutations. (Fig. 1B). The levels of the TFA α or β subunits were reduced in cells with mutations in the β-chain of TFP (T2) and (T3). The levels of TFA α or β subunits in fibroblasts with c.1528G > C α-subunit TFA mutation (T1) were not different than controls (Fig. 1C).
No significant changes in other proteins of this metabolic pathway, including the long and medium-chain acyl CoA dehydrogenases (LCAD, MCAD), and CPT1α by either the proteomic or Western blot quantification were quantified in cells with VLCAD and TFP mutations (Fig. 1C, and supplementary Fig. S2). Since the levels of CPT1α, which is localized in the outer mitochondrial membrane did not change, we assessed mitochondria morphology by confocal microscopy after staining cells with antibodies against CPT1α. Cellular fluorescence was acquired at the same intensity from 3 to 6 independent experiments for each cell line. Staining for CPT1α revealed typical mitochondrial network morphology and distribution in non-disease as well as in cells with VLCAD and TFP mutations (Fig. 1D and Supplementary Fig. S3). Quantification of the fluorescence intensity did not indicate significant differences between the non-disease controls and cells with either VLCAD or TFP mutations (Fig. 1D).
Collectively the data indicate that cells with VLCAD mutations not only have reduced levels of VLCAD but additional proteins that participate in LCFA oxidation. Cells with TFP mutations in the β subunits have a lower level of TFP. The reduction in protein levels accounts, in part, for the diminished capacity to oxidize LCFA. However, the cells with VLCAD and TFP mutations appear to have normal mitochondrial morphology. To further explore the mitochondrial in the cells with mutations we quantified mitochondrial function and dynamics.
3.2. Mitochondrial functions: respiration and calcium handling
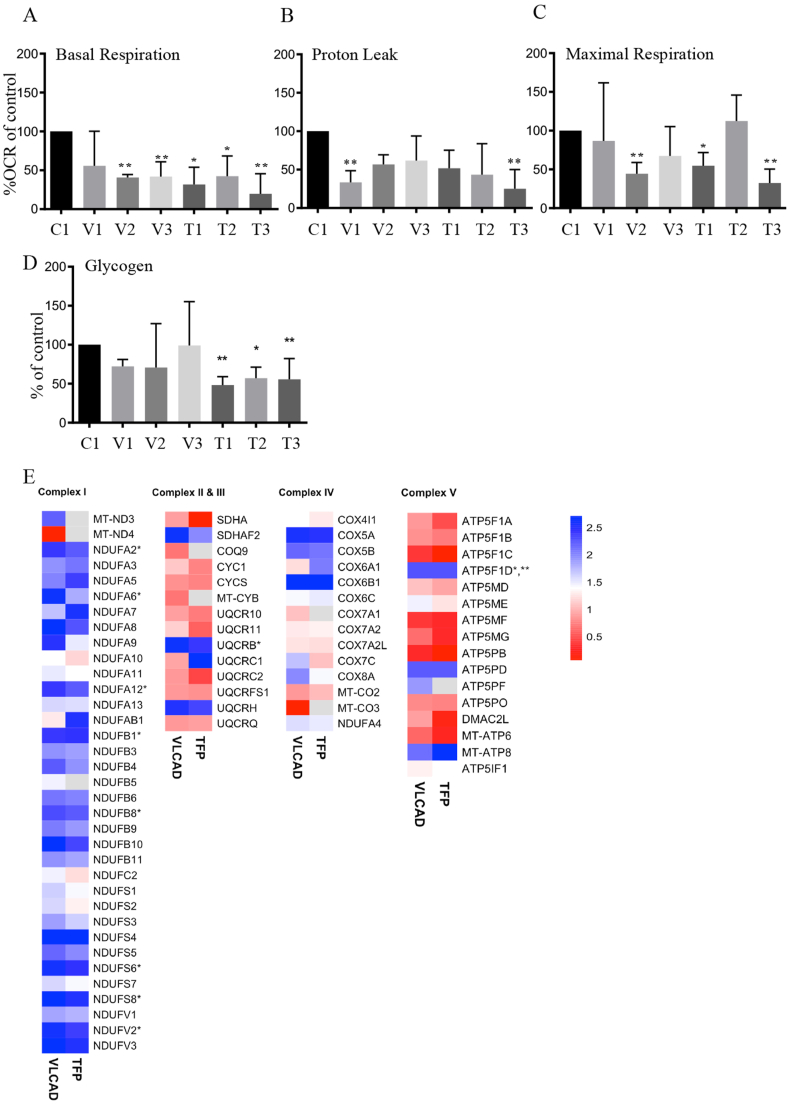
Mitochondrial oxygen consumption was quantified using the Oroboros O2K high-resolution respirometry system. Utilizing typical inhibitor-uncoupler protocols we quantified basal oxygen consumption, respiration coupled to ATP production, maximal respiration and non-mitochondrial oxygen consumption 1 h after culturing cells in glucose-free media to unmask potential deficiencies in oxidative phosphorylation. Fibroblasts with VLCAD and TFP mutations, except for cells with the homozygous c.364A > G VLCAD mutation (V1), displayed reduced basal mitochondrial respiration (Fig. 2A). Proton leak was not different than controls except for cells with the 364A > G VLCAD mutation (V1) and with the homozygous c.182G > A β-subunit TFA (T3) mutation, which had reduced proton leak. The maximal respiratory capacity was reduced in cells VLCAD with 887–888 deletion and exon 18 splice (V2) and in cells with c.1528G > C α-subunit TFA mutation (T1) and c.182G > A β-subunit TFA mutation (T3). The altered mitochondrial respiration resulted in lower ATP production in T1 and T3 cells (Supplementary Fig. S4). These findings are in part consistent with the previous reports in human fibroblasts with VLCAD and TFP mutations and indicate potential insufficiencies with oxidative phosphorylation [[19], [20], [21], [22], [23]]. In the absence of exogenous glucose, cells may convert stored glycogen to glucose to generate ATP and maintain a reduced cellular environment under stress. We quantified the levels of glycogen after glucose removal. Cells with TFP mutations had lower levels of glycogen as compared to non-disease fibroblasts (Fig. 2D). The lower levels of deposited glycogen in the cells with TFP mutations may indicate an increased demand and utilization of glucose in these cells to satisfy energetic needs.
Fig. 2.
Mitochondrial respiration in non-disease and mutant primary human fibroblasts. A-C. Mitochondrial oxygen consumption was quantified by high-resolution respirometry in the absence of glucose in the media. A. Basal respiration, B. Proton leak and C. Maximal respiration. Data are depicted as mean ± SD n = 3 different cell preparations for each line. *P < 0.05, **P < 0.01, by one-way ANOVA compared to control non-disease cells 1. To secure that the differences are not just to the comparison to the one control non-disease line we confirmed that mitochondrial oxygen consumption were not statistically different in three additional control fibroblasts as compared to control 1 (supplementary Fig. S4). D. Steady state quantification of glycogen levels. Glycogen was quantified 1 h after removal of glucose from the media. *P < 0.05, **P < 0.01, by one-way ANOVA compared to control non-disease cells 1. The levels of glycogen in non-disease, control fibroblasts 1 were similar to other three control non-disease fibroblasts (supplementary Fig. S4). E. Heat map depicting the changes in the relative levels of ETC proteins. *P < 0.05 protein levels that were different between VLCAD and controls ***P < 0.05 protein levels that were different between TFP and controls.
The levels of 74 proteins that participate in glycolysis, pyruvate metabolism and the TCA cycle were similar between cells with VLCAD or TFP mutations and non-disease control fibroblasts (Supplementary data file 1). However, a few changes in the protein levels were identified. The levels of β and γ enolases but not of α enolase in cells with VLCAD and β enolase in cells with TFP mutations were reduced as compared to controls suggesting a possible reduction in phosphoenolpyruvate production. The cells with VLCAD and TFP mutations had reduced levels of dihydrolipoamide succinyltransferase (E2) component of the 2-oxoglutarate dehydrogenase complex, which catalyzes the conversion of 2-oxoglutarate to succinyl-CoA and CO2. Cells with VLCAD mutations had also reduced levels of the 2-oxoglutarate dehydrogenase E1 component of the 2-oxoglutarate dehydrogenase complex indicating a possible reduction in the production of succinyl-CoA.
The proteomic analysis quantified the levels of 63 proteins of the mitochondrial electron transport chain (ETC) and as well as 16 proteins of complex V (ATPase). Increased levels of eight proteins in the NADH: ubiquinone oxidoreductase complex I, one protein in the ubiquinol-cytochrome c reductase complex III, and one ATPase protein were quantified in cells with VLCAD mutations (Fig. 2E). Furthermore, the proteomic analysis also indicated a significant increase in the levels of proteins that support and facilitate the assembly of ETC and the organization of the inner mitochondrial membrane. Significant increase in the relative levels in succinate dehydrogenase (SDHAF4), complex III (LYRM7) and cytochrome c oxidase (COA4, COX17) assembly factors was quantified in cells with VLCAD mutations (Supplementary Fig. S5 and supplementary data file 1). Additionally, a 2-fold increase in the levels of MICOS complex subunit 60 isoform 4, MICOS complex subunit 10 that secure intact mitochondrial cristae architecture were quantified in cells with VLCAD mutations (Supplementary Fig. S5 and supplementary data file 1). Increased levels of LYRM7, COX17, and MICOS complex subunit 25 were quantified in cells with TFP mutations (Supplementary Fig. S5 and supplementary data file 1). Quantification of 50 proteins that facilitate organization and functionality of mitochondria showed similar levels between cells with VLCAD or TFP mutations and non-disease control cells (Supplementary data file 1).
With regards to mitochondrial calcium handling, a slight but significant decrease in the levels of m-AAA protease-interacting protein 1 and calcium-binding mitochondrial carrier protein SCaMC-1 were quantified in cells with VLCAD mutations (Supplementary data file 1). However, cells with VLCAD mutations had a significant increase in the levels of mitochondrial proton/calcium exchanger protein. The levels of an additional 13 proteins participating in mitochondrial calcium regulation were not different between cells with VLCAD mutation and controls. The levels of all 16 proteins participating in calcium regulation were similar between cells with TFP mutations and non-disease control cells (Supplementary data file 1).
3.3. Antioxidant networks and stress responses
Previous studies indicated increased production of oxidants in fibroblasts with VLCAD and TFP mutations [22,23]. The ability of cells to withstand changes in the redox homeostasis derived from increased production of oxidants within mitochondria, cytosol and other cellular compartments is in part determined by a network of antioxidant proteins. The proteomic study offered the opportunity to evaluate the levels of proteins in all major antioxidant networks rather than individual proteins. The levels of 44 intracellular antioxidant proteins including superoxide dismutases (SOD), glutathione peroxidase, thioredoxins and peroxiredoxins were similar between cells with VLCAD and TFP mutations and non-disease control fibroblasts (Supplementary Fig. S5 and Supplementary data file 1). A significant reduction in the levels of extracellular SOD in cells with VLCAD mutations and catalase and glutathione S-transferase theta-1 in cells with TFP mutations were quantified.
The pentose phosphate pathway is a significant source of NADPH, which provides electrons for antioxidant systems and several biosynthetic pathways. The levels of glucose-6-phosphate 1-dehydrogenase and the four other principal enzymes in the pentose phosphate pathway except for GDH/6PGL endoplasmic bifunctional protein were not different between cells with VLCAD or TFP mutations and controls. The levels of the GDH/6PGL endoplasmic bifunctional protein were reduced in cells with VLCAD and TFP mutations suggesting a possible deficiency in NADPH production within the oxidizing environment of the endoplasmic reticulum (ER).
We also quantified the levels of 18 mitochondrial proteins that participate in mitochondrial stress responses including the Lon protease which degrades oxidatively modified peptides and regulate mitochondrial DNA replication. The cells with VLCAD mutation had reduced levels of the multifunctional cyclin-dependent kinase 1 and the cells with TFP mutations had increased levels of isoform-2 of the methionine-R-sulfoxide reductase B2, an enzyme that catalyzes the conversion of methionine sulfoxide to methionine. No other significant changes were quantified in cells with VLCAD and TFP as compared to non-disease control fibroblasts (Supplementary data file 1).
3.4. Mitochondrial dynamics: fusion, fission, mitophagy and distribution
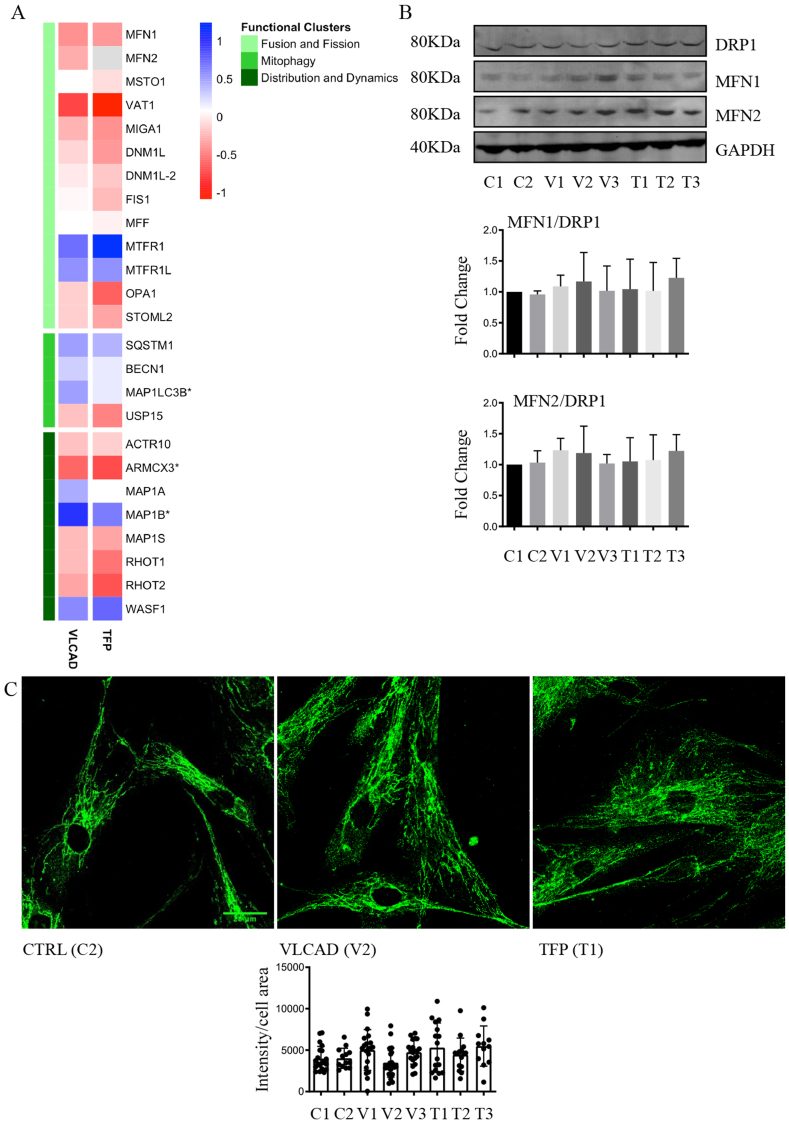
Fusion, fission and mitophagy are critical biochemical processes for mitochondrial biogenesis, morphology and function. The mass spectrometry-based proteomic analysis quantified the levels of 17 principal proteins that participate in fusion, fission and mitophagy and 8 proteins that regulate the movement and distribution of mitochondria within cells (Fig. 3A). No differences in the relative levels of proteins that participate in fusion and fission were quantified between VLCAD or TFP mutant cells and non-disease control fibroblasts (Fig. 3A). The proteomic data were corroborated by antibody-based detection of fusion mediators mitofusin-1 (MFN1) and mitofusin-2 (MFN2) as well as the pro-fission dynamin-related protein 1 (DRP1; also known as DNM1L, dynamin-1-like protein in the UniProt protein knowledgebase). Consistent with the proteomic data, no differences in the relative levels of these three GTPases was detected in cells with VLCAD or TFP mutations as compared to the non-disease control cells (Fig. 3B, original blots in supplementary Fig. S10). Except for an increase in the autophagy-related ubiquitin-like modifier (MAP1LC3B) in cells with VLCAD mutation, the levels of proteins that regulate mitophagy are similar between non-disease controls and cells with VLCAD and TFP mutations. Increased levels of microtubule-associated protein 1B and decreased levels of armadillo repeat-containing X-linked protein 3, which when degraded it regulates the mitochondrial distribution, were quantified in cells with VLCAD mutations. No other differences in the levels of proteins that orchestrate mitochondrial distribution were found between with VLCAD and TFP mutations and non-disease controls.
Fig. 3.
Assessment of mitochondrial fusion and fission.
A. Proteomic quantification of proteins participating in mitochondrial fusion, fission, mitophagy and mitochondrial localization. *P < 0.05 protein levels that were different between VLCAD or TFP and controls. B. Representative western blots (original blots are shown in supplementary Fig. S10) and quantification of MFN1/2 and DRP1. No significant changes in the relative levels of proteins that facilitate mitochondrial fusion (MFN1/2) and fission (DRP1) between non-disease (control) and mutant primary fibroblasts. Data are depicted as mean ± SD, n = 3. C. Representative confocal fluorescence images of cells stained with antibodies against MFN1 to visualize and quantify mitochondria networks. For the quantification of anti-MFN1 fluorescence intensities 4 independent experiments were performed, and quantification was acquired from 20 images for T2, T3, and 26–28 images for the rest of the cell lines.
Cells were also stained with antibodies against MFN1, which localizes in the outer mitochondrial membrane and facilitates mitochondrial clustering and fusion. Staining for MFN1 showed typical mitochondrial network morphology and localization. Quantification of fluorescence intensity did not reveal changes in MFN1 levels and localization between cells with VLCAD or TPF mutations and non-disease control fibroblasts (Fig. 3C and supplementary Fig. S3). Collectively the data indicate that the cells with VLCAD or TFP mutations do not have an apparent defect in the major biochemical processes that regulate mitochondria morphology and distribution.
3.5. Mitochondrial morphology in non-disease and mutant primary human fibroblasts
To further explore changes in cellular and mitochondria morphology we performed immunofluorescence for actin filaments, live-cell staining with Mito tracker green, and transmission electron microscopy. Cellular shape and cytoskeletal organization were assessed by phalloidin staining of actin filaments, which showed typical fibroblast cytoskeletal morphology. The average intensities for phalloidin were similar across all the fibroblasts indicating the preservation of cytoskeletal architecture (Supplementary Fig. S3). Mitochondrial localization and mass assessed by Mito tracker green were similar between cells with VLCAD and TFP mutations and controls supporting the imaging studies with CPT1α and MFN1 and the absence of significant perturbations in mitochondria morphology.
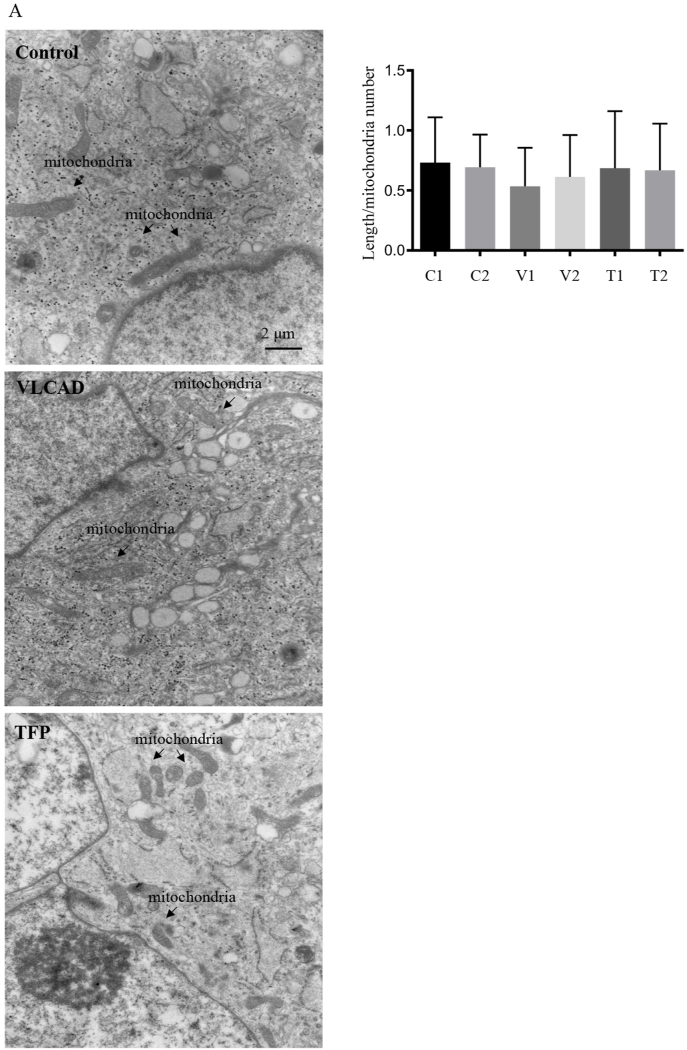
Transmission electron microscopy (TEM) and morphometric analysis of the images was used to evaluate mitochondrial and intracellular morphology (Fig. 4). Morphometric quantification of 10 images per cell line did not detect changes in the average length of mitochondria in the fibroblasts with VLCAD and TFP mutations as compared to non-disease control fibroblasts (Fig. 4). Since cells with VLCAD and TFP mutation may have reduced NADPH levels in the ER we quantified the volume occupied by the ER and distensions in the ER and Golgi apparatus. No significant differences in these morphometric parameters were quantified in cells with VLCAD or TFP mutations compared to non-disease controls (Supplementary Fig. S6). The TEM images indicated the presence of roughly circular glycogen granules, which we quantified biochemically (Fig. 4). Another feature of the TEM images was the presence of vacuoles, which may indicate fat accumulation. We stained for lipid droplets using Nile red. Quantification of the cells staining indicated the presence of lipid deposits but no significant differences between the control and cells with VLCAD or TFP mutations (Supplementary Fig. S6). Overall, the EM data is consistent with the fluorescence imaging and failed to show significant perturbations in mitochondrial and cellular architecture.
Fig. 4.
TEM images and morphometric quantification of mitochondria. A. Representative TEM images of non-disease and mutant primary human fibroblasts. B. Morphometric analysis of TEM images quantified the length of mitochondria from 10 images per cell line at a 25× magnification. Data are depicted as mean ± SD n = 3 independent cell cultures.
3.6. Alterations in the overall and mitochondrial proteomes
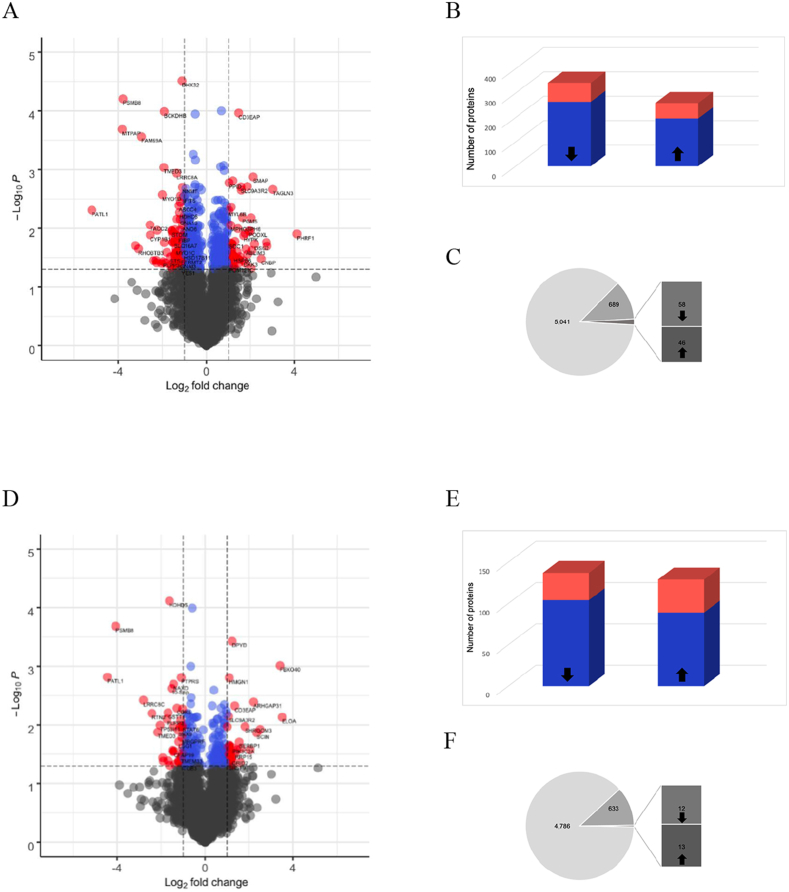
The mass spectrometry-based analysis quantified the relative levels of 5041 in cells with VLCAD mutations and 4786 in cells with TFP mutations as compared to non-disease control fibroblasts. The VLCAD mutant cells exhibited larger proteomic differences than the TFP mutant cells as compared to non-disease control. The relative levels of 601 proteins, representing 12% of the quantified proteome were different between VLCAD mutant lines and non-disease controls whereas 266 proteins representing 6% of the quantified proteome were different between TFP and controls (t-test p < 0.05, Fig. 5A and D and Supplementary data file 2). These differences are depicted in typical volcano plots, which depict the number of proteins that were statistically significant as well as those that had a greater than 2-fold decrease or increase in relative levels (Fig. 5A and D and quantification of proteins in Fig. 5B and E). Of the 601 proteins the levels 78 proteins decreased and 63 increased more than 2-fold in VLCAD mutant cells (Fig. 5B). In TFP mutant cells, of the 266 proteins, the relative levels of 33 proteins decreased and 40 increased changed more than 2-fold (Fig. 5E). Gene ontological analysis of the proteins with significant decrease in relative levels (p < 0.05) in cells with VLCAD mutations showed a reduction in metabolic processes (Supplementary data file 2). In the same cells, the proteins with a relative increase in levels were functionally clustered in pathway that participate in cellular organization or biogenesis, specifically mitochondrial respiratory chain assembly and respiration (Supplementary data file 2). A similar in cells with TFP mutations revealed that proteins with significantly reduced levels that participate in cellular organization, vesicular transport and metabolism whereas proteins with increased levels participate in RNA metabolism and actin-myosin filament biological processes.
Fig. 5.
Changes in the cellular and mitochondrial proteomes.
A and D. The proteomic analyses quantified and compared changes in the relative levels of proteins in non-disease control lines 1, 2 and 3 to cells with either VLCAD or TFP mutations. Typical volcano plots integrating the p-value and the magnitude change between cells with VLCAD mutations (A) and cells with TFP mutations (D) and non-disease control cells. The plots were constructed by log2 transformation, normalization by subtracting median values and application of t-test, p < 0.05. Gray dots indicate non-significant changes in the relative levels of individual proteins. Blue dots indicate the proteins with significant change (t-test, p <0.05) and red dots the proteins with significant change (t-test, p <0.05) of more than 2-fold. B and E. The number of proteins that were significantly decreased or increased in blue, and in red the number of proteins with 2-fold change or more between cells with VLCAD mutations (B) and cells with TFP mutations (E) and non-disease control cells. C and F. Mitochondrial proteins were curated using existing open source applications (Mitocarta 3.0, Uniprot knowledge base, GO). The data depicts the total number of proteins quantified, the number of mitochondria proteins and the fraction of mitochondrial proteins that were significantly different between cells with VLCAD mutations (C) and cells with TFP mutations (F) and non-disease control cells. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Cells with VLCAD and TFP mutations shared a core of 115 proteins that were statistically different as compared to non-diseased controls. The relative levels of 93 of the 115 proteins increased in both VLCAD and TFP, whereas 14 decreased and 8 were discordant. The 93 proteins with relative increase shared functional enrichment for RNA binding proteins and proteins localizing in focal adhesions and anchoring junctions (Supplementary data file 2) indicating adaptive changes that prioritize cellular structural integrity. This proteomic adaptation was evident by the greater than 5-fold increase in the levels of adseverin, a Ca2+-dependent actin filament-severing protein that regulates cytoskeletal dynamics [28], in cells with VLCAD and TFP mutations. Cells with VLCAD mutations had a greater than 6-fold increase in the levels of largen, a protein that regulates mitochondria and cell size [29]. Cells with TFP mutations had higher levels of E3 ubiquitin ligase F-box only protein 40 and shromm-3 that regulate cell shape [30].
The proteomic data were analyzed to select for mitochondrial proteins using the newly released Mitocarta 3.0, an inventory of 1136 manually curated human mitochondria proteins. Based on Mitocarta 3.0 we quantified 639 and 585 mitochondrial proteins in cells with mutations in VLCAD and TFP respectively. In addition, we generated a complementary inventory of mitochondrial proteins by submitting the cellular proteomes to Gene Ontology (GO) knowledgebase and extracting proteins with mitochondrial assignment using cellular component search. The resulting list was curated using the most current list of 1213 human proteins annotated as mitochondrial in the UniProtKB protein database and manually inspect to include only proteins with published evidence for mitochondrial localization under physiological conditions or upon stress. This resulted in additional 50 and 48 proteins in the cells with VLCAD and TFP mutations respectively. Taken together we quantified the levels of 689 and 633 mitochondrial proteins across all major functional pathways in cells with VLCAD and TFP mutations respectively (Supplementary Fig. S7 and Supplementary data file 2). Similar to the whole proteome, 14% of mitochondrial proteins, (58 decreased and 46 increased) were significantly different in cells with VLCAD mutations as compared to non-disease controls. Only 4% of the mitochondrial proteins (12 decreased and 13 increased) were significantly different in fibroblasts with TFP mutations (Fig. 5C and F). The mitochondrial proteins with reduced relative levels in cells with VLCAD mutations showed the expected reduction in functional clusters of fatty acid catabolic processes (Fig. 1 and Supplementary data file 2). On the other hand, the mitochondrial proteins with increased levels in VLCAD mutant cells were enriched for biological processes relating to mitochondria organization, ETC and ETC complex assembly (Fig. 2 and Supplementary data file 2).
These data are consistent with the biochemical and morphometric analyses indicating that despite the metabolic deficits, cells with VLCAD mutation and to a lesser extend cells with TFP mutations exhibit adaptive proteomic changes that aim to sustain cellular and mitochondria architecture.
4. Discussion
Long-chain fatty acid β-oxidation disorders are a collection of inherited autosomal recessive diseases affecting predominantly the heart, liver, and skeletal muscle [[1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14]]. Chronic energy insufficiency and accumulation of incomplete products of LCFA oxidation are two prevailing mutually nonexclusive pathogenic mechanisms that account in part for the clinical phenotypes. The possibility that mitochondria dysfunction beyond ATP production that disturbs mitochondrial networks, intracellular organization and redox homeostasis could be considered as additional mechanisms that contribute to organ failure. Here we confirmed the energetic deficits and by the use of proteomic analyses coupled with biochemical and morphometric data, provide evidence that despite the metabolic challenges the cells maintain structural integrity as well as mitochondrial morphology and function. Preservation of protein levels and proteomic adaption may account for the capacity of the cells to overcome the bioenergetic challenges.
Cells with VLCAD mutations had lower levels of VLCAD protein, which in addition to the VLCAD mutation, contributed to the significantly reduced capacity of these cells to metabolize palmitate. Lower levels of additional proteins in the LCFA oxidation pathway quantified in Fig. 1A in cells with VLCAD mutations may also contribute to the lower flux of palmitate in these cells. Cells with VLCAD mutations had also significant reductions in the levels of 2-oxoisovalerate dehydrogenase, alpha-aminoadipic semialdehyde synthase, and propionyl-CoA carboxylase alpha chain, which participate in amino acid metabolism. Although impaired amino acid metabolism has not been reported in humans with VLCAD mutations, mice deficient in long-chain acyl CoA dehydrogenase had impaired amino acid metabolism that contributed to hypoglycemia during fasting [31]. Consistent with previous studies cells with VLCAD and TFP mutations have altered mitochondrial respiration, which in cells with TFP mutations leads to reduced ATP production [[21], [22], [23]]. These mitochondrial bioenergetic deficits are not apparent under normal cell culture conditions but are revealed under stress conditions such as hypoglycemia in the absence [[20], [21], [22], [23]] or presence LCFA [19] in the culture media. Fibroblasts with VLCAD mutations have augmented glycolytic rate generating more pyruvate and lactate than non-disease control cells after re-introduction of glucose following a 90 min glucose deprivation [21]. Since both lactate and pyruvate are increased the ratio of lactate to pyruvate remains the same as controls. Similarly, cells with TFP mutations have increased glycolytic rate [21]. However, these cells generate more lactate than pyruvate and thus have an increased ratio of lactate to pyruvate indicating a defect in the tricarboxylic acid (TCA) cycle or oxidative phosphorylation [21]. This defect may be responsible for lower ATP generating capacity we quantified in cells with the most common c.1528G > C mutation in the TFP α-chain and with the homozygous c.182G > A β-chain TFP mutation. A reduction in the levels of succinyl-CoA due to lower levels of the 2-oxoglutarate dehydrogenase complex may explain a TCA cycle defect. Although this observation requires further investigation, it supports the hypothesis of TCA cycle intermediate depletion leading to an insufficient generation of ATP to meet metabolic demands in VLCAD and TFP mutations [32]. This hypothesis has been in part supported by the clinical phenotypic improvements in subjects with VLCAD and TFP mutations receiving medium-chain odd triglycerides that supply both acetyl-CoA and propionyl-CoA which is converted to succinyl-CoA that backfills the TCA cycle [33,34].
Although cells in culture may not reflect the energetic state in subjects with these disorders the data are in part consistent with metabolic studies in subjects with VLCAD and TFP deficiencies [[35], [36], [37], [38], [39]]. Residual VLCAD, and TFP activities and in part peroxisomal contributions appeared to be sufficient in metabolizing LCFA at rest [35,36]. However, subjects with VLCAD mutations failed to increase LCFA oxidation and relied on increased glycolysis to meet the metabolic demands of low-intensity prolonged exercise. Some subjects with VLCAD or TFP mutations have reduced exercise tolerance or reduced daily activities, which may relate to the inability to sustain ATP production relying exclusively on glycolysis [38,39]. This was evident in a study that compared carbohydrate versus medium-chain triglycerides (MCT) feeding before exercise in subjects with VLCAD, and TFP deficiencies [37]. MCT pre-treatment lowered glucose oxidation and decreased heart workload during exercise as compared to carbohydrate pre-treatment [37]. Other studies have suggested increased accumulation of incomplete products of LCFA oxidation, inflammation or a switch in muscle fiber composition in the absence of mitochondrial ATP production deficit as potential mechanisms for the reduced exercise tolerance of skeletal muscle in subjects with VLCAD mutations [38,39]. This is also a viable proposition since the cells with the three VLCAD mutations used in this study had preserved mitochondrial reserve capacity and increased levels of ETC proteins and proteins that facilitate the assembly of ETC complexes and inner mitochondrial membrane.
Histological and EM evaluation of biopsy tissues from subjects with TFP mutations indicated that the morphology of mitochondria is altered in severe disease [26]. Enlarged mitochondria were reported in liver biopsy along with a significant accumulation of lipid deposits. In four of seven subjects with the common c.1528G > C TFP α-chain mutation, mitochondria with increased size as well as the increased number of mitochondria were observed in muscle fibers. Some of the enlarged mitochondria had a swollen appearance with distorted cristae. Swollen mitochondria with normal cristae were also apparent. The mitochondria with altered appearance and number were more visible in atrophic muscle fibers [26]. Recently, a reduction in mitochondrial networks and fusion/fission defects were documented by live-cell microscopy and antibody-based protein detection in fibroblasts from 8 individuals with the c.1528G > C mutation in the α-subunit of TFP and 1 individual with c.881T > C/c.1438C > T VLCAD mutation [22]. The defect in fusion was evident by a reduction in the ratio of MFN2 to DNML 1 levels as well as the ratio of phosphorylated DNML1 to DNML1 [22]. Dynamic fission and fusion processes are critical as they control the size of mitochondria and allow the exchange of metabolites and mitochondrial DNA, securing optimized mitochondrial functionality. Although both MFN2 and DNML1 are important for regulating fusion and fission respectively, these processes required additional proteins and factors that co-regulate the dynamics of these opposing processes. Quantification of six proteins participating in fusion, six in fission and OPA1, a dynamin GTPase that regulates the equilibrium between fusion and fission did not show significant differences between cells with c.1528G > C TFP α-chain mutation and control fibroblasts. The proteomic data was corroborated by antibody-based protein detection. Moreover, the levels of proteins important for mitophagy, morphology, trafficking and distribution of mitochondria were not different between cells with TFP mutations and controls. Similarly, the levels of these proteins were not different in cells with the three different VLCAD mutations as compared to control fibroblasts. Cells with VLCAD mutations also had increased levels of proteins that facilitate tubular mitochondrial networks, cristae organization and assembly of respiratory complexes. The TEM analyses corroborated these findings and did not reveal any differences in mitochondria length in cells with VLCAD and TFP mutations as compared to controls. Moreover, quantification of endoplasmic reticulum (ER) distensions and interaction with mitochondria which were recently implicated in the regulation of glucose homeostasis failed to show any differences.
Collectively the data confirmed deficiencies in fatty acid oxidation and mitochondrial respiration in fibroblasts with VLCAD and TFP mutations. These metabolic deficiencies did not influence cellular architecture and size, as well as mitochondria morphology, distribution, size and functionality. In part, the capacity of the fibroblasts to maintain cell shape, intracellular architecture and mitochondrial morphology may relate to adaptive proteomic changes. However, with disease progression and increased severity of the metabolic deficits, these adaptive responses may not be sufficient to prevent deterioration of mitochondrial morphology and function. Therapeutics that restore mitochondrial fuel utilization and metabolic efficiency, as well as molecules that remove oxidants [40,41] may preserve mitochondrial functionality as it was documented in human fibroblasts with VLCAD mutations [23].
Availability of data and material
All data generated or analyzed during this study are included in this published article and its Supplementary files. The proteomic data are available via ProteomeXchange with identifier PXD024488.
Authors’ contributions
SR designed and conducted all the experiments, analyzed data, generated figures and edited the manuscript. GZ performed Western blot analysis, glycogen quantification and assisted with data analysis. IZ assisted with the flux assay and with the editing of the manuscript. LS, HF performed the proteomic analysis. AW and JV provided cells and edited the manuscript. HI planned, organized, and participated in the analysis and discussion of all the data writing and editing of the manuscript. All authors read and approved the final manuscript.
Funding
These studies were supported by the Gisela and Dennis Alter endowed chair in Pediatric Neonatology (HI).
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
The authors thank Drs. Ding Hua and Steve Seeholzer at the Protein and Proteomics Core Facility at the Children's Hospital of Philadelphia Research Institute, for their support and discussions regarding the proteomic analyses; Dr Orange, Columbia Vagelos College of Physicians and Surgeons for control fibroblasts, Alessia Angelin at the Center for Mitochondrial and Epigenomic Medicine for the use of Oroboros; and Elena Polishchuk at the Telethon Institute of Genetics and Medicine (TIGEM), Pozzuoli, Italy for advice regarding TEM image quantification. HI is the Gisela and Dennis Alter research professor at the Children's Hospital of Philadelphia Research Institute.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101923.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Houten S.M., Violante S., Ventura F.V., Wanders R.J.A. The biochemistry and physiology of mitochondrial fatty acid b-oxidation and its genetic disorders. Annu. Rev. Physiol. 2016;78:23–42. doi: 10.1146/annurev-physiol-021115-105045. [DOI] [PubMed] [Google Scholar]
- 2.Longo N., Frigeni M., Pasquali M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta. 2016;1863:2422–2435. doi: 10.1016/j.bbamcr.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett M.J. Pathophysiology of fatty acid oxidation disorders. J. Inherit. Metab. Dis. 2009;33:533–537. doi: 10.1007/s10545-010-9170-y. [DOI] [PubMed] [Google Scholar]
- 4.Strauss A.W., Powell C.K., Hale D.E., Anderson M.M. Molecular basis of human mitochondrial very-long-chain acyl-CoA dehydrogenase deficiency causing cardiomyopathy and sudden death in childhood. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10496–10500. doi: 10.1073/pnas.92.23.10496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andresen B.S., Vianey-Saban C., Bross P., Divry P. The mutational spectrum in very-long-chain acyl-CoA dehydrogenase deficiency. J. Inherit. Metab. Dis. 1996;19:169–172. doi: 10.1007/BF01799421. [DOI] [PubMed] [Google Scholar]
- 6.Andresen B.S., Olpin S., Poorthuis B.J., Scholte H.R. Clear correlation of genotype with disease phenotype in very-long-chain acyl-CoA dehydrogenase deficiency. Am. J. Hum. Genet. 1999;64:479–494. doi: 10.1086/302261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ushikubo S., Aoyama T., Kamijo T., Wanders R.J.A., Rinaldo P., Vockley J., Hashimoto T. Molecular characterization of mitochondrial trifunctional protein deficiency: formation of the enzyme complex is important for stabilization of both alpha- and beta-subunits. Am. J. Hum. Genet. 1996;58:979–988. [PMC free article] [PubMed] [Google Scholar]
- 8.IJlst L., Ruiter J.P.N., Hoovers J.M.N., Jakobs M.E., Wanders R.J.A. Common missense mutation G1528C in long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency: characterization and expression of the mutant protein, mutation analysis on genomic DNA and chromosomal localization of the mitochondrial trifunctional protein alpha subunit gene. J. Clin. Invest. 1996;98:1028–1033. doi: 10.1172/JCI118863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spiekerkoetter U., Khuchua Z., Yue Z., Bennett M.J. General mitochondrial trifunctional protein (TFP) deficiency as a result of either alpha- or beta subunit mutations exhibits similar phenotypes because mutations in either subunit alter TFP complex expression and subunit turnover. Pediatr. Res. 2014;55:190–196. doi: 10.1203/01.PDR.0000103931.80055.06. [DOI] [PubMed] [Google Scholar]
- 10.De Biase I., Viau K.S., Liu A., Yuzyuk T. Diagnosis, treatment, and clinical outcome of patients with mitochondrial trifunctional protein/long-chain 3-Hydroxy Acyl-CoA dehydrogenase deficiency. JIMD Rep. 2017;31 doi: 10.1007/8904_2016_558. 63-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spiekerkoetter U., Lindner M., Santer R., Grotzke M., Baumgartner M.R. Treatment recommendations in long-chain fatty acid oxidation defects: consensus from a workshop. J. Inherit. Metab. Dis. 2009;32:498–505. doi: 10.1007/s10545-009-1126-8. [DOI] [PubMed] [Google Scholar]
- 12.Bertrand C, Largilliere C, Zabot M., Mathieu M, Vianey-Saban C. Very long chain acyl-CoA dehydrogenase deficiency: identification of a new inborn error of mitochondrial fatty acid oxidation in fibroblasts. Biochim. Biophys. Acta 1180: 327-329 (19930. [DOI] [PubMed]
- 13.Diekman E.F., Ferdinandusse S., van der Pol L., Waterham H.R., Ruiter J.P., Ijlst L. Fatty acid oxidation flux predicts the clinical severity of VLCAD deficiency. Genet. Med. 2015;17:989–994. doi: 10.1038/gim.2015.22. [DOI] [PubMed] [Google Scholar]
- 14.Schiff M., Mohsen A.-W., Karunanidhi A. Molecular and cellular pathology of very-long-chain acyl-CoA dehydrogenase deficiency. Mol. Genet. Metabol. 2013;109:21–27. doi: 10.1016/j.ymgme.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gobin-Limballe S., Djouadi F., Aubey F., Olpin S. Genetic basis for correction of very-long-chain acyl-coenzyme A dehydrogenase deficiency by bezafibrate in patient fibroblasts: toward a genotype-based therapy. Am. J. Hum. Genet. 2007;81:1133–1143. doi: 10.1086/522375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bastin J., Lopes-Costa A., Djouadi F. Exposure to resveratrol triggers pharmacological correction of fatty acid utilization in human fatty acid oxidation-deficient fibroblasts. Hum. Mol. Genet. 2011;20:2048–2057. doi: 10.1093/hmg/ddr089. [DOI] [PubMed] [Google Scholar]
- 17.Ndukwe Erlingsson U.C., Iacobazzi F., Liu A., Ardon O., Pasquali M., Longo N. The effect of valinomycin in fibroblasts from patients with fatty acid oxidation disorders. Biochem. Biophys. Res. Commun. 2013;437:637–641. doi: 10.1016/j.bbrc.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djouadi F., Habarou F., Le Bachelier C. Mitochondrial trifunctional protein deficiency in human cultured fibroblasts: effects of bezafibrate. J. Inherit. Metab. Dis. 2016;39:47–58. doi: 10.1007/s10545-015-9871-3. [DOI] [PubMed] [Google Scholar]
- 19.Lefort B., Gouache E., Acquaviva C., Tardieu M., Benoist J.F., Dumas J.F. Pharmacological inhibition of carnitine palmitoyltransferase 1 restores mitochondrial oxidative phosphorylation in human trifunctional protein deficient fibroblasts. Biochim. Biophys. Acta. 2017;1863:1292–1299. doi: 10.1016/j.bbadis.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Ventura F.V., Ruiter J.P., Ijlst L., de Almeida I.T., Wanders R.J. Inhibitory effect of 3-hydroxyacyl-CoAs and other long-chain fatty acid beta-oxidation intermediates on mitochondrial oxidative phosphorylation. J. Inherit. Metab. Dis. 1996;19:161–164. doi: 10.1007/BF01799419. [DOI] [PubMed] [Google Scholar]
- 21.Ventura F.V., Ruiter J.P.N., IJlst L., Tavares De Almeida I., Wanders R.J.A. Lactic acidosis in long-chain fatty acid b-oxidation disorders. J. Inherit. Metab. Dis. 1998;21:645–654. doi: 10.1023/a:1005480516801. [DOI] [PubMed] [Google Scholar]
- 22.Hagenbuchner J., Scholl-Buergi S., Karall D., Ausserlechner M.J. Very long-/and long chain-3-hydroxy acyl-CoA dehydrogenase deficiency correlates with deregulation of the mitochondrial fusion/fission machinery. Sci. Rep. 2018;8:3254–3263. doi: 10.1038/s41598-018-21519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seminotti B., Leipnitz G., Karunanidhi A. Mitochondrial energetics is impaired in very long-chain acyl-CoA dehydrogenase deficiency and can be rescued by treatment with mitochondria-targeted electron scavengers. Hum. Mol. Genet. 2019;28:928–941. doi: 10.1093/hmg/ddy403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das A.M. Secondary respiratory chain defect in a boy with long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency: possible diagnostic pitfalls. Eur. J. Pediatr. 2000;159:243–246. doi: 10.1007/s004310050063. [DOI] [PubMed] [Google Scholar]
- 25.Rocchicciolo F., Wanders R., Aubourg P. Deficiency of long-chain 3-hydroxyacyl-CoA dehydrogenase: a cause of lethal myopathy and cardiomyopathy in early childhood. Pediatr. Res. 1990;28:657–662. doi: 10.1203/00006450-199012000-00023. [DOI] [PubMed] [Google Scholar]
- 26.Tyni T., Majander A., Kalimo H., Rapola J., Pihko H. Pathology of skeletal muscle and impaired respiratory chain function in long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency with the G1528C mutation. Neuromuscul. Disord. 1996;6:327–337. doi: 10.1016/0960-8966(96)00352-5. [DOI] [PubMed] [Google Scholar]
- 27.Ugras S., Daniels M.J., Fazelinia H., Gould N.S. Induction of the immunoproteasome subunit Lmp7 links proteostasis and immunity in α-synuclein aggregation disorders. EBioMedicine. 2018;31:307–319. doi: 10.1016/j.ebiom.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chumnarnsilpa S., Robinson R.C., Grimes J.M., Leyrat C. Calcium-controlled conformational choreography in the N-terminal half of adseverin. Nat. Commun. 2015;6:8254. doi: 10.1038/ncomms9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto K., Gandin V., Sasaki M. Largen: a molecular regulator of mammalian cell size control. Mol. Cell. 2014;53:904–915. doi: 10.1016/j.molcel.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 30.Plageman T.F., Jr., Zacharias A.L., Gage P.J., Lang R.A. Shroom3 and a Pitx2-N-cadherin pathway function cooperatively to generate asymmetric cell shape changes during gut morphogenesis. Dev. Biol. 2011;357:227–234. doi: 10.1016/j.ydbio.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houten SM Herrema H., Te Brinke H. Impaired amino acid metabolism contributes to fasting-induced hypoglycemia in fatty acid oxidation defects. Hum. Mol. Genet. 2013;22:5249–5261. doi: 10.1093/hmg/ddt382. [DOI] [PubMed] [Google Scholar]
- 32.Roe C.R., Sweetman L., Roe D.S., David F. Treatment of cardiomyopathy and rhabdomyolysis in long-chain fat oxidation disorders using an anaplerotic odd-chain triglyceride. J. Clin. Invest. 2002;110:259–269. doi: 10.1172/JCI15311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vockley J., Burton B., Berry G.T., Longo N. UX007 for the treatment of long chain-fatty acid oxidation disorders: safety and efficacy in children and adults following 24weeks of treatment. Mol. Genet. Metabol. 2017;120:370–377. doi: 10.1016/j.ymgme.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Gillingham M.B., Heitner S.B., Martin J., Rose S. Trihepatanoin versus trioctanoin for long-chain fatty acid oxidation disorders: a double blinded, randomized controlled trial. J. Inherit. Metab. Dis. 2017;40:831–843. doi: 10.1007/s10545-017-0085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ørngreen M.C., Sacchetti M., van Engelen B.G., Vissing J. Fuel Utilization in patients with very long-chain acyl-CoA dehydrogenase deficiency. Ann. Neurol. 2004;56:279–283. doi: 10.1002/ana.20168. [DOI] [PubMed] [Google Scholar]
- 36.Ørngreen M.C., Duno M., Ejstrup R., Christensen E. Fuel utilization in subjects with carnitine palmitoyltransferase 2 gene mutations. Ann. Neurol. 2005;57:60–66. doi: 10.1002/ana.20320. [DOI] [PubMed] [Google Scholar]
- 37.Behrend A.M., Harding C.O., Shoemaker J.D., Matern D. Substrate oxidation and cardiac performance during exercise in disorders of long chain fatty acid oxidation. Mol. Genet. Metabol. 2012;105:110–115. doi: 10.1016/j.ymgme.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diekman E.F., Visser G., Schmitz J.P., Nievelstein R.A. Altered energetics of exercise explain risk of rhabdomyolysis in very long-chain Acyl-CoA dehydrogenase deficiency. PloS One. 2016;11 doi: 10.1371/journal.pone.0147818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diekman E.F., Van der Pol W.P., Nievelstein R.A.J. Muscle MRI in patients with long-chain fatty acid oxidation disorders. J. Inherit. Metab. Dis. 2014;37:405–413. doi: 10.1007/s10545-013-9666-3. [DOI] [PubMed] [Google Scholar]
- 40.Olsen R.K., Cornelius N., Gregersen N. Genetic and cellular modifiers of oxidative stress: what can we learn from fatty acid oxidation defects? Mol. Genet. Metabol. 2013;110(Suppl):S31–S39. doi: 10.1016/j.ymgme.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Ghosh S., Wicks S.E., Vandanmagsar B., Mendoza T.M. Extensive metabolic remodeling after limiting mitochondrial lipid burden is consistent with an improved metabolic health profile. J. Biol. Chem. 2019;294:12313–12327. doi: 10.1074/jbc.RA118.006074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary files. The proteomic data are available via ProteomeXchange with identifier PXD024488.