Abstract
Molecular interactions between two different classes of β-lactamase enzymes and outer membrane protein A (OmpA) were studied by in vivo chemical cross-linking of a multi-drug-resistant strain of Acinetobacter baumannii AB5075. Class A β-lactamase blaGES-11 and Class D β-lactamase Oxa23, responsible for hydrolysis of different types of β-lactam antibiotics, were found to be cross-linked to similar lysine sites of the periplasmic domain of outer membrane protein OmpA, despite low sequence homology between the two enzymes. The findings from in vivo XL-MS suggest that the interacting surfaces between both β-lactamase enzymes and OmpA are conserved during molecular evolution, and the OmpA C-terminus domain serves an important function of anchoring different types of β-lactamase enzymes in the periplasmic space.
Keywords: in vivo cross-linking, Oxa23, blaGES-11, OmpA, AB5075
Graphical Abstract
INTRODUCTION
In vivo cross-linking (XL)-MS is a useful tool to study protein conformations, protein complex architectures, and molecular interactions under their native environment.1 In the past decade, our laboratory has developed a class of lysine-targeting chemical cross-linker molecules, named protein interaction reporter (PIR) cross-linker,2 and Real-time Analysis for Cross-linked peptide Technology (ReACT),3 as well as associated bioinformatic tools4,5 for in vivo applications of XL-MS on a variety of biological systems including bacterial cells,3,6,7 human cancer cells,8,9 subcellular organelle such as mitochondria,10 and tissue samples.11 In a previous study,12 we applied in vivo XL-MS to a multi-drug-resistant (MDR) strain of Acinetobacter baumannii, AB5075, which is a contemporary clinical isolate from a patient in the U.S. military health care system and has recently been established as a model strain for the evaluation of pathogenesis and antimicrobial treatments.13 In vivo XL-MS of AB5075 revealed multiple novel molecular interactions among outer membrane and periplasmic proteins, especially the interactions between a key resistance factor, Class D β-lactamase Oxa-23 encoded on the chromosome, and several outer membrane porin proteins such as OmpA, OmpW and CarO. These results provided new structural insights on how the enzyme Oxa-23 is localized at the entry port of antibiotics and other molecules through interaction with the porin proteins.12 Another study by Ambrosi et al.14 also showed that oxacillinases, including Oxa-23 and Oxa-51, copurified with outer membrane proteins such as OmpA, in other carbapenem-resistant A. baumannii strains, which provides independent validation of OmpA-β-lactamase interactions without using cross-linking reagents.
With increased in vivo XL-MS experiments using AB5075, the XL-MS protein interaction network of AB5075 was revealed with increasing detail, and we discovered that another import resistance factor, the Class A β-lactamase blaGES-11 encoded on the resistance island in one of the plasmids, also interacts with the C-terminal domain of the most abundant outer membrane protein OmpA. Both Oxa-23 and blaGES-11originate from the penicillin-binding proteins, the β-lactam antibiotic targets, and key enzymes involved in peptidoglycan biosynthesis.15 The two β-lactamase enzymes share some three-dimensional structural similarity and utilize a conserved serine residue as catalysis center despite low sequence identity.15 Composed of an N-terminal β-barrel domain spanning the cell outer membrane and C-terminal domain in the periplasmic space, OmpA is an integral membrane involved in a variety of biological processes, such as ion transport,16,17 maintenance of cell envelope structural integrity,18 and invasion of host cells.19,20 By comparing the cross-linked sites between the two β-lactamase enzymes and the OmpA C-terminal domain, we found that not only the three-dimensional folding of the enzymes but also their interaction surfaces with OmpA periplasmic domain are conserved, despite the striking primary sequence and β-lactam substrate differences. These findings have extended our knowledge on the interplay between β-lactamase enzymes and outer membrane porin proteins, as well as the conservation of interaction surfaces during molecular evolution. In addition, localization of multiple β-lactamase enzymes at the OmpA C-terminal domain could increase resistance to multiple antibiotics and be an important feature of MDR phenotypes.
EXPERIMENTAL SECTION
Agar Diffusion Assay.
Sensitivity of wild-type AB5075 and transposon insertion mutants against antibiotic drugs was evaluated by agar diffusion assay measurements. Overnight LB media A. baumannii cell cultures were diluted to OD600 nm = 0.2 and then spread onto LB agar plates with sterile cotton swabs. The antibiotic disks (BBL Sensi-Disc, Becton, Dick-inson and Company, Franklin Lakes, NJ) including aztreonam (30 μg/mL), ceftazidime (30 μg/mL), ertapenem (10 μg/mL), and meropenem (10 μg/mL) were placed onto the bacterial lawn. Cell growth on an agar plate with an antibiotic disk was allowed for 24 h at 37 °C. The annular radius (mm) of the inhibition zone was measured for drug sensitivity comparison. The transposon insertion mutants were obtained from the Manoil Lab at the University of Washington.21
In Vivo Cross-Linking of AB5075 and Analysis of Cross-Linked Peptides.
In vivo cross-linking of live AB5075 cells using the PIR cross-linker and LC−MS analysis of cross-linked peptide pairs were similar as previously described.6,12,22 Briefly, the harvested bacteria cells were cross-linked with biotin-aspartate proline-n-hydroxyphthalimide,2,22 lysed, and digested by trypsin. The digested sample were desalted, fractionated, and affinity enriched prior to LC-ReACT analysis. Details of the sample processing, LC−MS instrument settings, and data analysis are described in the Supporting Information.
RESULTS AND DISCUSSION
From genomic analysis, the AB5075 genome harbors three unique β-lactamase genes, the Class A β-lactamase blaGES-11, Class D β-lactamase Oxa23, and putative β-lactamase HcpC, in addition to another 15 orthologous β-lactamase genes also present in ATCC19606, a traditional model A. baumannnii strain commonly used by many other researchers. By data-dependent MS analysis of the proteome of AB5075, we found among all the β-lactamase enzymes, Oxa23 and blaGES-11 are the most abundantly expressed, with the peptide-spectra match counts around 6-fold of the third most abundant β-lactamase. Our previous study showed that inactivation of AmpC or Oxa-69 in WT AB5075 by transposon insertion does not decrease the intrinsic resistance to meropenem and impenem.12 Here, we further investigated β-lactam sensitivity of transposon mutants of Oxa-23 and blaGES-11 by agar disk diffusion assay (Figure 1). The results show that transposon mutant of Oxa23 exhibits reduced sensitivity to meropenem and ertapenem while retaining the resistance to aztreonam and ceftazidime; the transposon mutant of blaGES-11 exhibits increased sensitivity to aztreonam and ceftazidime but retains the resistance to meropenem and ertapenem. From these observations, we can conclude that AB5075 gains resistance to the extended spectrum of β-lactam antibiotics by producing a versatile family of β-lactamase enzymes that are complementary in hydrolyzing different classes of β-lactams.
Figure 1.
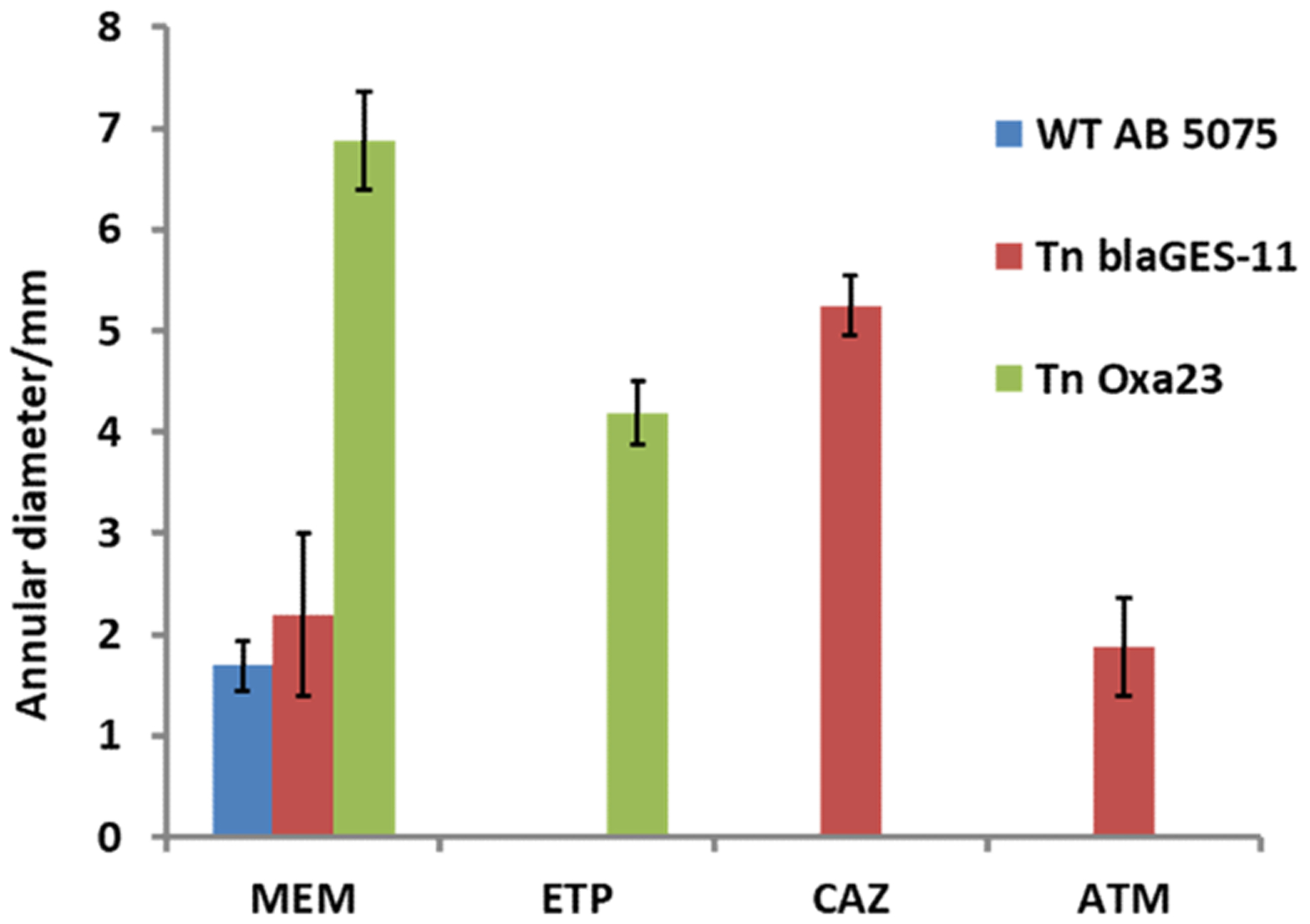
Antibiotic sensitivity comparison of WT AB5075 and transposon insertion mutants of blaGES-11 and Oxa23 in AB5075 by disc diffusion assay. The error bars denote standard deviation of measured annular diameter of growth inhibition zone from four biological replicates. Larger annular diameter of the growth inhibition zone indicates increased antibiotic sensitivity: MEM, meropenem, 10 μg/mL; ETP, ertapenem, 10 μg/mL; ATM, aztreonam, 30 μg/mL; CAZ, cefazidime, 30 μg/mL.
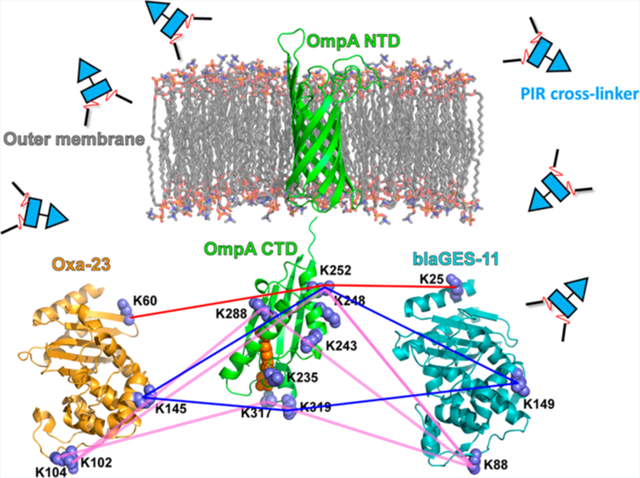
By in vivo XL-MS study, we found that blaGES-11 and Oxa23 are both extensively cross-linked to the periplasmic C-terminal OmpA domain. As an example for demonstration of PIR and ReACT applied in this study, Figure 2 shows the MS spectra corresponding to the identification of a cross-linked peptide pair between OmpA and Oxa23. All cross-linked peptide spectral matches for inter-protein crosslinks between two β-lactamases and OmpA C-terminus from three biological replicates of XL-ReACT MS experiments are summarized in Table S1. Table 1 lists the non-redundant interprotein cross-linked peptide pairs between the two β-lactamase enzymes and OmpA C-terminus. These results indicate that the two classes of β-lactamase enzymes are both localized in the periplasmic space between the outer membrane and the peptidoglycan layer of AB5075 through interaction with the C-terminal domain of OmpA. In spite of only 14% primary sequence identity, superimposition of crystal structures of Oxa23 (PDB: 4JF4) and blaGES-11 (PDB: 3V3R) reveals similarity in overall three dimensional folding of the two β-lactamase enzymes, rmsd = 2.4 Å) (Figure 3). Moreover, despite low conservation of lysine residues in blaGES-11 and Oxa-23, a highly similar interprotein linkage configuration with the OmpA C-terminal domain was observed. For instance, three pairs of lysine sites, K60 of Oxa23 and K25 of blaGES-11, K145 of Oxa23 and K149 of blaGES-11, and K104/K102 of Oxa23 and K88 of blaGES-11, located at similar relative positions within the three-dimensional surfaces of the two enzymes, as highlighted in Figure 3B, were found to be cross-linked to the same lysine sites on the OmpA C-terminus. Details of the cross-linking site interaction between the two β-lactamase enzymes and the OmpA C-terminal domain are depicted in Figure 4. Strikingly, these lysine residues located on the surface opposite to the antibiotic binding pocket (Figure 3B) were cross-linked to the same lysine sites in the OmpA C-terminal domain, despite very different local primary sequences. For example, K60 of Oxa23 located between strands β1 and β2 was identified to be cross-linked to K252 of OmpA, as was K25 in helix α1 of blaGES-11 (Figure 4A); K145 in helix α6 of Oxa23 and K149 in helix α6 of blaGE-11 were both cross-linked to K252 and K319 of OmpA (Figure 4B); K102 and K104 located in the α3−α4 loop of Oxa23, and K88 located in the P-loop of blaGES-11 were identified as being cross-linked to K252, K288, and K317 of OmpA (Figure 4C). From these site interactions defined by XL-MS, we can infer the relative spatial orientations of the two β-lactamase enzymes toward OmpA. The interacting surfaces between the two β-lactamase enzymes and OmpA C-terminus are opposite to the substrate binding pocket, thus allowing OmpA interaction compatible with free access of antibiotics with β-lactamase active sites, specifically the S-x-x-K motif conserved in both classes of enzymes (Figure 3B). In this way, blaGES-11 and Oxa23 are concentrated in the periplasmic space, which could deactivate β-lactam molecules upon entry into the cell and block their access to their target proteins, the penicillin binding proteins involved in the biosynthesis of the peptidoglycan layer. Moreover, interactions of both lactamase enzymes with the OmpA C-terminal domain are compatible with OmpA interaction with peptidoglycan which occurs via a pocket at the distal end of the domain (Figure 4).18 These results also suggest that, in addition to its role in stabilizing the outer membrane envelope by binding with peptidoglycan, the OmpA C-terminal domain has an important function of localizing and stabilizing β-lactamase enzymes in the periplasmic space between the cell outer membrane and peptidoglycan layer.
Figure 2.
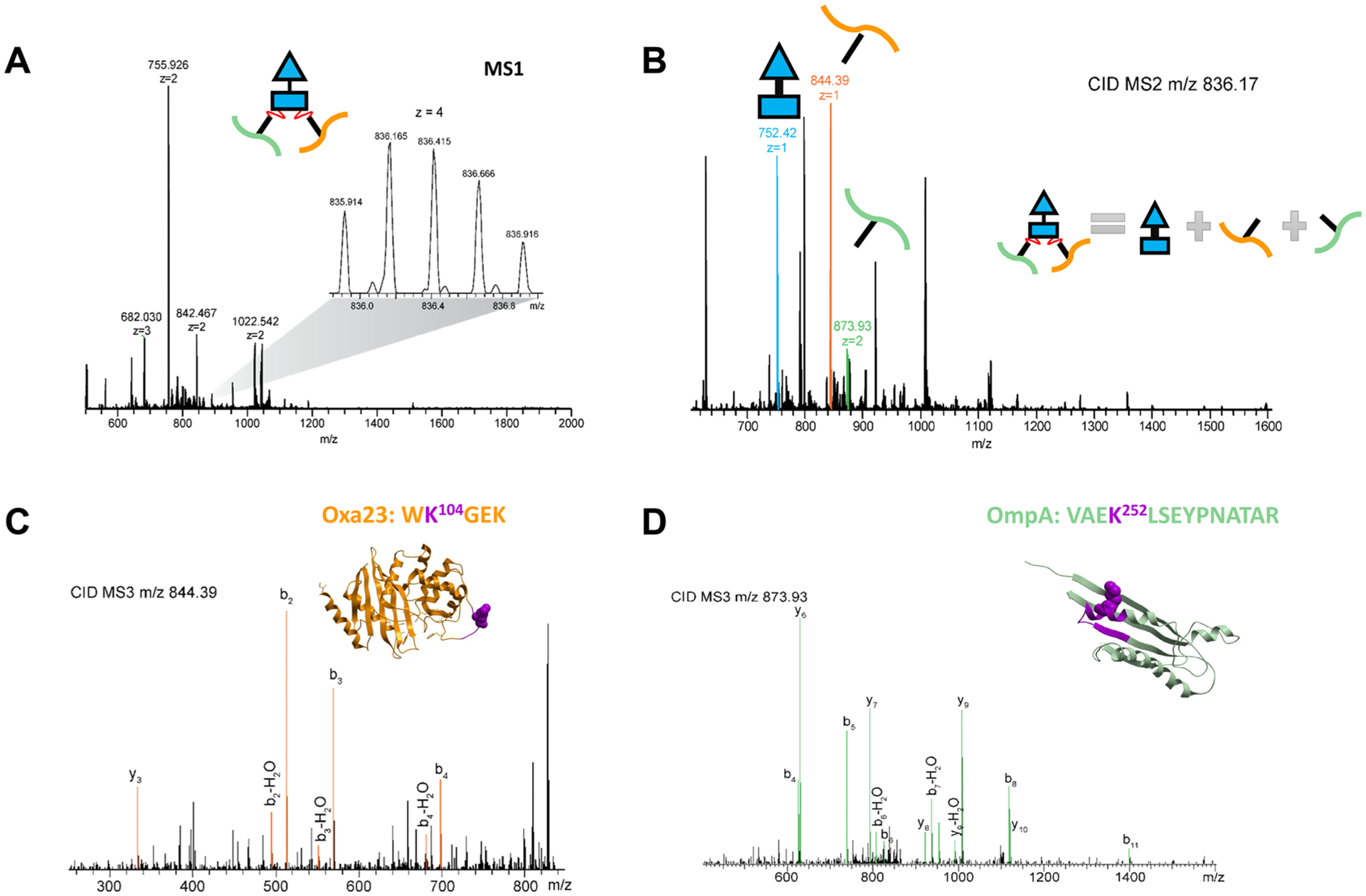
ReACT spectra for identification of cross-linking between Oxa23 K104 and OmpA K252. (A) Full MS survey scan of precursor ion; (B) CID MS/MS scan that discovers the cross-linking mass relationship; (C, D) CID MS/MS/MS scans for sequence identification of the two linear peptides released from the cross-linked precursor ion. The cross-linked peptide regions are highlighted in purple in the crystal structures of Oxa23 (PDB: 4JF4) and OmpA C-terminal domain (PDB: 3TD3).
Table 1.
List of Interprotein Cross-Linked Peptide Pairs between Oxa23 (uniport ID: Q9L4P2) and OmpA (uniprot ID: W6RU67), blaGES-11 (uniprot ID: C5HUY1), and OmpA from in Vivo Cross-Linking Experimentsa
| blaGES-11 peptide_OmpA peptide | Oxa-23 peptide_OmpA peptide |
|---|---|
| LTFK25TDLEK_ VAEK252LSEYPNATAR | K60INLYGNALSR_ VAEK252LSEYPNATAR |
|
K149IGDSVSR_ VAEK252LSEYPNATAR K149IGDSVSR TK319EGR |
IGLDLMQK145EVK_VAEK252LSEYPNATAR IGLDLMQK145EVK TK319EGR |
| K88LSYGPDMIVEWSPATER_VAEK252LSEYPNATAR | WK104GEK_ VAEK252LSEYPNATAR WK104GEKR_ VAEK252LSEYPNATAR TDINEIFK102WK_VAEK252LSEYPNATAR |
| K88LSYGPDMIVEWSPATER_ANSVK288SALVNEYNVDASR | TDINEIFK102WK_ANSVK288SALVNEYNVDASR |
| K88LSYGPDMIVEWSPATER_LSTQGFAWDQPIADNK317TK | WK104GEKR_ LSTQGFAWDQPIADNK317TK WK104GEK_ TK319EGR |
| LTFK25TDLEK_VFFDTNK235SNIKDQYKPEIAK LTFK25TDLEK_SNIKDQYKPEIAK248VAEK LTFK25TDLEK_ VAEK252LSEYPNATAR LTFK25TDLEK_ANSVK288SALVNEYNVDASR LTFK25TDLEK_LSTQGFAWDQPIADNK317TK LTFK25TDLEK_TK319EGR |
VTPIQEVEFVSQLAHTQLPFSEK194VQANVK_SNIKDQYKPEIAK248VAEK VTPIQEVEFVSQLAHTQLPFSEK194VQANVK_ANSVK288SALVNEYNVDASR TGWAMDIK224PQVGWLTGWVEQPDGK_VAEK252LSEYPNATAR WK104GEK_SNIKDQYKPEIAK248VAEK TDINEIFK102WK_SNIKDQYK243PEIAKVAEK VTPIQEVEFVSQLAHTQLPFSEK194VQANVK_VAEK252LSEYPNATAR |
The gray shaded linkages are highlighted in Figure 4.
Figure 3.
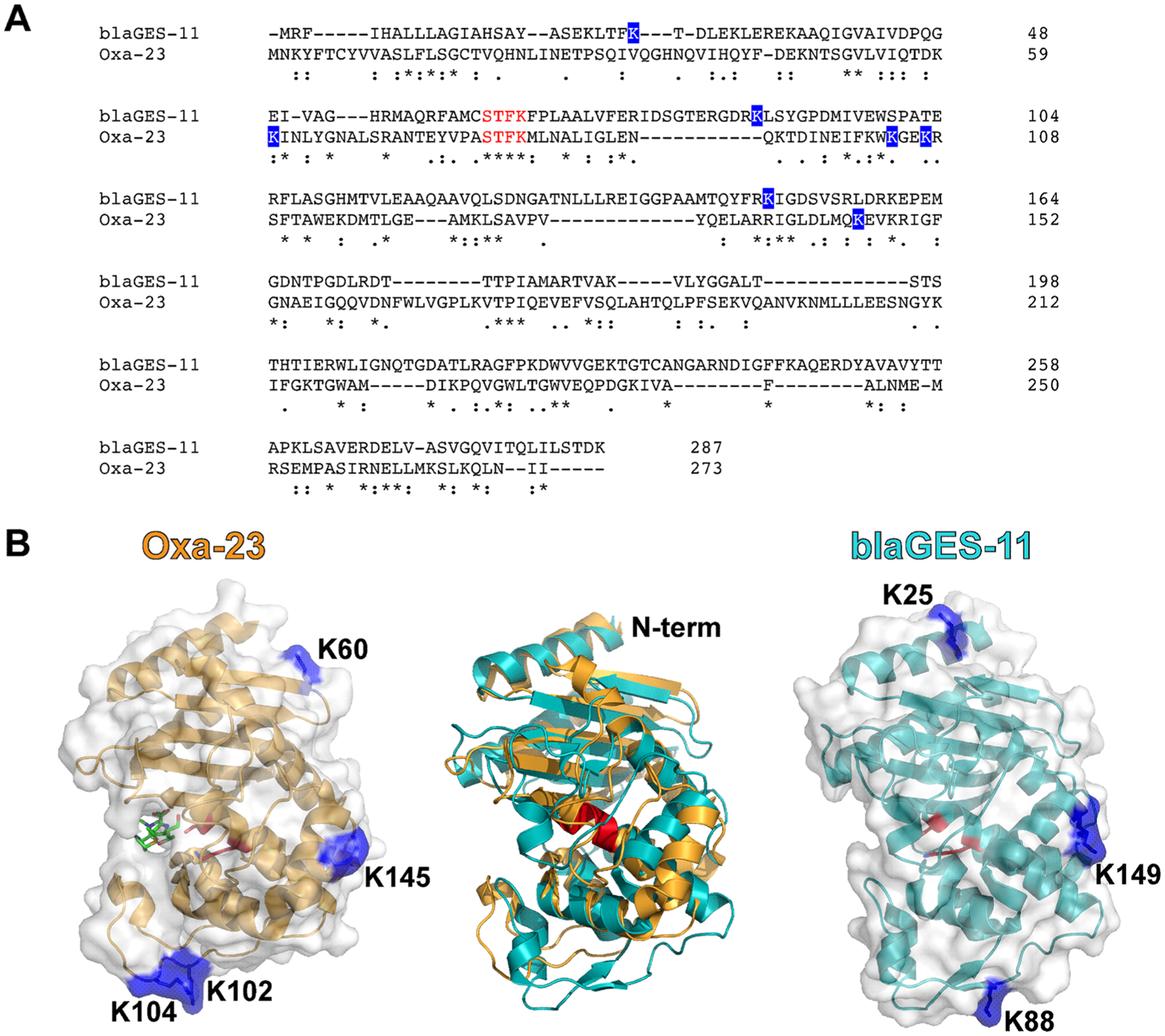
Sequence (A) and crystal structure (B) alignment of blaGES-11(PDB: 3V3R) and Oxa23 complexed with meropenem (PDB: 4JF4). Lysine residues crosslinked to the OmpA C-terminus and located in similar regions of 3D structures are highlighted in blue. The active site Ser and Lys residues in the conserved S-xx-K motif in both Class A and D β-lactamase enzymes are highlighted in red in the structures. The four consecutive conserved residues (STFK) near the substrate binding pocket are highlighted in red in the backbone alignment. Sequence alignment between blaGES-11 (UniprotID: C5HUY1) and Oxa23 (Uniprot ID: Q9L4P2) was performed by Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Figure 4.
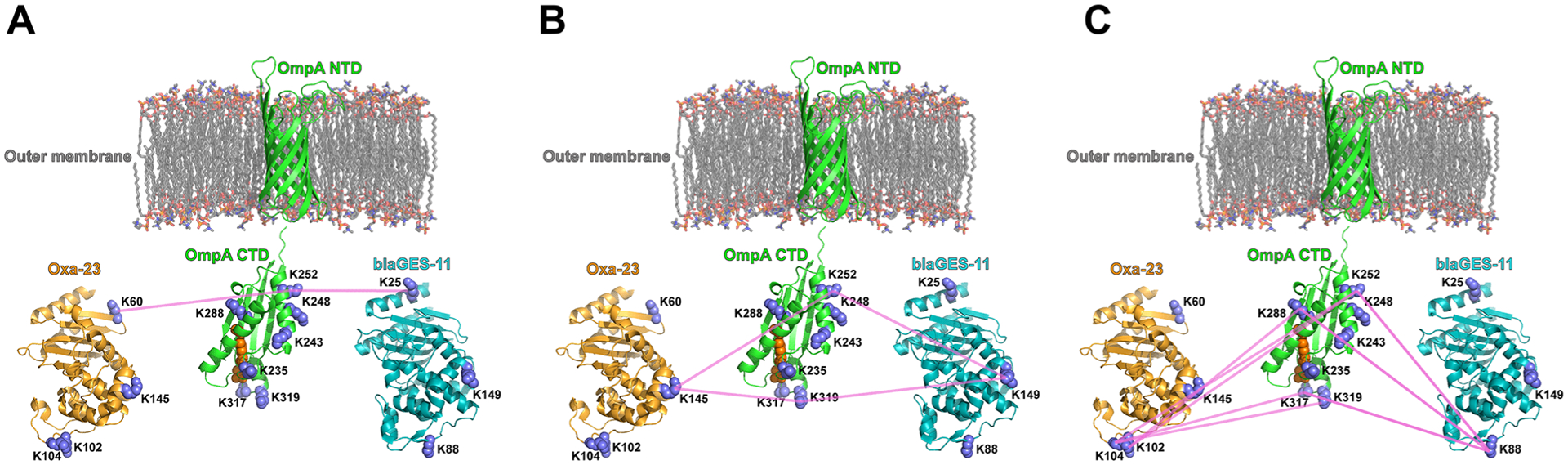
Conserved lysine site linkage between the two β-lactamase enzymes (PDB: 4JF4, 3V3R) and the OmpA C-terminal domain (PDB:3TD3). (A) Oxa23 K60 and blaGES-11 K25 are both crosslinked to OmpA K252; (B) Oxa23 K145 and blaGES-11 K149 are both crosslinked to OmpA K252 and OmpA K319; (C) blaGES-11 K88 is crosslinked to OmpA K252, K288, K317, and this same OmpA linkage was observed with K102/K104 of Oxa23. The OmpA N-terminal structure model (a.a. 23–191, Uniprot ID: W6RU67) was generated by Phyre2 using the intensive modeling mode.26 The residues for noncovalent interaction with peptidoglycan in the OmpA CTD domain, Asp268, Arg283, Ala284, are highlighted in orange.18
CONCLUSIONS
In vivo XL-MS of the multi-drug-resistant strain of A. baumannii, AB5075, provides new insight on how different classes of β-lactamase enzymes interact with the periplasmic domain of outer membrane protein OmpA. Despite little sequence homology, the Class D β-lactamase Oxa23 and Class A β -lactamase blaGES-11 have similar interacting surfaces with OmpA, which is revealed by conservation of cross-linking sites between the enzymes and OmpA C-terminal domain. These results also indicate that OmpA serves an important role in anchoring different types of β-lactamase enzymes to protect the bacterial cells from different classes of β-lactams. This finding is consistent with previous results indicating deletion of OmpA results in a significant decrease in minimum inhibitory concentration of multiple antibiotics in A. baumannii.23 Finally, our finding also may help explain the conclusion by Kwon et al. that OmpA contributes to the antimicrobial resistance of A. baumannii through the C-terminal domain.24 Deletion of the OmpA C-terminal domain not only impairs OmpA-peptidoglycan binding but also adversely affects the ability to localize β-lactamase enzymes at the site of this porin involved in antibiotic permeation.25 Through this study, we also demonstrated that general utility of in vivo XL-MS strategy exists for discovery of intermolecular linkage conservation of protein interaction partners, even for enzymes with considerable sequence divergence. Conserved linkage at common interactor sites may serve as a means of the future to not only discover common interactors but also reveal conserved structural features and molecular interfaces in a way not previously possible.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by grants from the US National Institutes of Health U19AI107775. We thank Dr. Manoil Lab, University of Washington, for providing the transposon mutants of AB5075.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.9b00021.
Experimental procedures (PDF)
ms2 data for β-lactamase Oxa23 (XLSX)
The authors declare no competing financial interest.
REFERENCES
- (1).Chavez JD; Bruce JE Chemical cross-linking with mass spectrometry: a tool for systems structural biology. Curr. Opin. Chem. Biol 2019, 48, 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Tang X; Bruce JE A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies. Mol. BioSyst 2010, 6, 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Weisbrod CR; Chavez JD; Eng JK; Yang L; Zheng C; Bruce JE In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J. Proteome Res 2013, 12, 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Schweppe DK; Zheng CX; Chavez JD; Navare AT; Wu X; Eng JK; Bruce JE XLinkDB 2.0: integrated, large-scale structural analysis of protein crosslinking data. Bioinformatics 2016, 32, 2716–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zheng CX; Weisbrod CR; Chavez JD; Eng JK; Sharma V; Wu X; Bruce JE XLink-DB: Database and Software Tools for Storing and Visualizing Protein Interaction Topology Data. Journal of Proteome Research 2013, 12, 1989–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zhong X; Navare AT; Chavez JD; Eng JK; Schweppe DK; Bruce JE Large-Scale and Targeted Quantitative Cross-Linking MS Using Isotope-Labeled Protein Interaction Reporter (PIR) Cross-Linkers. J. Proteome Res 2017, 16, 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Navare AT; Chavez JD; Zheng C; Weisbrod CR; Eng JK; Siehnel R; Singh PK; Manoil C; Bruce JE Probing the protein interaction network of Pseudomonas aeruginosa cells by chemical cross-linking mass spectrometry. Structure 2015, 23, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chavez JD; Schweppe DK; Eng JK; Bruce JE In Vivo Conformational Dynamics of Hsp90 and Its Interactors. Cell Chem. Biol 2016, 23, 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chavez JD; Schweppe DK; Eng JK; Zheng CX; Taipale A; Zhang YY; Takara K; Bruce JE Quantitative interactome analysis reveals a chemoresistant edgotype. Nat. Commun 2015, DOI: 10.1038/ncomms8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Schweppe DK; Chavez JD; Lee CF; Caudal A; Kruse SE; Stuppard R; Marcinek DJ; Shadel GS; Tian R; Bruce JE Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 1732–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chavez JD; Lee CF; Caudal A; Keller A; Tian R; Bruce JE Chemical Crosslinking Mass Spectrometry Analysis of Protein Conformations and Supercomplexes in Heart Tissue. Cell Syst. 2018, 6, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wu X; Chavez JD; Schweppe DK; Zheng C; Weisbrod CR; Eng JK; Murali A; Lee SA; Ramage E; Gallagher LA; Kulasekara HD; Edrozo ME; Kamischke CN; Brittnacher MJ; Miller SI; Singh PK; Manoil C; Bruce JE In vivo protein interaction network analysis reveals porin-localized antibiotic inactivation in Acinetobacter baumannii strain AB5075. Nat. Commun 2016, 7, 13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jacobs AC; Thompson MG; Black CC; Kessler JL; Clark LP; McQueary CN; Gancz HY; Corey BW; Moon JK; Si Y; Owen MT; Hallock JD; Kwak YI; Summers A; Li CZ; Rasko DA; Penwell WF; Honnold CL; Wise MC; Waterman PE; Lesho EP; Stewart RL; Actis LA; Palys TJ; Craft DW; Zurawski DV AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. mBio 2014, 5, e01076–01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ambrosi C; Scribano D; Aleandri M; Zagaglia C; Di Francesco L; Putignani L; Palamara AT Acinetobacter baumannii Virulence Traits: A Comparative Study of a Novel Sequence Type with Other Italian Endemic International Clones. Front. Microbiol 2017, 8, 1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jeon JH; Lee JH; Lee JJ; Park KS; Karim AM; Lee C-R; Jeong BC; Lee SH Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int. J. Mol. Sci 2015, 16, 9654–9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Jyothisri K; Deepak V; Rajeswari MR Purification and characterization of a major 40 kDa outer membrane protein of Acinetobacter baumannii. FEBS Lett. 1999, 443, 57–60. [DOI] [PubMed] [Google Scholar]
- (17).Sugawara E; Nikaido H OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. Journal of bacteriology. 2012, 194, 4089–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Park JS; Lee WC; Yeo KJ; Ryu KS; Kumarasiri M; Hesek D; Lee M; Mobashery S; Song JH; Kim SI; Lee JC; Cheong C; Jeon YH; Kim HY Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J. 2012, 26, 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Schweppe DK; Harding C; Chavez JD; Wu X; Ramage E; Singh PK; Manoil C; Bruce JE Host-Microbe Protein Interactions during Bacterial Infection. Chem. Biol 2015, 22, 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Choi CH; Lee JS; Lee YC; Park TI; Lee JC Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol. 2008, 8, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Gallagher LA; Ramage E; Weiss EJ; Radey M; Hayden HS; Held KG; Huse HK; Zurawski DV; Brittnacher MJ; Manoil C Resources for Genetic and Genomic Analysis of Emerging Pathogen Acinetobacter baumannii. J. Bacteriol 2015, 197, 2027–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Chavez JD; Mohr JP; Mathay M; Zhong X; Keller A; Bruce JE Systems structural biology measurements by in vivo cross-linking with mass spectrometry. Nat. Protoc 2019, 14, 2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Smani Y; Fabrega A; Roca I; Sanchez-Encinales V; Vila J; Pachon J Role of OmpA in the Multidrug Resistance Phenotype of Acinetobacter baumannii. Antimicrob. Agents Chemother 2014, 58, 1806–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kwon HI; Kim S; Oh MH; Na SH; Kim YJ; Jeon YH; Lee JC Outer membrane protein A contributes to antimicrobial resistance of Acinetobacter baumannii through the OmpA-like domain. J. Antimicrob. Chemother 2017, 72, 3012–3015. [DOI] [PubMed] [Google Scholar]
- (25).Iyer R; Moussa SH; Durand-Reville TF; Tommasi R; Miller A Acinetobacter baumannii OmpA Is a Selective Antibiotic Permeant Porin. ACS Infect. Dis 2018, 4, 373–381. [DOI] [PubMed] [Google Scholar]
- (26).Kelley LA; Mezulis S; Yates CM; Wass MN; Sternberg MJE The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc 2015, 10, 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


