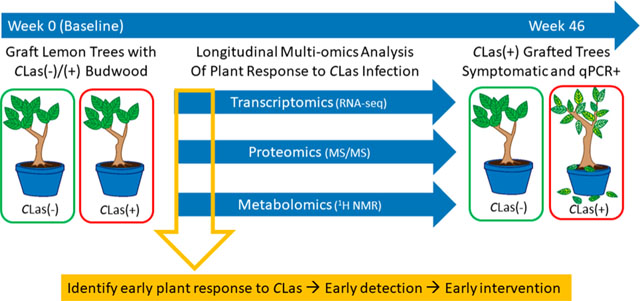
Abstract
Presymptomatic detection of citrus trees infected with Candidatus Liberibacter asiaticus (CLas), the bacterial pathogen associated with Huanglongbing (HLB; citrus greening disease), is critical to controlling the spread of the disease. To test whether infected citrus trees produce systemic signals that may be used for indirect disease detection, lemon (Citrus limon) plants were graft-inoculated with either CLas-infected or control (CLas-) budwood, and leaf samples were longitudinally collected over 46 weeks and analyzed for plant changes associated with CLas infection. RNA, protein, and metabolite samples extracted from leaves were analyzed using RNA-Seq, mass spectrometry, and 1H NMR spectroscopy, respectively. Significant differences in specific transcripts, proteins, and metabolites were observed between CLas-infected and control plants as early as 2 weeks post graft (wpg). The most dramatic differences between the transcriptome and proteome of CLas-infected and control plants were observed at 10 wpg, including coordinated increases in transcripts and proteins of citrus orthologs of known plant defense genes. This integrated approach to quantifying plant molecular changes in leaves of CLas-infected plants supports the development of diagnostic technology for presymptomatic or early disease detection as part of efforts to control the spread of HLB into uninfected citrus groves.
Keywords: citrus greening disease, Candidatus Liberibacter asiaticus, huanglongbing, transcriptomics, proteomics, metabolomics, RNA-seq, mass spectrometry, proton nuclear magnetic resonance
Graphical Abstract
INTRODUCTION
Detection of specific plant changes in response to pathogen infection (indirect detection) is an alternative to disease diagnostic strategies which rely on direct detection of the pathogen associated with the disease. All commercial citrus varieties are susceptible to Huanglongbing (HLB, citrus greening disease), a bacterial disease which leads to reduced fruit quality and quantity and early tree death, and which has spread throughout the world, including all citrus growing states of the U.S.1 A sensitive and specific quantitative polymerase chain reaction (qPCR) assay has been widely used for direct detection of Candidatus Liberibacter asiaticus (CLas), the bacterial pathogen associated with HLB, in citrus leaves and in the Asian citrus psyllid (Diaphorina citri), the insect vector of the pathogen.2 While this assay has been extensively validated and is highly robust, the uneven distribution of the bacteria in the tree creates a sampling problem, in which many leaves from a diseased tree may have undetectable levels of CLas.3 Furthermore, when CLas is transmitted to a healthy tree, the pathogen moves through the phloem and replicates to high levels in root tissue, while the titer in the foliar tissue can remain below the limit of qPCR detection for a year or more.4 During the time that infected trees are presymptomatic and the pathogen titer in foliar tissue is below the limit of currently available direct detection technologies, the insect vector is still capable of acquiring CLas from the flush tissue, or the youngest growth, of these trees, and rapidly spreading the pathogen throughout previously healthy groves.5 Horizontal transmission of CLas to psyllid nymphs via the flush tissue, or the youngest growth, occurs in just a single insect generation. This means the pathogen can rapidly spread within a grove even before a plant becomes systemically infected.
Indirect detection diagnostic technology relies on the ability to monitor specific molecular changes in the plant that occur upon infection, including changes in the expression of genes or proteins, and in the production of metabolites. Unique gene expression profiles associated with infection by Xylella fastidiosa, the causative agent of Pierce’s disease, have been identified in grapevines.6 Human peptide biomarkers identified in serum, urine, and saliva have been associated with heart disease, cancer, and other diseases.7 Metabolomic analysis of citrus fruit resulted in the identification of metabolite-based biomarkers whose abundance varies depending on whether the fruit was derived from healthy or CLas-infected trees.8 Advances in genomics, proteomics, and metabolomics technologies have dramatically increased the feasibility of obtaining detailed molecular profiles of complex samples, allowing classification of unknown samples as healthy or diseased depending on the observed biomarker profile.
Several studies have analyzed the effect of CLas in citrus using transcriptomics, proteomics, and metabolomics approaches. Proton nuclear magnetic resonance (1H NMR) metabolomics has revealed metabolic profiles associated with diseased fruit,8–10 RNA-Seq has been used for transcriptome analysis of fruit, roots, and leaves, and proteomic analysis has been applied to root and leaf samples of healthy and CLas-infected trees.11–19 To understand the factors underlying premature fruit drop from diseased trees,20 transcriptome analysis of the calyx abscission zones in navel oranges was performed, revealing that CLas infection was associated with dramatic upregulation of transcripts associated with ethylene and jasmonic acid metabolism.21 Direct ionization mass spectrometry was used to sample healthy and symptomatic diseased trees to identify seasonal variation in HLB biomarker metabolites.22 With the exception of the final case, each of these studies has analyzed differences between healthy and infected trees at a single time point.
In this study, lemon (Citrus limon) plants were grafted with either control (CLas−) or infected (CLas+) budwood. Over the course of the subsequent 46 weeks, leaf samples were collected from a total of 11 control grafted and 11 CLas grafted plants on a biweekly basis. Leaf samples were analyzed for CLas using the USDA qPCR protocol2,23 to quantify pathogen titer over time. Leaf samples were also collected for transcriptomic, proteomic, and metabolomics analyses, to evaluate specific plant changes associated with response to CLas which may be exploited for diagnostic purposes.
Detection of presymptomatic infected trees is a critical component of the extensive effort underway to contain the spread of HLB. This is particularly relevant to states such as California, where the insect vector has dramatically expanded its range since its first appearance in 2008. In 2012, the first HLB-positive citrus tree was identified in California, in Hacienda Heights, and since that time the number of diseased trees identified in surveys led by the California Department of Food and Agriculture has steadily increased. In 2018, nearly 700 diseased trees were newly identified in California, although to date no diseased trees have been found in commercial groves in that state, only in residential areas. Proactive measures are required to protect the livelihood of citrus growers by removing infected trees at the earliest possible stage to prevent the spread of the pathogen by the Asian citrus psyllid.
EXPERIMENTAL SECTION
Citrus Growth Conditions, Grafting, and Sample Collection
All research involving plants was conducted in accordance with state and federal guidelines regulating the culture, transport, and disposal of plants and plant material infected with the plant bacterial pathogen Candidatus Liberibacter asiaticus. The plants used in the experiments were Lisbon lemon scion (VI380, Limonoira 8A; California Citrus Clonal Protection Program) on Carrizo rootstock (rootstock was started from seed). The plants were maintained in round 7.6 L pots (22 cm diameter and 21 cm height) in an insect-free greenhouse at 27 °C (±1.5 °C) with supplemental lighting (high-pressure sodium lights; 16 h light:8 h dark) at the University of California, Davis, Contained Research Facility. Humidity was not controlled. The plants were watered as needed, and at each watering, the plants were fertilized with a complete fertilizer (5–12–26) that contained micronutrients. The plants were approximately 6 months old when they were graft-inoculated (control graft: N = 11; infected graft: N = 11). To inoculate each plant, one branch was selected to receive three T-bud grafts, and grafts were placed as close to the main stem as possible on the branch. The plant material used for the grafts were buds taken from Lisbon lemon source plants that either had repeatedly tested negative (control plants) or had repeatedly tested positive for CLas (Hacienda Heights isolate) using qPCR2,23 to determine the presence of CLas in plant tissue. The Ct values for the source plants were 40 for the control plants and ranged from 21 to 23 for the CLas plants. At approximately monthly intervals after grafting, leaf samples were collected at random from each plant and DNA was extracted from dissected midribs and analyzed by qPCR for the presence of CLas, using the USDA standard protocol for CLas detection.2
Citrus RNA-Seq Sample Preparation and Sequencing
RNA was extracted using a modified RNeasy (Qiagen) protocol from one flash frozen leaf sampled from each plant at each time point. Leaves were cryoground by mortar and pestle. A modified RLT lysis buffer [4 M guanidine isothiocyanate, 0.2 M sodium acetate pH 5.2, 25 mM EDTA, 2.5% (w/v) PVP-40, 1% (v/v) β-mercaptoethanol, 10% volume of 20% sarkosyl] was added to the ground tissue along with an equivolume amount of chloroform. Following vortexing and centrifugation, the upper phase was mixed with ethanol and transferred to an RNA purification cartridge, and washed and eluted according to standard protocols. RNA samples were visualized using gel electrophoresis to confirm minimal degradation, and RNA concentration was quantified by Nanodrop. Five μg of RNA from each sample were subjected to polyA enrichment, and the enriched RNA samples were used to construct barcoded strand-specific libraries.24 The 24 barcoded samples were pooled and 100 bp paired end sequencing was performed in high output mode using one lane of the Illumina HiSeq 2000/2500 instrument.
RNA-Seq Sequence Assembly
All RNA-Seq fastq files were mapped to both the Citrus clementina v1.025 and Citrus sinensis v1.025 genome sequence files using tophat v2.0.12.26 In addition, a de novo transcript assembly was constructed with all reads using Trinity r20140717.27 More reads mapped to the C. clementina genome than the C. sinensis genome (80% and 56.7% respectively) or the de novo assembly (70%). Because the de novo assembly was also fragmented (N50 = 385 bp) and contained a low number of transcripts (12 902), the C. clementina genome was used as a reference for all further analyses. Cufflinks 2.2.128 was used to identify new transcripts not in the C. clementina v1.0 gene annotation file. These were validated and putative function was assigned using BLAST matches and InterproScan. Predicted protein sequences from the transcripts which did not map to C. clementina were annotated using protein descriptions from the Arabidopsis thaliana best blast hit. Predicted protein sequences were parsed to remove any sequences derived from contigs less than 200 nucleotides in length, and sequences containing an internal stop codon. This quality filtering resulted in a final set of 828 novel C. limon predicted protein sequences. All biosample and sequence data from the Cufflinks assembly were submitted to GenBank and can be accessed at Bioproject ID PRJNA348468. The Trinity assembly was submitted to Figshare (DOI: 10.6084/m9.figshare.5671585), along with 11 protein sequences predicted from the C. limon RNA-Seq cufflinks assembly which were used for proteomics data analysis but did not pass GenBank submission criteria.
RNA-Seq Differential Expression Analysis
A citrus cDNA transcriptome database was created by clustering transcripts from the C. clementina and C. sinensis genomes, C. limon GenBank ESTs, and the sequences from C. limon Trinity assembly at 100% identity, resulting in a nonredundant set of 90 656 transcripts after removing 3966 duplicates. CD-HIT29 was used for this step with the following parameters: -c 0.95 -M 10000 -T 40 -d 0. Trimmomatic-0.3630 was used for quality trimming RNA-Seq reads with the following parameters: LEADING:5 ILLUMINACLIP:Tru-Seq3-PE-2.fa:2:30:10 LEADING:5 TRAILING:5 SLIDING-WINDOW:4:10 MINLEN:60. Quality trimmed RNA-Seq reads were aligned to the transcriptome database using two different algorithms, Hisat231 and RSEM.32 RSEM was run on paired-end data with bowtie2 sensitive parameters: -forward-prob 1-bowtie2-sensitivity-level very_sensitive-bowtie2-mis-match-rate 0.5. Hisat2–2.0.4 was used with the following parameters: -mm-threads 30-time-fr-dta-no-spliced-alignment. The percentage of reads aligned using Hisat2 (77–89%) was much higher than using RSEM (24–55%), so the Hisat2 results were used for further analysis. Stringtie33 was applied (parameters: -p 40 -e -B -l MSTRG -p 40) to reconstruct the lemon transcriptome from the alignment of RNA-Seq reads to the citrus transcriptome database. Within stringtie, the prepDE.py script was used to generate the read counts for RNA-Seq reads that map to a particular transcript in the Lisbon lemon transcriptome. edgeR34 was used to identify the transcripts differentially expressed between CLas and control grafted plants at the four time points. All transcripts that had less than one count per million in fewer than three replicates were excluded from differential expression analysis. The exact test for differential expression for negative binomially distributed counts35 was implemented in edgeR with the FDR threshold of 0.05. Transcripts with a q-value < 0.05, and with a fold difference between treatment groups greater than two, were classified as differentially expressed. The GO enrichment was performed with the topGO package using the weight01 algorithm.36 1058 of the 1588 differentially expressed transcripts and 45 101 of the total 90 656 transcripts were assigned GO terms using Interproscan.
Citrus Protein Extraction and Peptide Mass Spectrometry Sample Preparation
Proteins were extracted from one leaf sample per plant for each time point. Leaf samples were flash frozen in liquid nitrogen when collected, and were ground in liquid nitrogen by mortar and pestle. Protein precipitation solvent (10% trichloroacetic acid in acetone with 2% beta-mercaptoethanol) was made fresh and kept on ice until use. Precipitation solvent (10 mL/mg wet leaf weight) was added to ground leaf samples in conical screw cap tubes. Samples were vortexed and stored overnight at −20 °C. Precipitated protein pellets were washed three times with 10 mL ice-cold acetone. During the third acetone resuspension, protein precipitate solution was transferred into 1.5 mL microcentrifuge tubes and centrifuged. After decanting the supernatant from the final wash, the pellet was dried to completion. Pellets were resuspended in 500 μL protein reconstitution solvent [8 M urea, 50 mM triethylammonium bicarbonate (TEAB) in water], and samples were incubated in an agitator (TOMY microtube mixer MT-360) with a small stir bar at room temperature overnight. Samples were centrifuged at full speed to pellet insoluble material, and the supernatant was collected for protein analysis.
Sample protein concentration was measured using the Quick Start Bradford Protein Assay (Bio-Rad). Gel electrophoresis was used as a quality control check to validate the Bradford results. Ten μg of protein sample was adjusted to a volume of 25 μL using phosphate buffered saline solution. Twenty-five microliters of 2× Laemmli Sample Buffer (Bio-Rad) containing 5% beta-mercaptoethanol was added to each protein sample. Samples were incubated at 70 °C for 10 min and run on a 10% Mini-PROTEAN TGX Precast gel (Bio-Rad) at 80 V for 2 h, using SDS-PAGE running buffer and a Precision Plus Protein Kaleidoscope standard (Bio-Rad). After running, the gels were removed from their casing and transferred to a plastic container to stain using Invitrogen NOVEX Colloidal Blue Staining Kit, following instructions for Tris-Glycine gels (Life Technologies). Gels were stained overnight on a rocker. To destain, the gels were rocked in Milli-Q water for a few hours, replacing the water as needed. Once destained, gels were scanned and samples were then compared. Protein samples were reduced with tris-carboxy-ethyl phosphine (TCEP): 5 μL of 200 mM TCEP was added to 40 μg of protein in 100 μL TEAB and incubated at 55 °C for 1 h. Samples were briefly centrifuged and were cooled to room temperature. Cysteine alkylation was performed by adding 5 μL of 375 mM iodoacetamide to each sample. Samples were vortexed, centrifuged, and incubated for 1 h at room temperature in the dark. If necessary, TEAB was added to samples prior to trypsin digestion to reduce urea concentration to <1 M. Sequencing grade, modified trypsin (Promega, Madison, WI) was added to each sample (trypsin:protein ratio 1:40, by weight), and samples were vortexed, centrifuged, and incubated overnight at 30 °C.
Trypsin-digested samples were dried and resuspended in 500ul 0.1% formic acid in water. Samples are acidified to a pH ≤ 3 by adding 5 μL full strength formic acid. Waters Sep-Pak C18 1 cm3 vacuum cartridges (cat#: WAT054955) are used with a Phenomenex vacuum manifold, and pressure is kept between 4 and 5 in Hg. Columns were conditioned with 3 mL 100% acetonitrile, followed by 3 mL 0.1% formic acid. Columns were briefly dried, then samples were added and run through. Columns were then washed with 3 mL 0.1% formic acid, and briefly dried. Samples were eluted off the column into a collection tube by adding 500 μL 80% acetonitrile, 20% (0.1% formic acid). Column was dried to ensure all eluent is collected. Cleaned samples were dried down before mass spec analysis.
Mass spectrometry data acquisition and analysis
Reversed phase chromatographic separation of peptide samples was performed using an EASY-nLC 1000 system (Thermo Scientific) equipped with a 3 cm × 100 μm trapping column and a 60 cm × 75 μm analytical column both packed with 5 μm Reprosil C8 particles with 120 Å pores (Dr. Maisch GmbH). Dried-down tryptic peptides were reconstituted in 60 μL of 0.1% formic acid in water, and for each injection 3 μL of the peptide samples were loaded onto the trapping column using 20 μL of solvent A (water containing 0.1% formic acid) at a flow rate of 2 μL/min. Reversed phase separation over the analytical column was performed by applying a linear gradient from 98% solvent A and 2% solvent B (acetonitrile containing 0.1% formic acid) to 60% solvent A and 40% solvent B over 120 min at a flow rate of 300 nL/min. Eluting peptides were ionized by electrospray ionization by applying a voltage of 2.2 kV to a laser pulled tip at the tip of the analytical column. Mass spectrometry was performed using Q-Exactive Plus mass spectrometer (Thermo Scientific) operated using a data dependent analysis method with the following settings: a high resolution MS1 scan from 400 to 2000 m/z at resolving power of 70 000 @ 200 m/z, automatic gain control (AGC) target of 106 ions, maximum ion time of 100 ms. Up to the 20 most abundant ions in the MS1 were selected for MS2 with an isolation window of 1.6 m/z and a normalized collision energy of 35. MS2 scans covered the range for 200–2000 m/z at a resolving power of 35 000 with an AGC target of 50 000 ions and a maximum ion time of 50 ms. Ions with one, six or greater, or undetermined charge states were excluded from MS2. Ions selected for MS2 were dynamically excluded for 30 s.
Thermo *.raw files were converted into Mascot Generic Format (.mgf) using MSConvert in Proteowizard Version 3.0.9393 (64 bit). A nonredundant protein database containing 166 858 proteins (including forward and reverse sequences) was created using predicted proteins from the sequenced genomes of C. clementina, C. sinensis, and CLas, proteins from other citrus species from GenBank, and predicted lemon proteins from the cufflinks assembly of the RNA-seq reads. Mascot Daemon 2.3.2 (Matrix Science, Boston, MA) was used to submit .mgf files for Mascot searching against the citrus protein database. The search database included a set of 112 common contaminant proteins from yeast, bacteria, humans, and other animals. The database included the reverse sequence of all proteins as a decoy database for approximation of the false discovery rate. MS/MS search parameters included parent ion mass tolerance: 25 PPM (monoisotopic); fragment ion mass tolerance: 0.80 Da (monoisotopic); fixed modifications (cysteine: carbamidomethyl), variable modifications (aspar-agine, glutamine: deamidated; methionine: oxidation), and maximum one missed cleavage. Files with the *.dat extension resulting from Mascot searching were loaded into Scaffold Q+ 4.6.1 (version 4.6.1, Proteome Software, Portland, OR) and used to calculate normalized spectral counts for each protein from each sample. Scaffold protein and peptide thresholds were set at 95%, with a minimum peptide number of two per protein—this resulted in a protein false discovery rate (FDR) of 0.3% and a peptide FDR of 0.0%. Cluster mode was used in Scaffold to group proteins with shared peptides and calculate weighted spectral counts. The mass spectrometry data have been deposited to the PRIDE archive37,38 via the PRIDE partner repository with the data set identifiers PXD005905, PXD006010, and PXD006011. During initial analysis of weighted spectral count data, proteins which had less than one spectral count in all biological samples were removed. Statistical analysis of weighted spectral count data for identification of proteins differentially abundant between sample categories was performed in Scaffold with the Fisher’s exact test (significance level p < 0.05 using the Benjamini-Hochberg multiple test correction).
1H NMR Sample Preparation
Several leaf disks approximately 6.35 mm (1/4 in.) surveying the entire leaf area (approximately 10–20 cm, depending on leaf size) were taken for every leaf collected for metabolomics and transferred to 2 mL tubes and lyophilized for 24 h (Labconco FreeZone Plus). The lyophilized leaf material was ground with one 3.5 mm glass bead using a Biospec Mini-Beadbeater for 2 min. Metabolites were extracted from the resulting ground material with 10 mM phosphate buffer heated to 90 °C in a 1:20 ratio based on the dry weight of the leaf sample used, and mixed for 15 min at 90 °C at 1000 rpm (Eppendorf ThermoMixer C). Samples were then centrifuged at 4 °C for 15 min at 14 krcf. 585 μL of the resulting supernatant was collected and 65 μL of internal standard containing 5 mM 3-(trimethylsilyl)-1-propanesulfonic acid-d6 (DSS-d6) was added. 600 μL of the mixture was added to 5 mm NMR tubes, and stored at 4 °C until NMR data acquisition (within 24 h of sample preparation).
1H NMR Data Acquisition and Analysis
1H NMR data was collected for all control (n = 11) and treatment (n = 11) samples at 0, 2, 8, 10, 12, 14, 16, 18, 20, 22, and 46 wpg, but data analysis was conducted on all control (n = 11) and all qPCR+ treatment plants (n = 6; plant IDs: 61, 39, 75, 78, 57, 64) at each time point, with the exception of 12 wpg (control n = 6, treatment n = 6) and 18 wpg (control n = 11, treatment n = 5).
NMR spectra were obtained as previously described8 with a Bruker Advance 600 MHz NMR spectrometer equipped with a SampleJet. Spectra were acquired using the Bruker “noe-sypr1d” experiment with the following acquisition parameters: 12 ppm sweep width, 2.5 s acquisition time with 2.5 s relaxation delay, and a 100 ms mixing time. Water saturation was applied during the relaxation delay and mixing time. Spectra were zero-filled with 128 000 data points and an exponential apodization function corresponding to a line broadening of 0.5 Hz was applied. Metabolites were identified and quantified using Chenomx NMR suite v7.6 and metabolite concentrations were corrected for dilution, and metabolite concentrations were normalized to the dry weight of the sample used.
The Euclidean distance matrix was calculated on log10 transformed metabolite data in R (v3.4.1) using the package vegan. To visualize differences between the infected and control samples, the distance matrix was projected using nonmetric multidimensional scaling (NMDS). To make sure that each group had the same variance, multivariate homogeneity of the control and healthy data was tested using the vegan function betadisper; no significant difference in the dispersion was observed between control and infected plants at any time point (p < 0.05). To test for similarities between the matrices of treatment and control groups, permutational MANOVA (PERMANOVA) was performed using the vegan function adonis2 with alpha = 0.05 and the number of permutations was 999, and treatment (CLas or control), time (wpg) and the interaction of treatment and time were included as explanatory variables, stratifying by the plant to control for repeated sampling. Because the interaction between time and treatment was significant (p < 0.05) in the initial model, separate models were run for each time point to assess whether the treatment effect was significant throughout the study. At time points when differences between the centroids of treatment and control groups significantly differed (via PERMANOVA, p < 0.05), Mann-Whitney U testing was conducted on the untransformed metabolite concentrations to determine which metabolites contributed to the differences using the wilcox_test function from the R package coin. P-values of metabolites were adjusted with Benjamini-Hochberg (q-value) to correct for multiple testing and control for Type I errors. Because Bonferroni procedures (such as Benjamini-Hochberg) assume independence and may increase the probability of making a Type II error, effect size (r) was also calculated ((r = Z/√N) where Z is the Z score from the wilcox_test output and N is the total sample size). The effect size is considered large if |r| ≥ 0.539–42).
Data Availability
The data sets supporting the results of this article are available in publicly accessible repositories. Transcriptomics: All biosample and sequence data from the Cufflinks assembly were submitted to GenBank and can be accessed at Bioproject ID PRJNA348468. The Trinity assembly, and a fasta file containing 11 protein sequences predicted from the C. limon RNA-Seq cufflinks assembly, which were used for proteomics data analysis but did not pass GenBank submission criteria, have been submitted to Figshare (DOI: 10.6084/m9.figshare.5671585). Proteomics: The mass spectrometry proteomics data have been deposited to the PRIDE Archive (http://www.ebi.ac.uk/pride/archive)37,38 via the PRIDE partner repository with the data set identifiers PXD005905, PXD006010, and PXD006011. Metabolomics: The 1H NMR raw spectra and metadata have been made publicly available at citrusgreening.org and can be accessed using this link: https://citrusgreening.org/metabolomics_host/index.
RESULTS AND DISCUSSION
CLas qPCR Analysis of Grafted Plants
Leaf samples were collected from the 22 grafted lemon plants (11 control graft, 11 HLB graft) at time points between 10 and 45 weeks post graft (wpg) to measure the titer of CLas in sampled tissues. For qPCR analysis, multiple leaf samples were collected from different regions of the plants and pooled. Out of the 11 plants receiving the HLB positive graft, CLas DNA was detected in six plants at multiple consecutive time points using qPCR (Figure 1)—these were used as the CLas treatment group in downstream analyses. A plant was considered “positive” for CLas when the Ct value <37. The remaining five plants receiving grafts originating from CLas-infected plants were eliminated from analysis: two plants tested qPCR negative at all time points, and three plants tested negative at all but one time point (a borderline positive Ct value was recorded). It was not possible to determine whether graft inoculation of CLas failed in these plants, or whether the inoculation was successful but the plants never became qPCR positive. While we focused our analysis on plants which ultimately tested positive for CLas by qPCR, plants which were grafted with CLas budwood but did not become qPCR positive may provide valuable insight into the plant response to infection.
Figure 1.
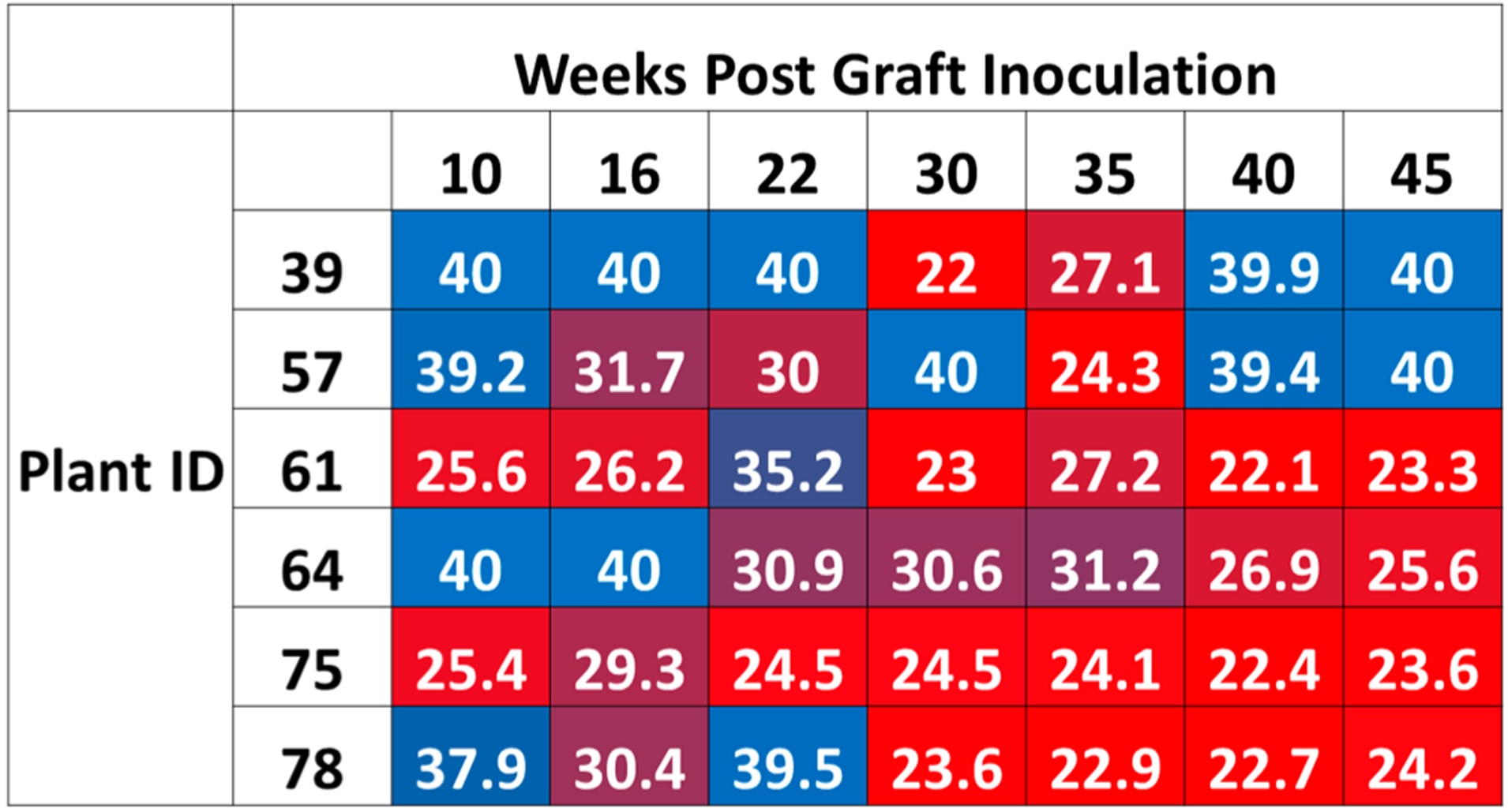
CLas qPCR data obtained from CLas grafted lemon trees. Cycle threshold (Ct) values are given at each time point for each of the five infected lemon trees analyzed in this study. In this heat map representation, red indicates high CLas titer, while blue indicates that the CLas titer is at or below the limit of detection.
RNA-Seq Differential Expression Analysis
A total of 24 RNA samples were extracted from leaf samples of CLas and control grafted plants (three plants for each treatment group) at four time points [0, 2, 10, and 14 wpg]. The UC Davis Contained Research Facility plant IDs of the infected plants used are 61, 75, and 78. The plant IDs of the healthy plants used are 38, 68, and 77. Following library construction and Illumina sequencing, RNA-Seq reads were mapped to a comprehensive database of citrus genes. Differentially expressed transcripts were identified between CLas-infected and healthy plants at each time point. The time point where the most differentially expressed transcripts between control and CLas grafted plants were found was 10 wpg (850; 464 CLas upregulated, 386 downregulated). At 14 wpg, 300 transcripts were identified as differentially expressed between control and CLas plants (215 CLas upregulated, 85 downregulated), and 313 differentially expressed transcripts were found at 2 wpg (135 CLas upregulated, 178 downregulated). Baseline (week 0) RNA samples were collected from lemon plants to identify any differences between the plants prior to CLas inoculation. While a relatively large number of transcripts were identified as different at 0 wpg (179; 136 CLas upregulated, 43 downregulated), nearly all of these transcripts represent sequences which are found at very low abundance in one class of plants and are undetected in the other, and they were not found to be differentially expressed at any other time points. Read count values for differentially expressed transcripts identified at all time points are given in Supporting Table S1. The read count values for all transcripts across all time points is available at Figshare (DOI: 10.6084/m9.figshare.5671585).
Gene Ontology (GO) enrichment analysis was performed on all transcripts identified as differentially expressed at all time points. The distribution of GO terms represented among the differentially expressed transcripts was compared to the distribution of GO terms predicted from all citrus sequences in the database used for RNA-Seq data analysis. The GO categories most enriched in the differentially expressed transcripts are shown in Table 1. The category of molecular and biological function most enriched in transcripts found to be differentially expressed between CLas and control grafted plants is S-adenosylmethionine (SAM) metabolism. SAM is a methyl group donor which is involved in a wide range of methyltransferase reactions acting on protein, nucleic acid, lipid, and secondary metabolite targets.43 In addition, SAM is the precursor for biosynthesis of the plant hormones ethylene and polyamines.
Table 1.
Gene Ontology (GO) Enrichment Analysis of Transcripts Differentially Expressed between CLas and Control Grafted Plantsa
| enrichment rank | biological function | molecular function | ||
|---|---|---|---|---|
| GO ID | term | GO ID | term | |
| 1 | GO:0006556 | S-adenosylmethionine biosynthesis | GO:0004478 | methionine adenosyltransferase activity |
| 2 | GO:0006879 | cellular iron ion homeostasis | GO:0008199 | ferric iron binding |
| 3 | GO:0006826 | iron ion transport | GO:0016161 | beta-amylase activity |
| 4 | GO:0006662 | glycerol ether metabolic process | GO:0047746 | chlorophyllase activity |
| 5 | GO:0000272 | polysaccharide catabolic process | GO:0016634 | oxidoreductase activity |
| 6 | GO:0015996 | chlorophyll catabolic process | GO:0015035 | protein disulfide oxidoreductase activity |
| 7 | GO:0010411 | xyloglucan metabolic process | GO:0004066 | asparagine synthase |
| 8 | GO:0016558 | protein import into peroxisome matrix | GO:1990837 | sequence-specific double-stranded DNA binding |
| 9 | GO:0009765 | photosynthesis, light harvesting | GO:0003849 | 3-deoxy-7-phosphoheptulonate synthase |
| 10 | GO:0006529 | asparagine biosynthetic process | GO:0005509 | calcium ion binding |
The 10 biological function and molecular function GO terms most enriched in differentially expressed transcripts, as compared to all citrus transcripts in the database.
Transcripts associated with a total of eight reactions connected to SAM metabolism were found to be differentially expressed between CLas and control grafted plants (Figure 2, Table 2). At 2 wpg, upregulation of methionine-gamma-lyase is predicted to direct methionine flux toward isoleucine biosynthesis, while at weeks 10 and 14 upregulation of S-adenosylmethionine synthetase redirects methionine toward SAM synthesis (Figure 2). At 10 and 14 wpg, transcripts encoding aminocylopropane-1-carboxylate (ACC) oxidase, a key enzyme in the production of ethylene from SAM, were upregulated in CLas grafted plants. Upregulation of SAM decarboxylase, which converts SAM into the precursor of polyamine metabolism, was observed in CLas grafted plants 14 wpg. The only SAM-related transcripts downregulated in CLas compared to control plants are two SAM dependent methyltransferases, one downregulated at 2 wpg and the other at 10 wpg. The methyltransferase downregulated at 10 wpg is the citrus ortholog of the transcript encoding the lignin biosynthetic enzyme caffeoyl-CoA-methyltransferase. The downregulation of this methyltransferase suggests that the function of SAM as a one-carbon donor may be less pertinent in the response of citrus to CLas infection than its function as a hormone precursor.
Figure 2.
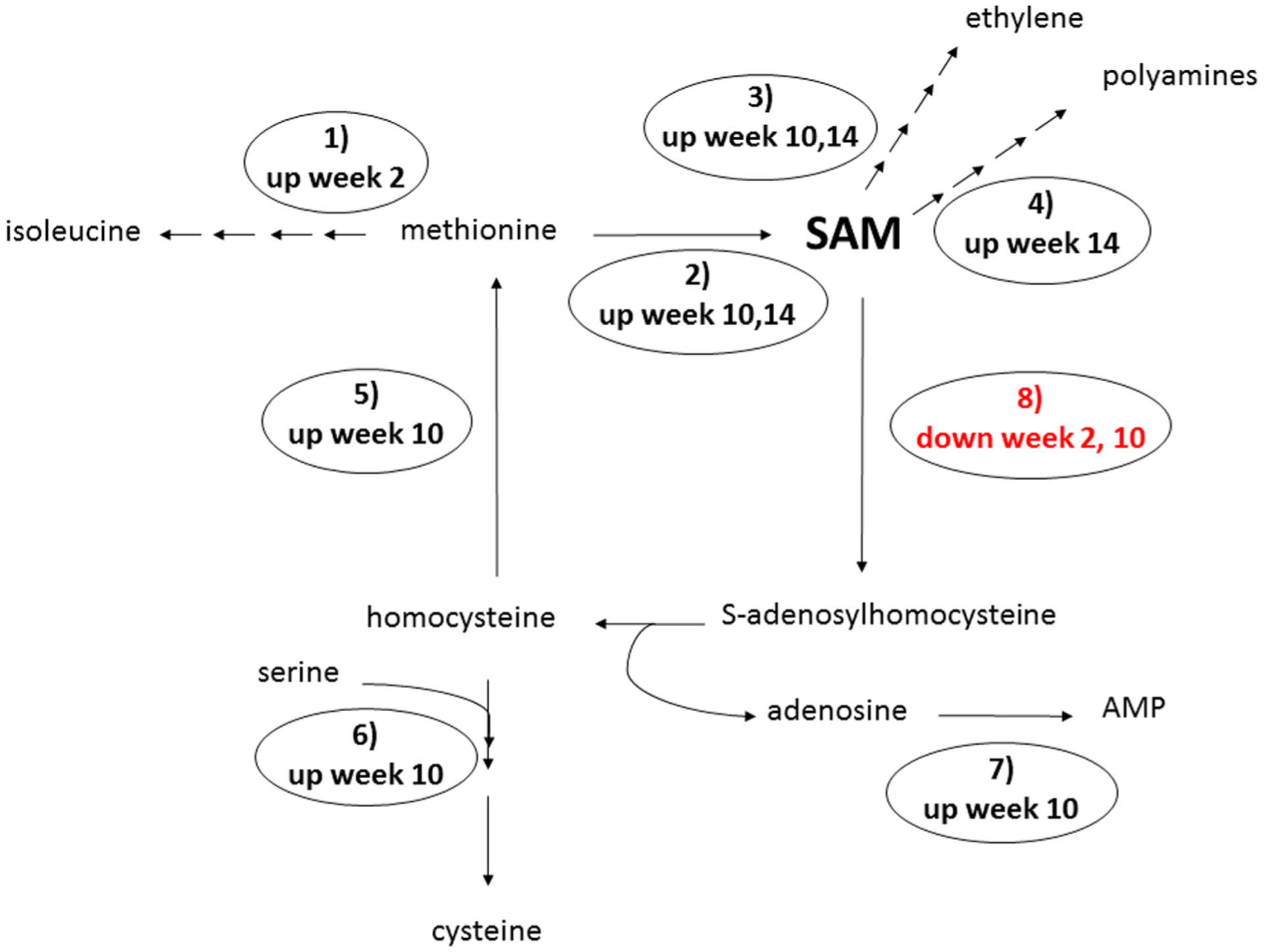
Differentially expressed lemon leaf transcripts involved in S-adenosylmethionine (SAM) metabolism identified between CLas and control grafted plants. At 2 weeks post graft (wpg), methionine-γ-lyase (1) transcripts are upregulated in CLas plants, directing flux of methionine toward isoleucine biosynthesis. At 10 wpg, three SAM synthetase (2) transcripts are upregulated in CLas plants, redirecting methionine flux toward production of SAM. A fourth SAM synthetase is upregulated in CLas plants at 14 wpg. ACC oxidase (3) catalyzes ethylene production from SAM, and is upregulated in CLas plants at 10 and 14 wpg. SAM decarboxylase (4), which regulates polyamine biosynthesis from SAM, is upregulated in CLas plants at 14 wpg. Homocysteine methyltransferase (5), cystathionine-β-synthase (6), and adenosine kinase (7) are involved in metabolism of products of the SAM cycle, and are all upregulated in CLas grafted plants 10 wpg. Two SAM-dependent methyltransferases (8) are downregulated in response to CLas, one at 2 wpg and the other at 10 wpg.
Table 2.
Citrus Transcripts Related to S-Adenosylmethionine Metabolism Differentially Expressed between CLas and Control Grafted Lemon Plantsa
| gene ID | description | week post graft CLas response | CLas | control |
|---|---|---|---|---|
| XM_006476228.2 | methionine gamma-lyase | 2 CLas up | 44.25 | 14.2 |
| XM_006476228.2 | methionine gamma-lyase | 2 CLas up | 26.53 | 5.33 |
| XM_006419196.1 | S-adenosylmethionine synthetase | 10 CLas up | 24.17 | 11.03 |
| XM_006428205.1 | S-adenosylmethionine synthetase | 10 CLas up | 126.2 | 55.32 |
| XM_006488687.2 | S-adenosylmethionine synthetase | 10 CLas up | 5.86 | 2.11 |
| XM_006492333.2 | S-adenosylmethionine synthetase 2 | 14 CLas up | 65.53 | 30.37 |
| XM_006419937.1 | homocysteine S-methyltransferase 3 | 10 CLas up | 22.96 | 8.22 |
| XM_006422911.1 | cystathionine-beta-synthase | 10 CLas up | 17.98 | 5.34 |
| XM_006486971.2 | cystathionine-beta-synthase | 10 CLas up | 2.25 | 0 |
| XM_006436119.1 | adenosine kinase | 10 CLas up | 15.54 | 4.0 |
| XM_006447590.1 | aminocylopropane-1-carboxylate (ACC) oxidase | 10 CLas up | 147.2 | 40.1 |
| XM_006447589.1 | aminocylopropane-1-carboxylate (ACC) oxidase | 14 CLas up | 30.05 | 0 |
| XM_006453476.1 | S-adenosylmethionine decarboxylase family protein | 14 CLas up | 5.25 | 1.07 |
| XM_006490953.2 | S-adenosyl-l-methionine-dependent methyltransferase | 2 CLas down | 0 | 2.88 |
| XM_006427197.1 | S-adenosyl-l-methionine-dependent methyltransferase | 10 CLas down | 57.02 | 120.2 |
Average RNA-Seq read counts from samples from three biological replicate plants per treatment per time point.
The upregulation of transcripts involved in ethylene and polyamine biosynthesis following graft inoculation is consistent with published reports of altered hormone metabolism in CLas-infected plants.44 In addition, transcripts involved in ethylene perception are upregulated in CLas grafted plants, along with several other transcripts encoding enzymes involved in hormone biosynthesis and perception (Table 3). Jasmonic acid associated transcripts upregulated in CLas grafted plants at 10 wpg include those encoding allene oxide synthase, allene oxide cyclase, and lipoxygenase 3 (jasmonic acid biosynthesis), and several jasmonate-ZIM domain (JAZ) proteins. JAZ proteins have been shown in maize and Arabidopsis to play a critical role in jasmonic acid signaling, and in regulating crosstalk between plant hormones during the response to pests and pathogens.45 At 10 wpg, two transcripts associated with hormone catabolism show opposite patterns of expression: cytokinin oxidase is downregulated, while gibberelin 2-oxidase (GA2ox) is upregulated. GA2ox plays a critical regulatory role in plant development by inactivating the plant hormone gibberellin, and at the same time point when GA2ox transcripts are upregulated, transcripts encoding two gibberellin regulated proteins are downregulated. A transcript encoding an auxin responsive protein is also upregulated in CLas-infected plants at 10 wpg (Table 3).20 Changes in plant hormone levels during the response to CLas infection may promote accelerated abscission of unripe fruit.
Table 3.
Citrus Hormone Metabolism and Signaling Transcripts Differentially Expressed between CLas and Control Grafted Lemon Plantsa
| gene ID | description | weeks post graft CLas response | CLas | control |
|---|---|---|---|---|
| XM_006449381.1 | ethylene response factor | 10 up | 20.4 | 5.21 |
| XM_006467633.2 | ethylene response factor | 10 up | 12.2 | 3.06 |
| XM_006477062.2 | ethylene responsive element binding factor | 10 up | 34.5 | 4.85 |
| XM_006432477.1 | ethylene-insensitive 3 (EIN3) | 10 up | 2.25 | 0 |
| XM_006482231.2 | allene oxide cyclase 4 | 2 down | 0 | 4.38 |
| XM_006437669.1 | allene oxide cyclase 3 | 10 up | 13.1 | 2.4 |
| XM_006434833.1 | allene oxide synthase | 10 up | 1.65 | 0.2 |
| XM_006426618.1 | lipoxygenase 3 | 10 up | 123.4 | 42.1 |
| XM_006465842.2 | lipoxygenase 3 | 10 up | 10.3 | 2.5 |
| XM_006438782.1 | jasmonate-zim-domain protein | 10 up | 13.5 | 2.09 |
| orange1.1 g028898m | jasmonate-zim-domain protein | 10 up | 50.7 | 7.67 |
| XM_006452782.1 | jasmonate-zim-domain protein | 10 up | 7.49 | 0.46 |
| XM_006449649.1 | jasmonate-zim-domain protein | 10 up | 5.39 | 0.3 |
| XM_006474637.2 | jasmonate-zim domain protein | 10 up | 2.61 | 0.09 |
| XM_006426643.1 | jasmonate-zim-domain protein | 10 up | 19.5 | 0.41 |
| XM_006474637.2 | jasmonate-zim-domain protein | 10 up | 9.29 | 0.77 |
| XM_006422814.1 | Auxin-responsive GH3 family protein | 10 up | 5.58 | 0.35 |
| XM_006449629.1 | gibberellin 2-oxidase | 10 up | 2.13 | 0.47 |
| XM_006430958.1 | Gibberellin-regulated family protein | 10 down | 1.59 | 6.07 |
| XM_006422805.1 | Gibberellin-regulated family protein | 10 down | 0 | 2.52 |
| XM_006435008.1 | cytokinin oxidase 5 | 10 down | 7.8 | 33.8 |
Average read counts from RNaseq analysis of three biological replicate plants per treatment per time point. See Table 2 for read counts for ACC oxidase and SAM dehydrogenase.
Other functional categories enriched for transcripts differentially expressed between CLas and control grafted lemon plants are involved in metabolism of carbohydrates and amino acids, photosynthesis and chlorophyll catabolism, iron binding, and the response to oxidative stress. Polysaccharide catabolism and xyloglucan metabolism are two specific biological function categories enriched among the differentially expressed transcripts (Table 1). Xyloglucan is a major cell wall hemicellulose composed of glucose, xylose, galactose, and fucose monomers, and changes in xyloglucan metabolism are expected to have a significant impact on the structure and function of the plant cell wall.
Mass Spectrometry-Based Quantitative Proteomics
Five plants in which CLas was reliably detected by qPCR were selected for whole proteome analysis, along with five control grafted plants which tested negative for CLas over the length of the experiment. The UC Davis Contained Research Facility plant IDs of the infected plants are 39, 61, 64, 75, and 78. The plant IDs of the healthy plants are 38, 44, 45, 51, 56, and 68 (plant 44 was analyzed in place of plant 56 at one of the three time points). Peptides prepared from leaf samples collected from ten plants at three time points post graft (2, 10, 14 wpg) were analyzed by high resolution mass spectrometry, and peptides derived from 3108 citrus proteins were identified using Mascot46 and Scaffold47 (Supporting Table S2). A combined total of 122 proteins were found to be differentially abundant at the three time points analyzed, with the majority (85 proteins) identified at 10 wpg (Supporting Table S3).
In leaf samples collected at 2 wpg, six proteins were identified as differentially abundant (three at higher levels, three at lower levels) in CLas grafted plants compared to control grafted plants (Table 4).
Table 4.
Lemon Proteins Differentially Abundant between Control and CLas Infected Plants 2 Weeks after Inoculationa
| protein ID | protein description | CLas response | CLas | control |
|---|---|---|---|---|
| Q09MH0.1 | Ribulose bisphosphate carboxylase large chain Short RuBisCO large subunit | up | 581.2 | 442.4 |
| Q09MF1.1 | Photosystem II CP47 chlorophyll apoprotein | up | 57.4 | 35.2 |
| KDO56915.1 | Photosystem I subunit H2 | up | 26.4 | 12.6 |
| XP_006481103.1 | Ferritin-3, chloroplastic-like | down | 14.9 | 25.5 |
| XP_006441440.1 | Kunitz family trypsin and protease inhibitor | down | 1.8 | 7.2 |
| KDO37535.1 | heat shock protein 21 | down | 7.3 | 18.2 |
Average spectral count of lemon proteins found to be differentially abundant between control and CLas infected plants at 2 weeks post inoculation.
The three proteins found at higher levels in CLas-infected plants are involved in photosynthesis. The three proteins found at lower levels in CLas-infected plants at 2 wpg have documented roles in pathogen defense—an iron binding ferritin, a Kunitz trypsin protease inhibitor, and the citrus ortholog of Arabidopsis hsp21, a plastid-encoded heat shock protein which has been shown to protect chloroplasts from oxidative stress.48 This downregulation of defense-associated proteins suggests that at this early stage of infection the bacterium may be suppressing plant defenses.
At 10 wpg, 85 proteins were identified as differentially abundant between CLas and control grafted plants, and at 14 wpg, 29 differentially abundant proteins were identified (Supporting Table S3). For all differentially expressed proteins, an associated gene ontology term best capturing its biological function was selected using Uniprot,49 and the distribution of protein functional classes was charted for four groups of proteins—upregulated in CLas plants at 10 and 14 weeks, and downregulated in CLas plants at 10 and 14 weeks (Figure 3). At weeks 10 and 14, in contrast to week 2 (Table 4), a ferritin protein was found to be significantly more abundant in CLas plants compared to control grafted plants (Supporting Table S3). At 10 wpg, a total of 12 protease (endopeptidase) inhibitors were found to be more abundant in CLas grafted plants than controls (Figure 3). Photosystem components involved in the light dependent reactions of photosynthesis were the next most abundant class of proteins observed to be upregulated in CLas grafted plants at the 10 week time point (Figure 3). In contrast, at this same time point, proteins involved in the reductive pentose phosphate cycle (Calvin cycle) and photorespiration comprise the largest classes of proteins downregulated in CLas grafted plants, suggesting that the light dependent (photosystem) reactions of photosynthesis are decoupled from the light independent (Calvin cycle) reactions. Chloroplast light-dependent signaling has recently received attention for its role in the response to pathogen attack.50 Photorespiration is believed to protect against oxidative stress through dissipation of excessive reduction potential generated during the light-dependent photosynthetic reactions, and photorespiratory mutants have been found to experience elevated oxidative stress.51,52 Multiple citrus proteins with predicted function in response to oxidative stress are upregulated in CLas grafted plants at 10 and 14 wpg (Figure 3). Photorespiration exerts a major impact on plant nitrogen metabolism through production of ammonia, which is reassimilated by glutamine synthetase.53
Figure 3.
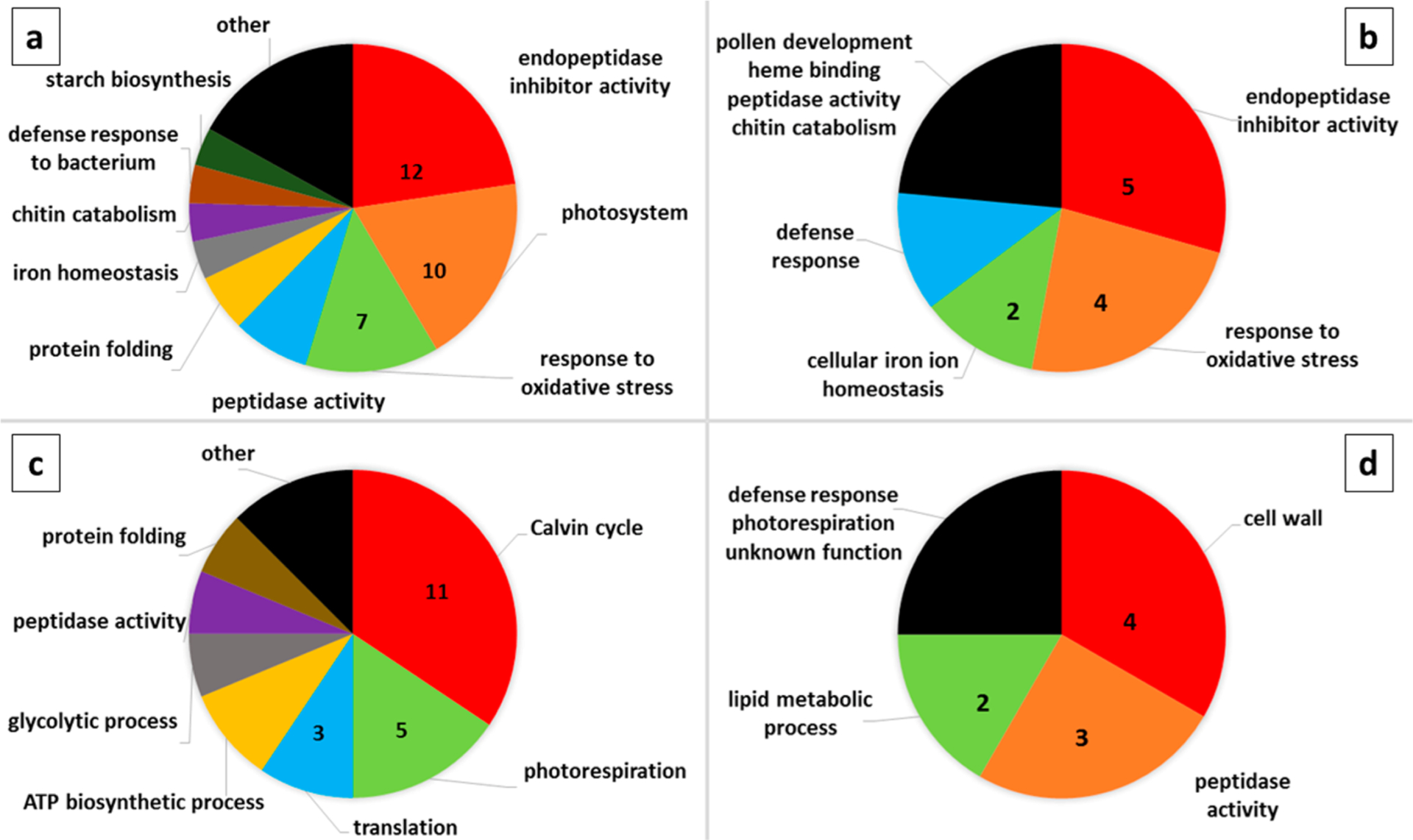
Pie chart representation of the proportion of lemon proteins with shared predicted biological function, out of all proteins found to be differentially abundant between healthy and CLas infected plants at that time point. The number of differentially abundant proteins belonging to the three most common functional classes are indicated for each of the four groups: more abundant in CLas infected plants at (a) 10 weeks post graft, and (b) 14 weeks post graft; and less abundant in CLas infected plants at (c) 10 weeks post graft, and (d) 14 weeks post graft. The black segment of each pie chart includes the functional classes represented by one protein.
At 14 wpg, two serine peptidases and one aspartic peptidase were downregulated in CLas grafted plants, as were lipid metabolism and cell wall proteins (Figure 3). Chitinase (XP_006442848.1) and β−1,3-glucanase (XP_006421176.1), plant hydrolases which have been characterized as a component of the plant response to pathogens and to ethylene signaling in multiple pathosystems,54,55 were both detected at significantly higher levels in CLas-infected plants 14 wpg (Figure 3). β−1,3-Glucanase functions in callose degradation, and has been shown to play a critical role in the maintenance of plasmodesmata, where the synthesis and degradation of callose are balanced to control cell-to-cell transport.56 In CLas-infected citrus, excessive callose deposition blocking plasmadesmata has been associated with the development of HLB symptoms.57 O-Glycosyl hydrolase 17 has been identified as a plasmodesmata localized protein with callose degrading activity in Arabidopsis,58 and the citrus ortholog of this protein (XP_006424832.1) was found to be significantly upregulated in CLas-infected plants at 10 wpg (Supporting Table S3). The protein differences detected between healthy and CLas-infected plants at 10 and 14 weeks provide insight into elements of the plant response to infection, including the activation of defense responses and alterations in metabolism and physiology resulting from disruption of the plant vascular system by this phloem-localized bacterium.
1H NMR-Based Metabolomics
Metabolite extracts were prepared from leaf samples collected from CLas and control grafted lemon plants throughout the duration of the longitudinal study. Samples from all six of the CLas grafted plants which were consistently qPCR positive (Figure 1) were analyzed alongside samples from all 11 control plants. Baseline samples (pregrafting) and samples from 2, 8, 10, 12, 14, 16, 18, 20, 22, and 46 wpg were analyzed. Targeted profiling of 25 metabolites was performed using 1H NMR spectroscopy. Analyzed metabolites included sugars, amino acids, and other primary and secondary metabolites. Metabolite concentration was normalized according to an internal standard added during sample preparation and corrected for dilution. The median concentration for each metabolite at each time point in control and CLas-infected plants is given in Supporting Table S4.
The differences in the metabolome between infected and healthy control samples were assessed using nonmetric multidimensional scaling (NMDS) and PERMANOVA. NMDS59 is a rank order-based method that can be used to visualize multidimensional data sets onto a predefined number of dimensions (two dimensions were used in this analysis). PERMANOVA was performed on all the metabolomics data postgrafting (i.e., excluding baseline), and indicated a significant difference in metabolite composition due to the interaction between treatment and time (pseudo-F = 1.44 and p < 0.05). PERMANOVA was conducted at each time point and revealed significant differences in the metabolome between control and infected plants at 2 and 22 wpg (pseudo-F values = 2.77 and 2.34, respectively; p < 0.05). Differences in the metabolome between control and CLas-infected leaves approached significance at 46 wpg (p = 0.052, pseudo F = 2.06). Indeed, these differences could be visualized in the NMDS ordination plots at 2, 10, 12, 22, and 46 wpg (although the clustering at 10 and 12 wpg did not reach statistical significance) (Figure 4).
Figure 4.
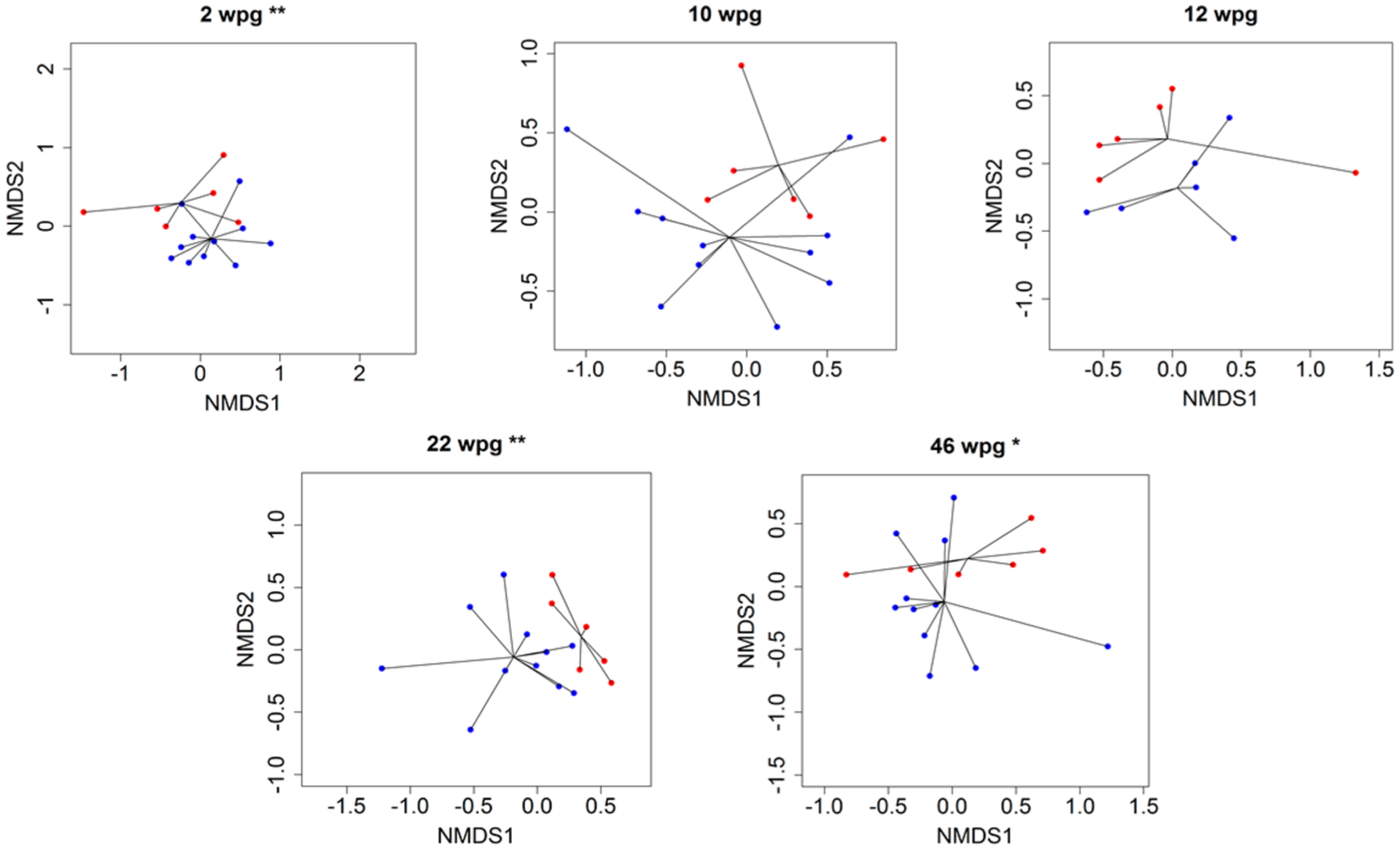
Nonmetric multidimensional scaling (NMDS) ordination plots based on Euclidean distance matrices of metabolomic samples at 2, 10, 12, 22, and 46 weeks postgrafting for control (blue) and red (CLas+) samples. Lines are extending from the centroid of each group. Asterisks indicate significant differences between the centroids of the two clusters **p ≤ 0.05; *p ≤ 0.1.
At time points where PERMANOVA indicated significant differences between control and CLas-infected plants, univariate testing was conducted to understand specific metabolites that differed between the groups. Because of the uneven and small sample sizes (control n = 11, CLas n = 6), Mann-Whitney U, a nonparametric test, was used.60,61 At 2 wpg, concentrations of alanine, sucrose, GABA, galactose, histidine, isoleucine, phenylalanine, uridine, proline betaine, lysine, and valine were significantly lower in infected plants compared to control (q < 0.1, |r| ≥ 0.5, Table 5). At 2 wpg the median concentration for most metabolites was lower in infected plants compared to controls (Figure 5). Correspondingly, protein changes between CLas and control plants were also identified at 2 wpg, where three proteins associated with plant defense were found at lower levels in CLas grafted plants (Table 4).
Table 5.
Metabolites Significantly Differing between Healthy and CLas Infected Plants at Time Points Selected Using PERMANOVAa
| weeks post graft | metabolite | p value | FDR (q value) | |r| | CLas response |
|---|---|---|---|---|---|
| 2 | Alanine | 0.007 | 0.061 | 0.634 | down |
| 2 | Sucrose | 0.007 | 0.061 | 0.634 | down |
| 2 | 4-Aminobutyrate (GABA) | 0.010 | 0.061 | 0.609 | down |
| 2 | Galactose | 0.015 | 0.061 | 0.585 | down |
| 2 | Histidine | 0.015 | 0.061 | 0.585 | down |
| 2 | Isoleucine | 0.015 | 0.061 | 0.585 | down |
| 2 | Phenylalanine | 0.027 | 0.076 | 0.536 | down |
| 2 | Uridine | 0.027 | 0.076 | 0.536 | down |
| 2 | Proline Betaine | 0.027 | 0.076 | 0.536 | down |
| 2 | Lysine | 0.037 | 0.083 | 0.512 | down |
| 2 | Valine | 0.037 | 0.083 | 0.512 | down |
| 22 | Asparagine | 0.002 | 0.048 | 0.707 | down |
| 22 | Alanine | 0.005 | 0.061 | 0.658 | down |
| 22 | Unknown-1 | 0.015 | 0.121 | 0.585 | down |
| 22 | Uridine | 0.027 | 0.171 | 0.536 | down |
| 46 | Glucose | 0.003 | 0.077 | 0.683 | up |
| 46 | 4-Aminobutyrate (GABA) | 0.037 | 0.304 | 0.512 | down |
| 46 | Myoinositol | 0.037 | 0.304 | 0.512 | up |
Metabolites significantly differing between healthy and CLas infected plants (Mann-Whitney U test; p ≤ 0.1) at time points selected based on PERMANOVA results. The q value is the FDR-corrected p value. |r| is the effect size.
Figure 5.
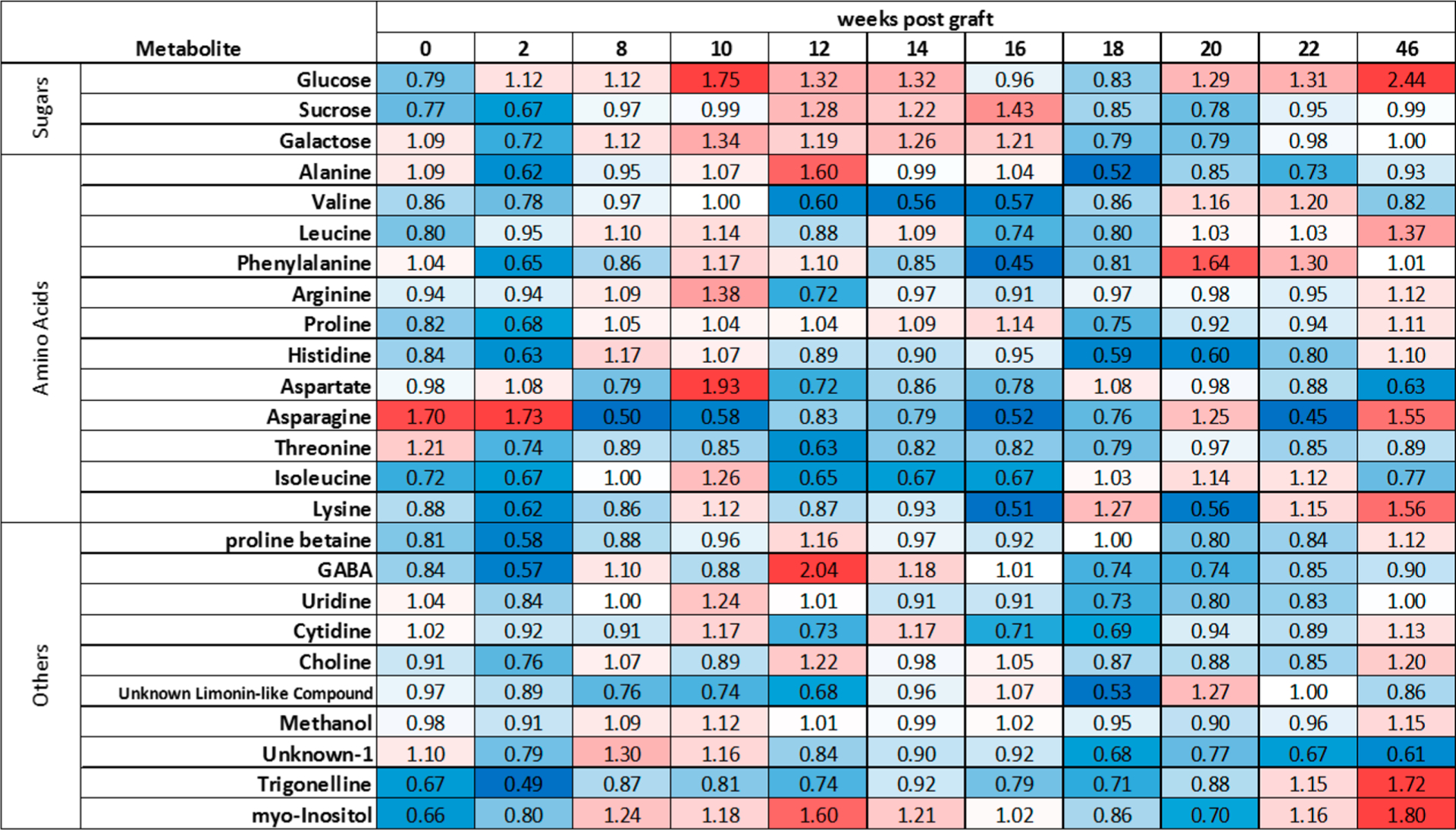
Visual depiction of metabolite data. Median fold change of the average metabolite concentrations (infected relative to control) at each time point weeks post graft. Red indicates a higher median concentration in the infected plants, blue indicates a higher concentration in the control plants, white indicates equal concentration between conditions. More intense colors indicate a larger magnitude of difference.
At 22 wpg, asparagine and alanine had lower median concentrations in infected plants compared to controls (q < 0.1, |r| ≥ 0.5), and uridine and unknown-1 were significantly different prior to multiple test correction (p ≤ 0.5) and had large effect sizes (|r| ≥ 0.5, Table 5). At 46 wpg, the median glucose concentration was significantly higher in infected plants compared to controls ((q < 0.1, |r| ≥ 0.5). GABA and myoinositol had lower and higher median concentrations, respectively, in infected plants relative to control (significant prior to multiple test correction (p < 0.05)) and had large effect sizes (|r| ≥ 0.5) at 46 wpg.
The median concentration of glucose and galactose (10, 12, 14 wpg) and sucrose (12, 14 wpg) were higher in CLas-infected leaves compared to controls at consecutive time points (Figure 5). Asparagine and aspartate were overall lower in infected plants throughout the experiment with the notable exception of week 10, where aspartate levels were 2-fold higher in infected plants. Although the median concentration of many metabolites were lower in treatment plants compared to controls at baseline (Figure 5), clustering between control and infected plants was not observed in NMDS, and PERMANOVA did not indicate significant differences between the two groups at baseline.
Integration of Transcriptomic, Proteomic, and Metabolomics Data Sets
Comparative analysis of differentially expressed transcripts, proteins, and metabolites identified between CLas and control grafted lemon plants provides the opportunity for integrated analysis of citrus response to infection over time. The 1588 differentially expressed transcripts were blasted (blastx e-value cutoff 1 × 10−10) against a database containing all nonredundant protein sequences in NCBI GenBank. The blast report containing the top 10 protein hits for each differentially expressed transcript is found in Supporting Table S5. We found that for 14 of the 103 differentially expressed proteins, the corresponding transcripts were also found to be differentially expressed (Table 6). Nine of these proteins are protease inhibitors, which were upregulated at 10 and 14 wpg—the corresponding transcripts were all upregulated in CLas grafted plants at 14 wpg. Among these is a miraculin-type protease inhibitor, which has been found to be both upregulated and downregulated in CLas-infected plants in previous transcriptomic and proteomic studies.13,15,16 In a parallel longitudinal study of graft-inoculated navel oranges, miraculin-like protease inhibitor gene and protein expression was found to be significantly affected by CLas infection.62
Table 6.
List of Proteins Found by Mass Spectrometry to Be Differentially Abundant between CLas and Control Plants, for Which the Corresponding Transcripts Were Found by RNAseq to Be Differentially Expressed
| protein description | GenBank ID | CLas protein response | CLas Transcript response |
|---|---|---|---|
| Serine protease inhibitor, potato inhibitor I-type | XP_006421576.1 | up week 10 | up week 14 |
| Serine protease inhibitor, potato inhibitor I-type | XP_006421578.1 | up week 10 | up week 14 |
| basic Chitinase | XP_006422475.1 | up week 10 | up week 14 |
| O-glycosyl hydrolases family 17 protein | XP_006424832.1 | up week 10 | up week 10 |
| Sec14p-like phosphatidylinositol transfer family | XP_006434672.1 | up week 10 | down week 14 |
| Kunitz family trypsin and protease inhibitor | XP_006441461.1 | up week 10 | up week 14 |
| Kunitz family trypsin and protease inhibitor | XP_006441462.1 | up week 10 | up week 14 |
| Kunitz family trypsin and protease inhibitor | XP_006442410.1 | up weeks 10, 14 | up week 14 |
| Kunitz family trypsin and protease inhibitor | XP_006477857.1 | up weeks 10, 14 | up week 14 |
| Kunitz family trypsin and protease inhibitor | XP_006478132.1 | up week 10 | up week 14 |
| miraculin-like | XP_006478135.1 | up week 10 | up week 14 |
| ferritin-3, chloroplastic-like | XP_006481103.1 | up weeks 10, 14; down week 2 | down weeks10, 14 |
| ferritin-3, chloroplastic-like | XP_006486262.1 | up weeks 10, 14 | up week 14 |
| inhibitor of trypsin and hageman factor-like | XP_006495465.1 | up weeks 10, 14 | up week 14 |
Another citrus sequence for which the transcript and protein were found to be upregulated by CLas is the callose degrading protein O-glycosyl hydrolase 17 (Table 6). Callose deposition is a common component of the plant defense response to pests and pathogens,63,64 and the observed protein and transcript upregulation of O-glycosyl hydrolase 17 in CLas-infected plants may serve to regulate callose levels. Transcripts and proteins from two separate ferritin-3 proteins were both found to be responsive to CLas infection, highlighting the function of plant regulation of iron availability as a component of the citrus response to bacterial infection. While ferritin-3 (XP_006486262.1) transcript and protein levels were both found to be upregulated by CLas, ferritin-3 (XP_006481103.1) exhibits a more complicated CLas response. Ferritin-3 (XP_006481103.1) protein was less abundant in CLas grafted plants at 2 wpg, but was found at greater abundance in CLas grafted compared to control plants at 10 and 14 wpg, while the corresponding transcript was downregulated in CLas plants at 10 and 14 wpg. On the basis of these results, ferritin-3 (XP_006481103.1) appears to be subject to dynamic regulation in citrus in response to CLas, with the bacteria potentially influencing the release of iron in the plant for their use. Iron is a critical enzymatic cofactor which bacteria have developed sophisticated mechanisms to scavenge from their hosts in plant and animal pathosystems.65–67 Phosphatidylinositol transfer proteins function in signaling and membrane trafficking,68 and a protein in this family was found to be more abundant at 10 wpg in infected compared to healthy plants, while at the same time point the corresponding transcript was found to be downregulated in infected compared to healthy plants. This pattern of expression suggests that these proteins are subject to tight regulation and possibly feedback inhibition, with increased protein levels leading to downregulation of transcription (Table 6).
At 10 wpg, all three analyses revealed major differences between infected and control plants; the greatest number of differentially expressed/abundant transcripts and proteins was observed at 10 wpg. Proteomics data revealed coordinated downregulation of photorespiration and Calvin cycle proteins at this time point, while several proteins involved in light-dependent photosynthetic reactions were concurrently upregulated (Figure 3). Photorespiration serves as an important pathway for the mitigation of photooxidative damage,69 and the downregulation of this pathway during the response to CLas may result in the accumulation of reactive oxygen species.52 Accordingly, at this time point several transcripts and proteins involved in response to oxidative stress are induced in the plant. Ammonia produced during photorespiration represents a crucial component of plant nitrogen metabolism, accounting for ten times more nitrogen flux in plants than nitrogen uptake from soil.70 Serine hydroxymethyltransferase and glycine decarboxylase act together during photorespiration to convert serine into glycine, and both of these proteins are found at greatly reduced levels in CLas compared to control leaf protein samples (Table 7, Figure 6). The primary system for reassimilation of photorespiratory ammonia is through glutamine synthetase/ferredoxin-dependent glutamate synthase (GS/GOGAT),70 and glutamate synthase proteins were found to be downregulated at week at 10 and 14 wpg. Glutamate is found in high levels in plant phloem sap, and CLas may disrupt glutamate transport in infected plants. Glutamate decarboxylase transcripts were found by RNA-Seq to be upregulated at 10 wpg—this leads to the production of gamma-aminobutyric acid (GABA), which has been shown to accumulate in plants in response to pathogen attack71 (Table 7, Figure 6). From our metabolomics analysis, the median GABA concentration was found to be slightly lower in CLas-infected compared to control plants at 10 wpg, but by 12 wpg, twice as much GABA was found in infected compared to control plants (Figure 5).
Table 7.
Average Spectral/Read Count Values for All CLas regulated Proteins/Transcripts Discussed in Figure 6
| analyte | ID | description | weeks post graft CLas response | CLas | control |
|---|---|---|---|---|---|
| protein | XP_006494086.1 | ferredoxin-dependent glutamate synthase 1 | 10 down | 4.191 | 25.36 |
| protein | XP_006421109.1 | glutamate synthase 1 | 10 down | 0.856 | 5.67 |
| protein | XP_006494086.1 | ferredoxin-dependent glutamate synthase 1 | 14 down | 37.22 | 55.02 |
| protein | KDO78681.1 | serine transhydroxymethyltransferase | 10 down | 12.91 | 20.75 |
| protein | XP_006426225.1 | serine transhydroxymethyltransferase 1 | 10 down | 27.36 | 40.47 |
| protein | XP_006450834.1 | glycine decarboxylase P-protein 1 | 10 down | 19.08 | 35.11 |
| transcript | XM_006478363.2 | aspartate kinase-homoserine dehydrogenase | 10 down | 0.865 | 3.645 |
| transcript | XM_006488487.2 | glutamine-dependent asparagine synthase 1 | 14 down | 11.63 | 71.9 |
| transcript | XM_006425030.1 | glutamine-dependent asparagine synthase 1 | 14 down | 10.64 | 54.6 |
| transcript | XM_006447986.1 | glutamate decarboxylase 4 | 10 up | 4.759 | 0 |
| transcript | XP_006449748.1 | APS reductase 3 | 10 up | 74.75 | 16.77 |
| transcript | XM_006449686.1 | APS reductase 1 | 10 up | 2.583 | 0 |
| transcript | orange1.1 g005543m | sulfite reductase | 10 down | 0 | 39.58 |
Figure 6.
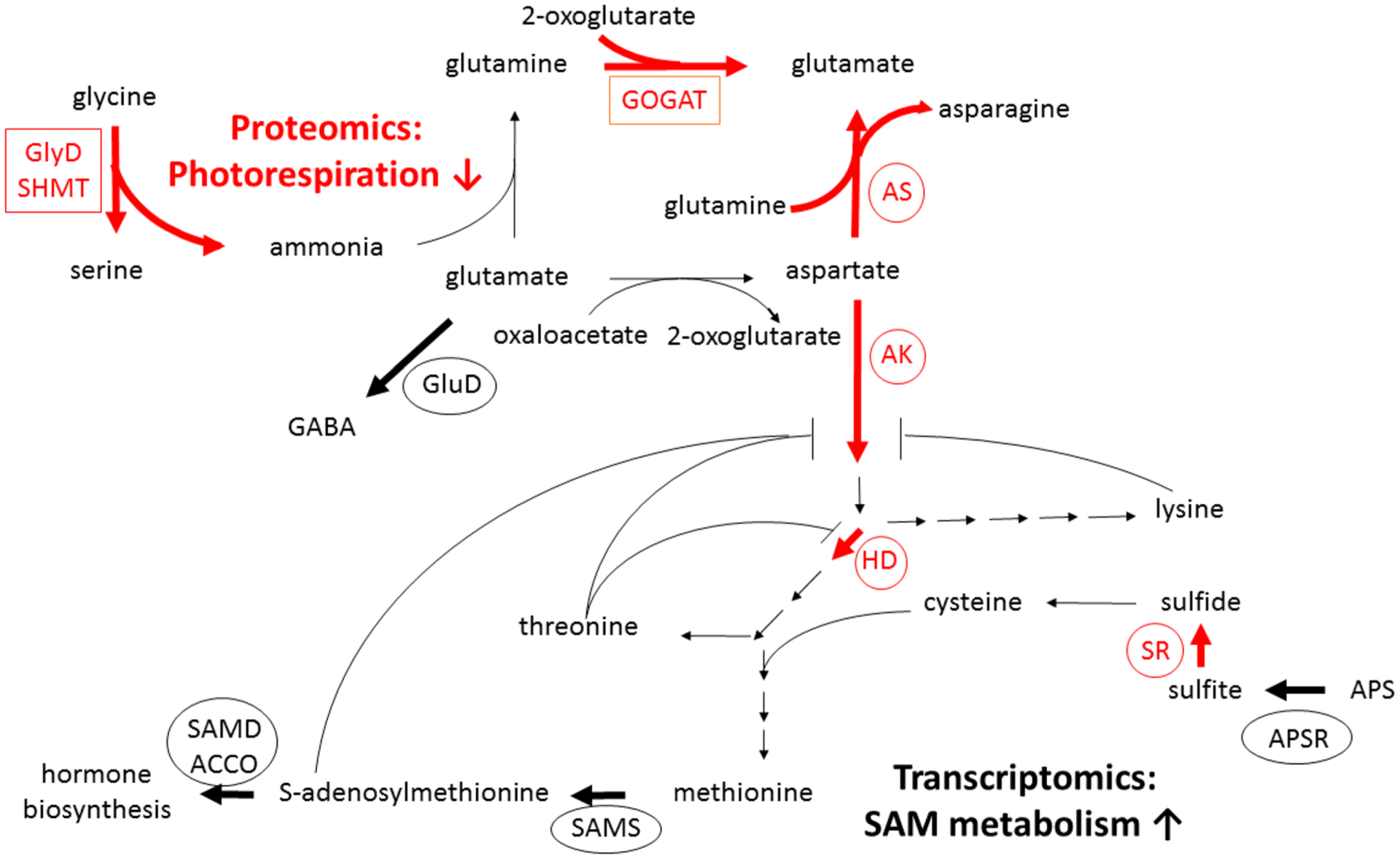
Integration of citrus response to CLas from transomic analysis. Selected nitrogen and sulfur metabolism transcripts (circled) or proteins (boxed) regulated by CLas at 10 and/or 14 weeks post graft infection are depicted by bold arrows (black: CLas up; red: CLas down). Proteomics: Downregulation of photorespiration proteins in CLas grafted trees: glycine decarboxylase (GlyD, week 10), serine hydroxymethyltransferase (SHMT, week 10), and glutamate synthase (GOGAT, weeks 10, 14). Transcriptomics: CLas upregulation of glutamate decarboxylase (GluD; GABA biosynthesis, week 10). Downregulation of aspartate metabolic enzymes asparagine synthetase (AS, week 14), aspartate kinase/homoserine dehydrogenase (AK/HD, week 10). Predicted AK/HD feedback inhibition by lysine, threonine, and/or S-adenosylmethionine (SAM) is depicted. Upregulation of SAM metabolism, including SAM synthetase (SAMS, week 10, 14) and SAM-derived hormone metabolism, including ACC oxidase (ACCO, weeks 10, 14) and SAM decarboxylase (SAMD, week 14). Upregulation of 5′-adenylsulfate reductase (APSR, week 10), downregulation of sulfite reductase (SR, week 10) impacts sulfate assimilation into cysteine.
Glutamate also serves as the precursor to the amino acids arginine, proline, and aspartate, the latter of which was found at nearly 2-fold higher levels in CLas-infected compared to control plants at 10 wpg (Figure 5). Aspartate is produced from glutamate and oxaloacetate by aspartate aminotransferase (AspAT): while transcript and protein levels of AspAT were not found to change in response to CLas, it is possible that the downregulation of the GS/GOGAT cycle redirects glutamate flux toward aspartate at 10 wpg, potentially as a result of feedback inhibition. Threonine, lysine, isoleucine, and SAM are derived from aspartate, and have been characterized as allosteric regulators of aspartate metabolism.72 Lysine and isoleucine levels were minimally elevated in CLas-infected plants compared to control plants at 10 wpg, while threonine levels were somewhat reduced (Figure 5). Downregulation of asparagine synthase transcripts suggests a reduction of conversion of aspartate to asparagine—this may result from a decrease in glutamine levels predicted from reduced photorespiration, and it reinforces the hypothesis that flux through aspartate is primarily directed toward SAM in CLas-infected plants (Table 7, Figure 6). The metabolomics data reveal that asparagine levels are approximately 2-fold less in CLas-infected compared to control plants at 8 and 10 wpg (Figure 5).
Biosynthesis of SAM and other sulfur-containing plant metabolites depends on the uptake of sulfate and its subsequent conversion to cysteine. One of the key enzymes for this conversion is 5′-adenylyl sulfate (APS) reductase73—two APS reductase transcripts are upregulated in response to CLas, while the sulfite reductase associated with the subsequent step in the sulfate reduction pathway is downregulated (Table 7, Figure 6). The coordinated regulation of sulfur assimilation and SAM is predicted to lead to biosynthesis of the plant hormones ethylene and polyamines.
In addition to the transcriptome data showing widespread involvement of hormone biosynthesis and perception in the C. limon response to CLas, proteomics data revealed the abscisic acid biosynthesis protein zeaxanthin epoxidase (XP_006470187.1) to be upregulated in CLas grafted plants at 10 wpg. Other differentially expressed citrus proteins of interest are orthologs of proteins with established roles in host-microbe interactions in other pathosystems. Remorin proteins are found at the plasmodesmata and plasma membrane and have been reported to participate in the plant immune response, controlling cell-to-cell movement of microbes.74 A remorin protein (XP_006421908.1) was found to be upregulated in CLas grafted plants at week 10, while a related remorin transcript (orange1.1 g029522m) was downregulated in CLas plants at the same time point. Fascilin-like arabinogalactan (FLA) proteins are cell wall glycoproteins which affect cell-cell interactions and the deposition and orientation of cellulose microfibrils in cell walls.75 Two transcripts encoding FLAs were found to be downregulated at 10 weeks post graft (XM_006469388.1, XM_006447747.2), and two related FLA proteins (XP_006491459.1, XP_006452590.1) were found to be downregulated at 14 wpg. FLAs are glycosylated by arabinogalactan, a polysaccharide consisting of arabinose and galactose monomers. Galactose is a major cell wall carbohydrate, as a component of xyloglucans as well as arabinogalactans.76 Elevated galactose and glucose levels in CLas compared to control plants detected by 1H NMR metabolomics at 10 wpg may result from changes in the composition of cell wall glycoproteins and polysaccharides. Several cell wall and lipid metabolism proteins potentially relevant to endocytosis were found to vary significantly between control and CLas grafted plants at 10 and 14 wpg (Figure 3, Supporting Table S3).
This study reveals details of coordinated changes in C. limon (Lisbon lemon) leaf transcripts, proteins, and metabolites over time following inoculation with CLas. These changes may be interpreted as components of a plant defense response to pathogen infection, or a reflection of the changes in the plant as it develops symptoms, declines in health, and succumbs to the disease. Field and greenhouse trials have classified a number of lemon varieties as HLB tolerant, including the Bearss lemon, the Volkamer lemon and the Eureka lemon.77–79 In these studies, the tolerant citrus hosts of CLas become infected with the pathogen, but infected tolerant plants exhibited relatively mild symptoms compared to sensitive citrus varieties. Eureka lemon was found in greenhouse trials to be highly tolerant to HLB, with minimal visual symptoms and vigorous growth. However, these tolerant Eureka lemon plants had high titers of CLas which were comparable with titers in sensitive hosts.78 In Eureka lemon field trials, visual symptoms of disease did develop, but unlike with sensitive sweet orange genotypes, these trees continued to retain their leaves and survived for the duration of the four year trial.79 A parallel study of the response of sweet orange (Citrus sinensis var. Osbeck) was performed alongside the study reported here.62 Comparative analysis of CLas titer and development of disease symptoms revealed that lemon plants became CLas positive and developed visual symptoms several weeks before sweet orange plants.62 Characterization of the differences in the transcripts, proteins, and metabolites induced by CLas in these different citrus species will provide insight into host plant-specific defense responses.
These data reveal changes in C. limon leaves in response to CLas infection which can be detected at the transcript, protein, and metabolite levels weeks before plants show symptoms or test positive for CLas by qPCR. Some of the molecular changes which we observed at the presymptomatic stage of infection are early indicators of physiological changes which are manifested weeks and months later in visual symptoms in diseased plants. Phytohormones serve as primary mediators of the plant response to pathogens,80 and the signaling pathways which they activate affect diverse physiological processes including fruit abscission and leaf chlorosis. Transcriptome and proteome data reveal differences in expression of genes and proteins involved in hormone biosynthesis and perception between presymptomatic CLas and control plants. Genes involved in ethylene biosynthesis, as well as genes encoding ethylene response factors, are upregulated at CLas-infected plants compared to controls. While the ripening of citrus, a nonclimacteric fruit, is not associated with increased ethylene production and respiration, application of exogenous ethylene has been found to promote citrus fruit abscission.81 Transcriptome and proteome evidence for changes in auxin, gibberellin, and abscisic acid metabolism in infected plants suggest a role of plant hormones in the early fruit drop observed in diseased plants. Another visual symptom of citrus greening disease is leaf blotchy mottle, the development of asymmetric chlorotic patches on the leaves of infected trees which is associated with loss of chlorophyll.82 One of the gene ontology terms most highly represented in the citrus transcripts differentially expressed between control and CLas plants is “chlorophyll catabolic process” (Table 1), and there are two chlorophyllase 1 and two chlorophyllase 2 transcripts found to be ~10-fold upregulated in infected leaves at 14 wpg (Supporting Table S1). These changes in gene expression related to chlorophyll metabolism may represent an early plant molecular response to infection which ultimately leads to the development of visual disease symptoms. These physiological changes in diseased plants may affect the biology of the citrus greening insect vector, Diaphorina citri, through changes in plant nutritional quality predicted from metabolite analysis, and through visual cues in leaf color changes. Yellow leaf color associated with blotchy mottle symptoms is thought to serve as a visual cue to D. citri83—this may reflect a vector manipulation strategy by CLas, where healthy insects are more attracted to diseased than healthy plants,84 increasing the likelihood that the pathogen will be acquired by the vector and transmitted to new plant hosts.
CONCLUSIONS
The citrus response to infection is dynamic and subject to regulation at multiple levels. Different components of interrelated pathways were found to be regulated at the transcriptional and translational levels. Integration of these data sets from multiple levels of analysis provides a holistic picture of the greenhouse-grown plant response to infection, connecting genes, proteins, and metabolites to pathways and whole-tree physiology. Detailed understanding of the plant response to infection can be used to inform breeding strategies to increase the effectiveness of the plant immune system in responding to the presence of CLas and preventing its spread within the plant. Candidate peptide and metabolite biomarkers of infection must be evaluated for their effectiveness in the field, their relevance to a range of citrus varieties, and their specificity to CLas. Development of automation to decrease sample preparation time and increase throughput of sample analysis will be necessary for deployment of validated biomarkers in the field, enabling presymptomatic detection and expediting removal of diseased trees as part of efforts to stop the spread of the pathogen.
Supplementary Material
Funding
Funding was provided by USDA CRIS Project #8062-22410-006-00D, Citrus Research Board Projects #5300-150 and #5300-155, and NIH grant 1S10RR011973-01.
ABBREVIATIONS
- ACC
aminocyclopropane-1-carboxylate
- ACCO
aminocyclopropane-1-carboxylate oxidase
- AK/HD
aspartate kinase/homoserine dehydrogenase
- APS
5′-adenylyl sulfate
- APSR
5′-adenylyl sulfate reductase
- AS
asparagine synthetase
- AspAT
aspartate aminotransferase
- CLas
Candidatus Liberibacter asiaticus
- FDR
false discovery rate
- GA2ox
gibberellin 2-oxidase
- GAGA
gamma-aminobutyric acid
- GluD
glutamate decarboxylase
- GlyD
glycine decarboxylase
- GS/GOGAT
glutamine synthetase/ferredoxin-dependent glutamate synthase
- 1H NMR
proton nuclear magnetic resonance
- HLB
huanglongbing
- JAZ
jasmonate-ZIM domain
- NMDS
nonmetric multidimensional scaling
- PCA
principal component analysis
- qPCR
quantitative polymerase chain reaction
- SAM
S-adenosylmethionine
- SAMS
S-adenosylmethionine synthetase
- SHMT
serine hydroxymethyltransferase
- SR
sulfite reductase
- TEAB
triethylammonium bicarbonate
- wpg
weeks post graft
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00802.
Table S1: Read count values for all transcripts identified as differentially expressed between control and CLas grafted plants at all time points; Differentially expressed citrus transcripts (XLSX)
Table S2: Spectral count data for all proteins identified in Citrus limon leaf samples; All proteomics spectral count data (XLSX)
Table S3: Citrus proteins identified as differentially abundant between control and CLas grafted plants at all time points; Differentially abundant citrus proteins (XLSX)
Table S4: Median, minimum, and maximum metabolite concentrations (nmol/g dry weight) for control and CLas grafted plants at each time point; All metabolite concentration data (XLSX)
Table S5: Blastx report; Citrus proteins with sequence similarity to differentially expressed citrus transcripts (XLSX)
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jproteome.9b00802
Contributor Information
John S. Ramsey, USDA Emerging Pests and Pathogens Research Unit, Robert W. Holley Center for Agriculture and Health, Ithaca, New York 14853, United States; Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, United States.
Elizabeth L. Chin, Department of Food Science and Technology, University of California, Davis, California 95616, United States
Juan D. Chavez, Department of Genome Sciences, University of Washington, Seattle, Washington 98195, United States
Surya Saha, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, United States.
Darya Mischuk, Department of Food Science and Technology, University of California, Davis, California 95616, United States.
Jaclyn Mahoney, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, United States.
Jared Mohr, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, United States; Department of Genome Sciences, University of Washington, Seattle, Washington 98195, United States.
Faith M. Robison, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, United States
Elizabeth Mitrovic, Contained Research Facility, University of California, Davis, California 95616, United States.
Xuefei Zhong, Department of Genome Sciences, University of Washington, Seattle, Washington 98195, United States.
MaryLou Polek, Citrus Research Board, Visalia, California 93291, United States; National Clonal Germplasm Repository for Citrus, Riverside, California 92507, United States.
Kris E. Godfrey, Contained Research Facility, University of California, Davis, California 95616, United States
James J. Giovannoni, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, United States; USDA Plant, Soil, and Nutrition Research Unit, Robert W. Holley Center for Agriculture and Health, Ithaca, New York 14853, United States; Plant Biology Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, United States
Lukas A. Mueller, Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, United States
Carolyn M. Slupsky, Department of Food Science and Technology, University of California, Davis, California 95616, United States.
James E. Bruce, Department of Genome Sciences, University of Washington, Seattle, Washington 98195, United States.
Michelle Heck, USDA Emerging Pests and Pathogens Research Unit, Robert W. Holley Center for Agriculture and Health, Ithaca, New York 14853, United States; Boyce Thompson Institute for Plant Research, Ithaca, New York 14853, United States; Plant Pathology and Plant-Microbe Biology Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, United States.
REFERENCES
- (1).Wang N The Citrus Huanglongbing Crisis and Potential Solutions. Mol. Plant 2019, 12 (5), 607–609. [DOI] [PubMed] [Google Scholar]
- (2).Li WB; Hartung JS; Levy L Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods 2006, 66 (1), 104–115. [DOI] [PubMed] [Google Scholar]
- (3).Johnson EG; Wu J; Bright DB; Graham JH Association of ‘Candidatus Liberibacter asiaticus’ root infection, but not phloem plugging with root loss on huanglongbing- affected trees prior to appearance of foliar symptoms. Plant Pathol. 2014, 63 (2), 290–298. [Google Scholar]
- (4).Louzada ES; Vazquez OE; Braswell WE; Yanev G; Devanaboina M; Kunta M Distribution of ‘Candidatus Liberibacter asiaticus’ Above and Below Ground in Texas Citrus. Phytopathology 2016, 106 (7), 702–709. [DOI] [PubMed] [Google Scholar]
- (5).Lee JA; Halbert SE; Dawson WO; Robertson CJ; Keesling JE; Singer BH Asymptomatic spread of huanglongbing and implications for disease control. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (24), 7605–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Choi HK; da Silva FG; Lim HJ; Iandolino A; Seo YS; Lee SW; Cook DR Diagnosis of Pierce’s Disease Using Biomarkers Specific to Xylella fastidiosa rRNA and Vitis vinifera Gene Expression. Phytopathology 2010, 100 (10), 1089–1099. [DOI] [PubMed] [Google Scholar]
- (7).Beasley-Green A Urine Proteomics in the Era of Mass Spectrometry. Int. Neurourol. J 2016, 20, 70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chin EL; Mishchuk DO; Breksa AP; Slupsky CM Metabolite Signature of Candidatus Liberibacter asiaticus Infection in Two Citrus Varieties. J. Agric. Food Chem 2014, 62 (28), 6585–6591. [DOI] [PubMed] [Google Scholar]
- (9).Slisz AM; Breksa AP; Mishchuk DO; McCollum G; Slupsky CM Metabolomic Analysis of Citrus Infection by ‘Candidatus Liberibacter’ Reveals Insight into Pathogenicity. J. Proteome Res 2012, 11 (8), 4223–4230. [DOI] [PubMed] [Google Scholar]
- (10).Hung WL; Wang Y Metabolite Profiling of Candidatus Liberibacter Infection in Hamlin Sweet Oranges. J. Agric. Food Chem 2018, 66 (15), 3983–3991. [DOI] [PubMed] [Google Scholar]
- (11).Martinelli F; Uratsu SL; Albrecht U; Reagan RL; Phu ML; Britton M; Buffalo V; Fass J; Leicht E; Zhao WX; Lin DW; D’Souza R; Davis CE; Bowman KD; Dandekar AM Transcriptome Profiling of Citrus Fruit Response to Huanglongbing Disease. PLoS One 2012, 7, No. e38039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zhong Y; Cheng CZ; Jiang NH; Jiang B; Zhang YY; Hu BML; Zeng JW; Yan HX; Yi GJ; Zhong GY Comparative Transcriptome and iTRAQ Proteome Analyses of Citrus Root Responses to Candidatus Liberibacter asiaticus Infection. PLoS One 2015, 10, No. e0126973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Nwugo CC; Lin H; Duan YP; Civerolo EL The effect of ‘Candidatus Liberibacter asiaticus’ infection on the proteomic profiles and nutritional status of pre-symptomatic and symptomatic grapefruit (Citrus paradisi) plants. BMC Plant Biol. 2013, 13, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Xu MR; Li Y; Zheng Z; Dai ZH; Tao Y; Deng XL Transcriptional Analyses of Mandarins Seriously Infected by ‘Candidatus Liberibacter asiaticus’. PLoS One 2015, 10, No. e0133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Nwugo CC; Duan YP; Lin H Study on Citrus Response to Huanglongbing Highlights a Down-Regulation of Defense-Related Proteins in Lemon Plants Upon ‘Ca. Liberibacter asiaticus’ Infection. PLoS One 2013, 8 (6), e67442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Fan J; Chen CX; Yu QB; Brlansky RH; Li ZG; Gmitter FG Comparative iTRAQ proteome and transcriptome analyses of sweet orange infected by “Candidatus Liberibacter asiaticus. Physiol. Plant 2011, 143 (3), 235–245. [DOI] [PubMed] [Google Scholar]
- (17).Boava LP; Cristofani-Yaly M; Machado MA Physiologic, Anatomic, and Gene Expression Changes in Citrus sunki, Poncirus trifoliata, and Their Hybrids After ‘Candidatus Liberibacter asiaticus’ Infection. Phytopathology 2017, 107 (5), 590–599. [DOI] [PubMed] [Google Scholar]
- (18).Aritua V; Achor D; Gmitter FG; Albrigo G; Wang N Transcriptional and Microscopic Analyses of Citrus Stem and Root Responses to Candidatus Liberibacter asiaticus Infection. PLoS One 2013, 8, No. e73742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Mafra V; Martins PK; Francisco CS; Ribeiro-Alves M; Freitas-Astua J; Machado MA Candidatus Liberibacter americanus induces significant reprogramming of the transcriptome of the susceptible citrus genotype. BMC Genomics 2013, 14, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Baldwin E; Plotto A; Bai J; Manthey J; Zhao W; Raithore S; Irey M Effect of Abscission Zone Formation on Orange (Citrus sinensis) Fruit/Juice Quality for Trees Affected by Huanglongbing (HLB). J. Agric. Food Chem 2018, 66 (11), 2877–2890. [DOI] [PubMed] [Google Scholar]
- (21).Zhao W; Baldwin EA; Bai JH; Plotto A; Irey M Comparative analysis of the transcriptomes of the calyx abscission zone of sweet orange insights into the huanglongbing-associated fruit abscission. Hortic. Res 2019, 6, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Li W; Yao YN; Wu L; Hu B Detection and Seasonal Variations of Huanglongbing Disease in Navel Orange Trees Using Direct Ionization Mass Spectrometry. J. Agric. Food Chem 2019, 67 (8), 2265–2271. [DOI] [PubMed] [Google Scholar]
- (23).Selvaraj V; Maheshwari Y; Hajeri S; Chen JC; McCollum TG; Yokomi R Development of a duplex droplet digital PCR assay for absolute quantitative detection of “Candidatus Liberibacter asiaticus. PLoS One 2018, 13, No. e0197184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhong S; Joung J; Zheng Y; Chen Y; Liu B; Shao Y; Xiang J; Fei Z; Giovannoni J High-Throughput Illumina Strand-Specific RNA Sequencing Library Preparation. Cold Spring Harbor Protocols 2011, 2011, pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- (25).Wu GA; Prochnik S; Jenkins J; Salse J; Hellsten U; Murat F; Perrier X; Ruiz M; Scalabrin S; Terol J; Takita MA; Labadie K; Poulain J; Couloux A; Jabbari K; Cattonaro F; Del Fabbro C; Pinosio S; Zuccolo A; Chapman J; Grimwood J; Tadeo FR; Estornell LH; Munoz-Sanz JV; Ibanez V; Herrero-Ortega A; Aleza P; Perez-Perez J; Ramon D; Brunel D; Luro F; Chen CX; Farmerie WG; Desany B; Kodira C; Mohiuddin M; Harkins T; Fredrikson K; Burns P; Lomsadze A; Borodovsky M; Reforgiato G; Freitas-Astua J; Quetier F; Navarro L; Roose M; Wincker P; Schmutz J; Morgante M; Machado MA; Talon M; Jaillon O; Ollitrault P; Gmitter F; Rokhsar D Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol 2014, 32 (7), 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kim D; Pertea G; Trapnell C; Pimentel H; Kelley R; Salzberg SL TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 2013, 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Grabherr MG; Haas BJ; Yassour M; Levin JZ; Thompson DA; Amit I; Adiconis X; Fan L; Raychowdhury R; Zeng QD; Chen ZH; Mauceli E; Hacohen N; Gnirke A; Rhind N; di Palma F; Birren BW; Nusbaum C; Lindblad-Toh K; Friedman N; Regev A Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol 2011, 29 (7), 644–U130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Trapnell C; Williams BA; Pertea G; Mortazavi A; Kwan G; van Baren MJ; Salzberg SL; Wold BJ; Pachter L Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 2010, 28 (5), 511xU174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Li WZ; Godzik A Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22 (13), 1658–1659. [DOI] [PubMed] [Google Scholar]
- (30).Bolger AM; Lohse M; Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30 (15), 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kim D; Langmead B; Salzberg S L HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 2015, 12 (4), 357–U121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Li B; Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Pertea M; Pertea GM; Antonescu CM; Chang TC; Mendell JT; Salzberg SL StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 2015, 33 (3), 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Anders S; Huber W Differential expression analysis for sequence count data. Genome Biol. 2010, DOI: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Robinson MD; Smyth GK Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 2007, 9 (2), 321–332. [DOI] [PubMed] [Google Scholar]
- (36).Alexa ARJ topGO: Enrichment Analysis for Gene Ontology. R package, version 2.26.0; 2016. [Google Scholar]
- (37).Vizcaino JA; Csordas A; del-Toro N; Dianes JA; Griss J; Lavidas I; Mayer G; Perez-Riverol Y; Reisinger F; Ternent T; Xu QW; Wang R; Hermjakob H 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44 (D1), D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Vizcaino JA; Deutsch EW; Wang R; Csordas A; Reisinger F; Rios D; Dianes JA; Sun Z; Farrah T; Bandeira N; Binz PA; Xenarios I; Eisenacher M; Mayer G; Gatto L; Campos A; Chalkley RJ; Kraus HJ; Albar JP; Martinez-Bartolome S; Apweiler R; Omenn GS; Martens L; Jones AR; Hermjakob H ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol 2014, 32 (3), 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Perneger TV What’s wrong with Bonferroni adjustments. BMJ: British Medical Journal 1998, 316 (7139), 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Nakagawa S A farewell to Bonferroni: the problems of low statistical power and publication bias. Behav. Ecol 2004, 15 (6), 1044–1045. [Google Scholar]
- (41).Benjamini Y; Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. Series B (Methodological) 1995, 57, 289–300. [Google Scholar]
- (42).Fritz CO; Morris PE; Richler JJ Effect size estimates: current use, calculations, and interpretation. J. Exp. Psychol. Gen 2012, 141 (1), 2. [DOI] [PubMed] [Google Scholar]
- (43).Roje S S-Adenosyl-L-methionine: Beyond the universal methyl group donor. Phytochemistry 2006, 67 (15), 1686–1698. [DOI] [PubMed] [Google Scholar]
- (44).Martinelli F; Reagan RL; Uratsu SL; Phu ML; Albrecht U; Zhao WX; Davis CE; Bowman KD; Dandekar AM Gene Regulatory Networks Elucidating Huanglongbing Disease Mechanisms. PLoS One 2013, 8, No. e74256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chung HS; Koo AJK; Gao XL; Jayanty S; Thines B; Jones AD; Howe GA Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008, 146 (3), 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Perkins DN; Pappin DJ; Creasy DM; Cottrell JS Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20 (18), 3551–67. [DOI] [PubMed] [Google Scholar]
- (47).Searle BC Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 2010, 10 (6), 1265–9. [DOI] [PubMed] [Google Scholar]
- (48).Zhong LL; Zhou W; Wang HJ; Ding SH; Lu QT; Wen XG; Peng LW; Zhang LX; Lu CM Chloroplast Small Heat Shock Protein HSP21 Interacts with Plastid Nucleoid Protein pTAC5 and Is Essential for Chloroplast Development in Arabidopsis under Heat Stress. Plant Cell 2013, 25 (8), 2925–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Bateman A; Martin MJ; O’Donovan C; Magrane M; Apweiler R; Alpi E; Antunes R; Ar-Ganiska J; Bely B; Bingley M; Bonilla C; Britto R; Bursteinas B; Chavali G; Cibrian-Uhalte E; Da Silva A; De Giorgi M; Dogan T; Fazzini F; Gane P; Cas-Tro LG; Garmiri P; Hatton-Ellis E; Hieta R; Huntley R; Legge D; Liu WD; Luo J; MacDougall A; Mutowo P; Nightin-Gale A; Orchard S; Pichler K; Poggioli D; Pundir S; Pureza L; Qi GY; Rosanoff S; Saidi R; Sawford T; Shypitsyna A; Turner E; Volynkin V; Wardell T; Watkins X; Watkins Cowley A; Figueira L; Li WZ; McWilliam H; Lopez R; Xenarios I; Bougueleret L; Bridge A; Poux S; Redaschi N; Aimo L; Argoud-Puy G; Auchincloss A; Axelsen K; Bansal P; Baratin D; Blatter MC; Boeckmann B; Bolleman J; Boutet E; Breuza L; Casal-Casas C; De Castro E; Coudert E; Cuche B; Doche M; Dornevil D; Duvaud S; Estreicher A; Famiglietti L; Feuermann M; Gasteiger E; Gehant S; Gerritsen V; Gos A; Gruaz-Gumowski N; Hinz U; Hulo C; Jungo F; Keller G; Lara V; Lemercier P; Lieberherr D; Lombardot T; Martin X; Masson P; Morgat A; Neto T; Nouspikel N; Paesano S; Pedruzzi I; Pilbout S; Pozzato M; Pruess M; Rivoire C; Roechert B; Schneider M; Sigrist C; Sonesson K; Staehli S; Stutz A; Sundaram S; Tognolli M; Verbregue L; Veuthey AL; Wu CH; Arighi CN; Arminski L; Chen CM; Chen YX; Garavelli JS; Huang HZ; Laiho KT; McGarvey P; Natale DA; Suzek BE; Vinayaka CR; Wang QH; Wang YQ; Yeh LS; Yerramalla MS; Zhang J; Consortium U UniProt: a hub for protein information. Nucleic Acids Res. 2015, 43 (D1), D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Delprato ML; Krapp AR; Carrillo N Green Light to Plant Responses to Pathogens: The Role of Chloroplast Light-Dependent Signaling in Biotic Stress. Photochem. Photobiol 2015, 91 (5), 1004–1011. [DOI] [PubMed] [Google Scholar]
- (51).Sweetlove LJ; Lytovchenko A; Morgan M; Nunes-Nesi A; Taylor NL; Baxter CJ; Eickmeier I; Fernie AR Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. U. S. A 2006, 103 (51), 19587–19592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Voss I; Sunil B; Scheibe R; Raghavendra AS Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol. 2013, 15 (4), 713–722. [DOI] [PubMed] [Google Scholar]
- (53).Jamai A; Salome PA; Schilling SH; Weber APM; McClung CR Arabidopsis Photorespiratory Serine Hydroxymethyl-transferase Activity Requires the Mitochondrial Accumulation of Ferredoxin-Dependent Glutamate Synthase. Plant Cell 2009, 21 (2), 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Mauch F; Staehelin LA Functional Implications of the Subcellular-Localization of Ethylene-Induced Chitinase and Beta-1,3-Glucanase in Bean-Leaves. Plant Cell 1989, 1 (4), 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Wu CT; Bradford KJ Class I Chitinase and beta-1,3-glucanase are differentially regulated by wounding, methyl jasmonate, ethylene, and gibberellin in tomato seeds and leaves. Plant Physiol. 2003, 133 (1), 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).De Storme N; Geelen D Callose homeostasis at plasmodesmata: molecular regulators and developmental relevance. Front. Plant Sci 2014, 5, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Koh EJ; Zhou LJ; Williams DS; Park J; Ding NY; Duan YP; Kang BH Callose deposition in the phloem plasmodesmata and inhibition of phloem transport in citrus leaves infected with “Candidatus Liberibacter asiaticus. Protoplasma 2012, 249 (3), 687–697. [DOI] [PubMed] [Google Scholar]
- (58).Gaudioso-Pedraza R; Benitez-Alfonso Y A phylogenetic approach to study the origin and evolution of plasmodesmata-localized glycosyl hydrolases family 17. Front. Plant Sci 2014, 5, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Marco-Ramell A; Palau-Rodriguez M; Alay A; Tulipani S; Urpi-Sarda M; Sanchez-Pla A; Andres-Lacueva C Evaluation and comparison of bioinformatic tools for the enrichment analysis of metabolomics data. BMC Bioinf. 2018, 19, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Chin E; Godfrey K; Polek M; Slupsky C H-1 NMR analysis of Citrus macrophylla subjected to Asian citrus psyllid (Diaphorina citri Kuwayama) feeding. Arthropod-Plant Inte 2017, 11 (6), 901–909. [Google Scholar]
- (61).O’Sullivan A; He X; McNiven EMS; Hinde K; Haggarty NW; Lonnerdal B; Slupsky CM Metabolomic Phenotyping Validates the Infant Rhesus Monkey as a Model of Human Infant Metabolism. J. Pediatr. Gastroenterol. Nutr 2013, 56 (4), 355–363. [DOI] [PubMed] [Google Scholar]
- (62).Chin EL; Ramsey JS; Mishchuk DO; Saha S; Foster E; Chavez JD; Howe K; Zhong X; Polek M; Godfrey KE; Mueller LA; Bruce JE; Heck M; Slupsky CM Longitudinal Transcriptomic, Proteomic, and Metabolomic Analyses of Citrus sinensis (L.) Osbeck Graft-Inoculated with “Candidatus Liberibacter asiaticus. J. Proteome Res 2020, 19 (2), 719–732. [DOI] [PubMed] [Google Scholar]
- (63).Jin L; Mackey DM Measuring Callose Deposition, an Indicator of Cell Wall Reinforcement, During Bacterial Infection in Arabidopsis. Methods Mol. Biol 2017, 1578, 195–205. [DOI] [PubMed] [Google Scholar]
- (64).Luna E; Pastor V; Robert J; Flors V; Mauch-Mani B; Ton J Callose deposition: a multifaceted plant defense response. Mol. Plant-Microbe Interact 2011, 24 (2), 183–93. [DOI] [PubMed] [Google Scholar]
- (65).Aznar A; Chen NWG; Thomine S; Dellagi A Immunity to plant pathogens and iron homeostasis. Plant Sci. 2015, 240, 90–97. [DOI] [PubMed] [Google Scholar]
- (66).Kruse A; Fattah-Hosseini S; Saha S; Johnson R; Warwick E; Sturgeon K; Mueller L; MacCoss MJ; Shatters RG; Heck MC Combining ‘omics and microscopy to visualize interactions between the Asian citrus psyllid vector and the Huanglongbing pathogen Candidatus Liberibacter asiaticus in the insect gut. PLoS One 2017, 12, No. e0179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Parrow NL; Fleming RE; Minnick MF Sequestration and Scavenging of Iron in Infection. Infect. Immun 2013, 81 (10), 3503–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Huang J; Ghosh R; Bankaitis VA Sec14-like phosphatidylinositol transfer proteins and the biological landscape of phosphoinositide signaling in plants. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2016, 1861 (9), 1352–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Moreno JI; Martin R; Castresana C Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J. 2005, 41 (3), 451–463. [DOI] [PubMed] [Google Scholar]
- (70).Coschigano KT; Melo-Oliveira R; Lim J; Coruzzi GM Arabidopsis gls mutants and distinct Fd-GOGAT genes: Implications for photorespiration and primary nitrogen assimilation. Plant Cell 1998, 10 (5), 741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Michaeli S; Fromm H Closing the loop on the GABA shunt in plants: are GABA metabolism and signaling entwined? Front. Plant Sci 2015, 6, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Jander G; Joshi V Recent Progress in Deciphering the Biosynthesis of Aspartate-Derived Amino Acids in Plants. Mol. Plant 2010, 3 (1), 54–65. [DOI] [PubMed] [Google Scholar]
- (73).Martin MN; Tarczynski MC; Shen B; Leustek T The role of 5 ‘-adenylylsulfate reductase in controlling sulfate reduction in plants. Photosynth. Res 2005, 86 (3), 309–323. [DOI] [PubMed] [Google Scholar]
- (74).Gui JS; Liu C; Shen JH; Li LG Grain setting defect1, Encoding a Remorin Protein, Affects the Grain Setting in Rice through Regulating Plasmodesmatal Conductance. Plant Physiol. 2014, 166 (3), 1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).MacMillan CP; Mansfield SD; Stachurski ZH; Evans R; Southerton SG Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J. 2010, 62 (4), 689–703. [DOI] [PubMed] [Google Scholar]
- (76).Pena MJ; Ryden P; Madson M; Smith AC; Carpita NC The galactose residues of xyloglucan are essential to maintain mechanical strength of the primary cell walls in Arabidopsis during growth. Plant Physiol. 2004, 134 (1), 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Deng HH; Achor D; Exteberria E; Yu QB; Du DL; Stanton D; Liang G; Gmitter FG Phloem Regeneration Is a Mechanism for Huanglongbing-Tolerance of “Bearss” Lemon and “LB8–9” Sugar Belle (R) Mandarin. Front. Plant Sci 2019, 10, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Folimonova SY; Robertson CJ; Garnsey SM; Gowda S; Dawson WO Examination of the Responses of Different Genotypes of Citrus to Huanglongbing (Citrus Greening) Under Different Conditions. Phytopathology 2009, 99 (12), 1346–1354. [DOI] [PubMed] [Google Scholar]
- (79).Ramadugu C; Keremane ML; Halbert SE; Duan YP; Roose ML; Stover E; Lee RF Long-Term Field Evaluation Reveals Huanglongbing Resistance in Citrus Relatives. Plant Dis. 2016, 100 (9), 1858–1869. [DOI] [PubMed] [Google Scholar]
- (80).Ma KW; Ma WB Phytohormone pathways as targets of pathogens to facilitate infection. Plant Mol. Biol 2016, 91 (6), 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Merelo P; Agusti J; Arbona V; Costa ML; Estornell LH; Gomez-Cadenas A; Coimbra S; Gomez MD; Perez-Amador MA; Domingo C; Talon M; Tadeo FR Cell Wall Remodeling in Abscission Zone Cells during Ethylene-Promoted Fruit Abscission in Citrus. Front. Plant Sci 2017, 8, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Sagaram M; Burns JK Leaf Chlorophyll Fluorescence Parameters and Huanglongbing. J. Am. Soc. Hortic. Sci 2009, 134 (2), 194–201. [Google Scholar]
- (83).Hall DG; Hentz MG; Ciomperlik MA A comparison of traps and stem tap sampling for monitoring adult asian citrus psyllid (Hemiptera: Psyllidae) in citrus. Fla. Entomol 2007, 90 (2), 327–334. [Google Scholar]
- (84).Ingwell LL; Eigenbrode SD; Bosque-Perez NA Plant viruses alter insect behavior to enhance their spread. Sci. Rep 2012, 2, 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the results of this article are available in publicly accessible repositories. Transcriptomics: All biosample and sequence data from the Cufflinks assembly were submitted to GenBank and can be accessed at Bioproject ID PRJNA348468. The Trinity assembly, and a fasta file containing 11 protein sequences predicted from the C. limon RNA-Seq cufflinks assembly, which were used for proteomics data analysis but did not pass GenBank submission criteria, have been submitted to Figshare (DOI: 10.6084/m9.figshare.5671585). Proteomics: The mass spectrometry proteomics data have been deposited to the PRIDE Archive (http://www.ebi.ac.uk/pride/archive)37,38 via the PRIDE partner repository with the data set identifiers PXD005905, PXD006010, and PXD006011. Metabolomics: The 1H NMR raw spectra and metadata have been made publicly available at citrusgreening.org and can be accessed using this link: https://citrusgreening.org/metabolomics_host/index.


