ABSTRACT
Hippo signaling mediates influences of cytoskeletal tension on organ growth. TRIP6 and LIMD1 have each been identified as being required for tension-dependent inhibition of the Hippo pathway LATS kinases and their recruitment to adherens junctions, but the relationship between TRIP6 and LIMD1 was unknown. Using siRNA-mediated gene knockdown, we show that TRIP6 is required for LIMD1 localization to adherens junctions, whereas LIMD1 is not required for TRIP6 localization. TRIP6, but not LIMD1, is also required for the recruitment of vinculin and VASP to adherens junctions. Knockdown of TRIP6 or vinculin, but not of LIMD1, also influences the localization of myosin and F-actin. In TRIP6 knockdown cells, actin stress fibers are lost apically but increased basally, and there is a corresponding increase in the recruitment of vinculin and VASP to basal focal adhesions. Our observations identify a role for TRIP6 in organizing F-actin and maintaining tension at adherens junctions that could account for its influence on LIMD1 and LATS. They also suggest that focal adhesions and adherens junctions compete for key proteins needed to maintain attachments to contractile F-actin.
KEY WORDS: Mechanotransduction, Tension, Hippo, Junction, LIMD1, Cytoskeleton
Summary: Clarification of the relationship between two tension-regulated proteins, LIMD1 and TRIP6, and identification of a role for TRIP6 in regulating the organization of the actin cytoskeleton.
INTRODUCTION
The Hippo signaling network controls organ growth and cell fate in a wide range of animals, and when dysregulated can contribute to oncogenesis (Misra and Irvine, 2018; Zanconato et al., 2016). Hippo signaling mediates its effects through regulation of the transcriptional co-activator proteins YAP1 and TAZ (Yorkie in Drosophila). YAP1 and TAZ (collectively, YAP proteins) are inhibited by Hippo signaling through phosphorylation by the LATS kinases LATS1 and LATS2 (Warts in Drosophila). Hippo signaling integrates diverse upstream inputs, including cytoskeletal tension. Distinct mechanisms through which cytoskeletal tension could modulate Hippo signaling have been suggested (Misra and Irvine, 2018; Sun and Irvine, 2016). One of the best characterized involves tension-dependent recruitment and inhibition of LATS kinases at adherens junctions (AJ). This was first discovered in Drosophila, in which the Ajuba LIM protein (Jub) is recruited to AJ under tension, and Jub then recruits and inhibits Warts (Rauskolb et al., 2019, 2014; Razzell et al., 2018). Tension-dependent inhibition and recruitment of LATS kinases to AJ has also been observed in mammalian cells, but two different proteins have been implicated in this recruitment: LIMD1 and TRIP6 (Dutta et al., 2018; Ibar et al., 2018).
Each of the three mammalian Ajuba family proteins, AJUBA, WTIP and LIMD1, co-localize with LATS kinases at AJ, and have been reported to be able to co-immunoprecipitate with LATS kinases, and to inhibit them when overexpressed (Das Thakur et al., 2010; Ibar et al., 2018; Jagannathan et al., 2016). However, LIMD1 is the Ajuba family protein most closely related to Drosophila Jub, and is uniquely required in MCF10A cells for the regulation of LATS at AJ under tension (Ibar et al., 2018). In the absence of LIMD1, or under conditions of low cytoskeletal tension, LATS kinases are not recruited to AJ, and LATS activity is increased. Localization of Ajuba family proteins to junctions requires α-catenin, and observations that Ajuba family proteins co-localize with and can co-precipitate α-catenin, together with identification of α-catenin mutations that constitutively recruit Jub, imply that they are recruited to junctions through interaction with α-catenin (Alégot et al., 2019; Ibar et al., 2018; Marie et al., 2003) (Fig. 1A). As α-catenin undergoes a tension-dependent conformational change (Kim et al., 2015; Yao et al., 2014; Yonemura et al., 2010), conformation-dependent binding to α-catenin could explain tension-dependent recruitment of Ajuba family proteins (Alégot et al., 2019; Ibar et al., 2018).
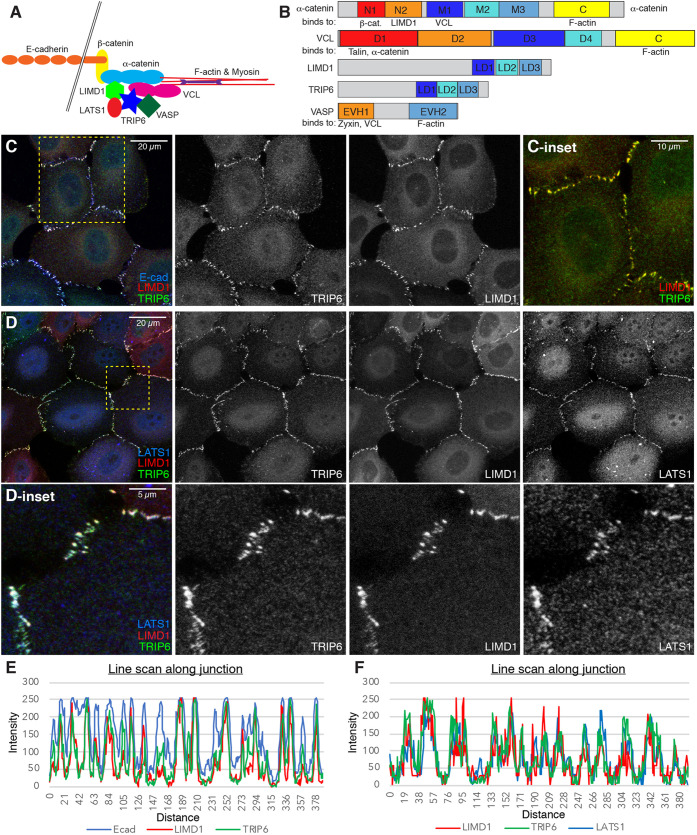
Fig. 1.
Co-localization of TRIP6, LIMD1 and LATS1 at AJ. (A) Schematic of proteins at AJ, illustrating interactions between proteins analyzed here. (B) Diagrams showing protein domains of proteins analyzed here and some of their binding partners. (C,D) MCF10A cells plated at low density and cultured for 48 h, and then stained for TRIP6, LIMD1 and E-cad (C) or LATS1 (D), as indicated. Images are z-projections through whole cells and are representatives of at least three biological replicates. Insets show higher magnification of the boxed regions. (E,F) Examples of line scans along junctions showing intensity distribution of cells shown in panels C and D.
TRIP6 is a member of the Zyxin family of LIM domain proteins, which like the Ajuba family proteins have three LIM domains at their C-terminus (Yi and Beckerle, 1998) (Fig. 1B). Zyxin, the best characterized family member, plays important roles in stabilizing and remodeling actin stress fibers subject to mechanical strain (Smith et al., 2014). A wide variety of roles for TRIP6 have been described, including effects on tumorigenesis, cell motility, telomere function, anti-apoptotic signaling and transcription, and TRIP6 has been reported to interact with a wide variety of binding partners (Lin and Lin, 2011; Willier et al., 2011). Notably, in epithelial cell lines, TRIP6 can co-immunoprecipitate with LATS kinases, co-localize with LATS kinases at junctions, and is required for localization of LATS kinases to junctions under tension (Dutta et al., 2018). TRIP6 was recruited to junctions under tension in a vinculin (VCL)-dependent process (Dutta et al., 2018). As VCL localizes to AJ through association with a tension-induced ‘open’ form of α-catenin (Kim et al., 2015; Yao et al., 2014; Yonemura et al., 2010), interaction with VCL could contribute to the tension-dependent recruitment of TRIP6 to AJ (Fig. 1A).
The observation that LIMD1 and TRIP6 both exhibit tension-dependent localization to AJ and are both required for recruitment and inhibition of LATS kinases at junctions, raised the question of how their effects on LATS kinases are related. By analyzing LIMD1 and TRIP6 localization in cells subject to siRNA-mediated knockdown of TRIP6 or LIMD1, respectively, we found that TRIP6 is required for the recruitment of LIMD1 to junctions, but LIMD1 is not required for the recruitment of TRIP6 to junctions. Analysis of markers of junctional tension revealed that TRIP6, but not LIMD1, is also required for normal tension at AJ, which could account for its role in recruiting LIMD1, while staining for F-actin revealed that apical F-actin organization is altered in the absence of TRIP6. Knockdown of TRIP6 also leads to an increase in basal F-actin stress fibers and an apparent re-localization of VCL and VASP from AJ to focal adhesions (FA), suggesting the existence of a competition between FA and AJ for proteins that stabilize tensile F-actin fibers, with TRIP6 promoting recruitment of these proteins to AJ.
RESULTS
TRIP6 is required for LIMD1 localization to AJ
To directly compare localization of LIMD1 and TRIP6, MCF10A cells were cultured under conditions that direct their localization to AJ. These proteins localize to AJ under conditions that promote high cytoskeletal tension at AJ, presumably because their localization depends upon a tension-induced conformation of α-catenin (Fig. 1A). Previous work has identified culture conditions that promote sufficient tension at AJ to establish their recruitment to AJ, including low cell density, activation of Rho or mechanical stretching of cells (Dutta et al., 2018; Ibar et al., 2018). Conversely, reduction of cytoskeletal tension by treatment with inhibitors of myosin or Rho-associated protein kinase (ROCK), or by culture at high cell density, prevents localization of LIMD1 and TRIP6 to junctions (Dutta et al., 2018; Ibar et al., 2018). We focused initially on low cell density conditions as a simple method to induce tension-dependent recruitment of TRIP6 and LIMD1 to junctions. Fixed cells at low cell density were stained with antisera against both proteins. This revealed extensive co-localization of LIMD1 and TRIP6 at AJ, where both proteins also co-localize with E-cadherin (E-cad) and with LATS1 (Fig. 1C-F).
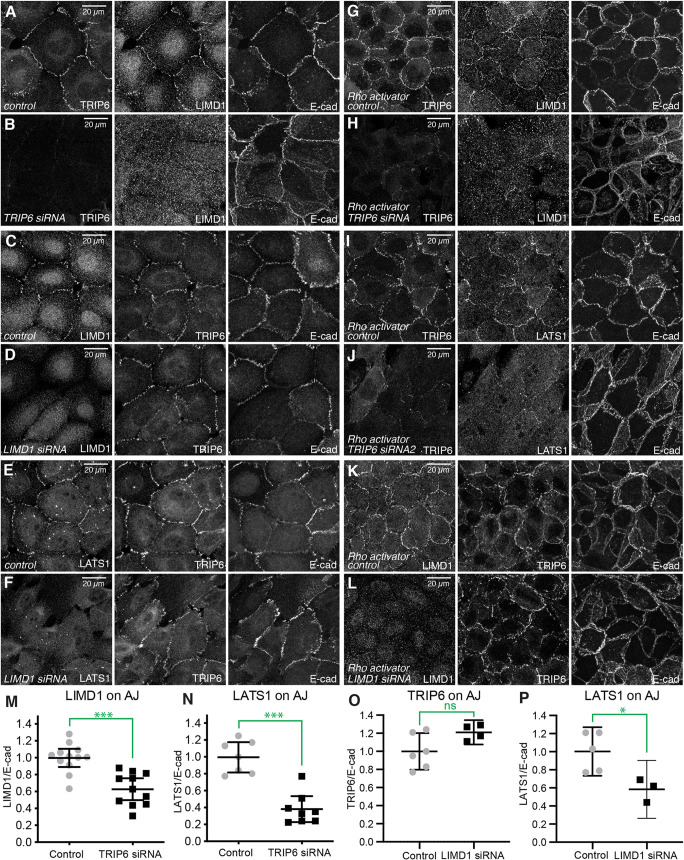
As TRIP6 and LIMD1 co-localize at AJ, we next investigated whether localization of one protein depends upon the other. As TRIP6 siRNA was not completely effective in all cells, we used TRIP6 immunostaining to identify cells with strong knockdown of protein expression, and also examined knockdown using two independent siRNAs (Fig. 2; Fig. S1). siRNA-mediated knockdown of TRIP6 substantially reduced LIMD1 and LATS1 localization to cell-cell junctions in cells cultured at low cell density, whereas E-cad staining remained similar to or only slightly weaker than that in control experiments (Fig. 2A,B,M,N; Fig. S1A-F). Western blotting did not reveal changes in E-cad or LIMD1 levels, although LATS1 levels were slightly reduced (Fig. S2). Thus, TRIP6 is required for normal localization of LIMD1 at AJ. To further confirm this result, we used an siRNA-insensitive TRIP6:GFP plasmid to rescue TRIP6 siRNA and observed that localization of LIMD1 to junctions is restored in cells transfected with TRIP6:GFP, but not in cells transfected with a control plasmid (Ecad:GFP) (Fig. S1G,H). Conversely, siRNA-mediated knockdown of LIMD1 did not reduce junctional localization of TRIP6, even though these same siRNA conditions clearly reduced LIMD1 levels (Fig. 2C,D,O; Fig. S2B). Moreover, functional knockdown of LIMD1 under these conditions was demonstrated by the loss of the junctional localization of LATS1 (Fig. 2E,F,P), which depends upon LIMD1 (Ibar et al., 2018).
Fig. 2.
TRIP6 is required for junctional localization of LIMD1. (A-F) MCF10A cells plated at low density and transfected with control (A,C,E), TRIP6 (B) or LIMD1 (D,F) siRNA, and cultured, fixed and stained for TRIP6, LIMD1, LATS1 or E-cad, as indicated. (G-L) MCF10A cells plated at high density and transfected with control (G,I,K), TRIP6 (H,J) or LIMD1 (L) siRNA, and cultured and then treated with 1 µg/ml Rho-activator-II for 3 h before fixing, and stained for TRIP6, LIMD1 or LATS1 or E-cad. Images are z-projections through whole cells and are representatives of at least three biological replicates. (M-P) Quantification of junctional levels of LIMD1, TRIP6 and LATS1 proteins normalized to junctional E-cad levels under both control and TRIP6 or LIMD1 knockdown conditions. Each dot represents results from a confocal image stack containing several cells, as in the examples. For LIMD1 analysis, N=12 for control and 11 for TRIP6 siRNA. For LATS1 analysis, N=7 for control and 8 for TRIP6 siRNA. For TRIP analysis, N=6 for control and 4 for LIMD1 siRNA. For LATS1 analysis, N=5 for control and 3 for LIMD1 siRNA. Data are mean±95% c.i. *P<0.05; ***P<0.001; ns, not significant (unpaired two-tailed t-tests).
To further investigate the relationship between TRIP6 and LIMD1, we examined them under a distinct set of culture conditions associated with their localization to AJ. Instead of low cell density, we used high cell density combined with treatment with Rho activator II (Schmidt et al., 1997). Activation of Rho increases F-actin and myosin activity, and promotes localization of LIMD1 and LATS1 to AJ even under high cell density conditions (Ibar et al., 2018). Junctional localization of LIMD1 and LATS1 was severely reduced by knockdown of TRIP6, even within Rho-activator-treated cells (Fig. 2G-J; Fig. S1I-L,O,P). Conversely, knockdown of LIMD1 did not decrease junctional localization of TRIP6 in Rho-activator-treated cells (Fig. 2K,L; Fig. S1N). Altogether, these observations imply a hierarchical relationship between junctional localization of LIMD1 and TRIP6, in which TRIP6 is localized to AJ independently of LIMD1, whereas LIMD1 localization to AJ requires TRIP6. Moreover, they suggest that TRIP6 acts downstream or in parallel to the influence of Rho activation on LIMD1 and LATS1 localization.
TRIP6 is required for cytoskeletal tension at AJ
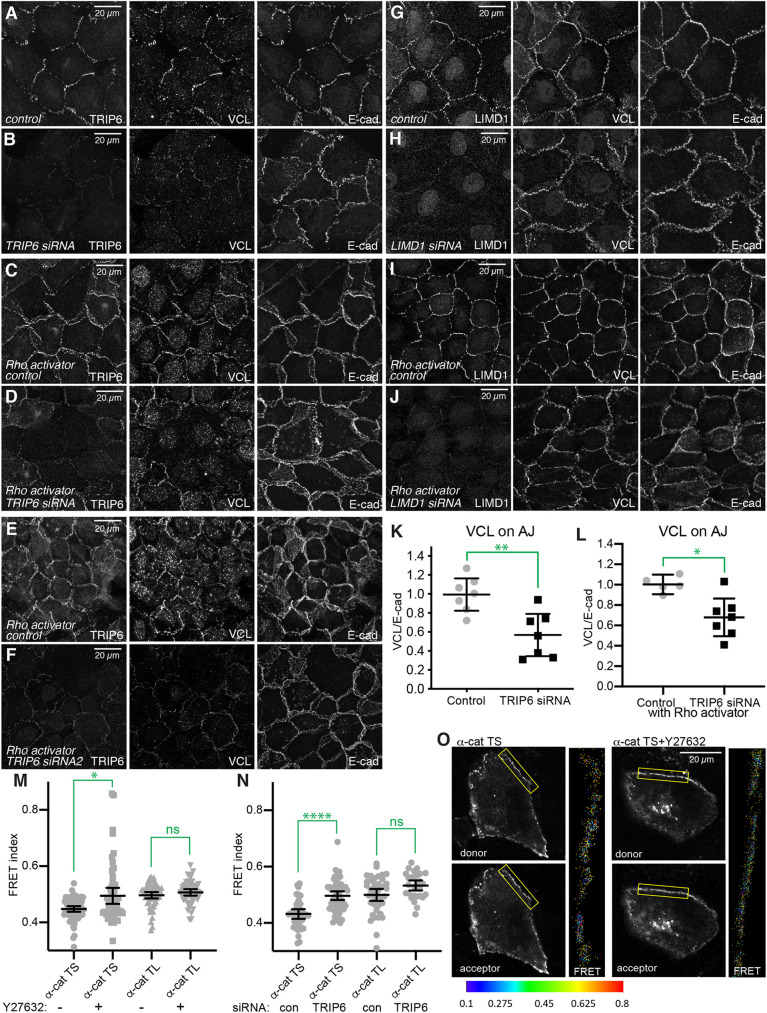
The requirement for TRIP6 to localize LIMD1 could in part reflect a physical association between them that recruits LIMD1. Indeed, LIMD1 and TRIP6 can be co-immunoprecipitated from cultured cells (Fig. S3A,B). However, as LIMD1 localization to AJ requires cytoskeletal tension, the requirement for TRIP6 might instead reflect an influence of TRIP6 on tension at AJ. To investigate this latter possibility, we examined VCL, as the localization of VCL to AJ depends upon a tension-induced conformational change in α-catenin (Kim et al., 2015; Yao et al., 2014; Yonemura et al., 2010). We observed that siRNA of TRIP6 diminished localization of VCL to AJ, whereas siRNA of LIMD1 had no noticeable effect, and the loss of VCL from junctions could be rescued by transfection with TRIP6:GFP (Fig. 3A-L; Fig. S3). VCL levels were not significantly decreased by TRIP6 siRNA, although a modest decrease in VCL levels was detected by western blotting in LIMD1 siRNA-treated cells (Fig. S6G). The requirement for TRIP6 in VCL localization to AJ was observed both under low cell density conditions and in cells at high density treated with Rho activator II (Fig. 3A-L). The finding that TRIP6 is required for junctional localization of VCL as well as of LIMD1 suggests that TRIP6 is required to maintain tension at AJ. We note that our observations differ from Dutta et al. (2018), who reported that recruitment of VCL to junctions was not affected by knockdown of TRIP6, possibly due to differences in experimental conditions.
Fig. 3.
TRIP6 is required for tension at AJ. (A-J) MCF10A cells plated at low (A,B,G,H) or high (C-F,I,J) density, and transfected with control (A,C,E,G,I), TRIP6 (B,D,F) or LIMD1 (H,J) siRNA 24 h after seeding. Cells grown at high density were then treated with 1 µg/ml Rho-activator-II for 3 h. Cells were fixed and stained for TRIP6 or LIMD1, VCL anti-rabbit (A-F) or VCL anti-mouse (G-J) and E-cad. Images are z-projections of confocal stacks and are representatives of at least three biological replicates. (K,L) Quantification of junctional levels of VCL normalized to junctional E-cad levels under control and TRIP6 knockdown conditions at low density (K) or high density with Rho-activator-II (L), respectively. Each dot represents results from a confocal image stack containing several cells. N=7 for control, 7 for TRIP6 siRNA, 5 for control with Rho activator and 7 for TRIP6 siRNA with Rho activator. (M,N) FRET index measurements for α-cat TS (tension-sensitive) or α-cat TL (tension-less) constructs with or without ROCK inhibitor (Y27632) treatment (M) or with control or TRIP6 siRNA (N), respectively. Each data point represents the average FRET index for all the pixels within the ROIs marked in an individual confocal image with one or two fluorescently labeled cells. N=64 (α-cat TS control), 55 (α-cat TS +Y27632), 57 (α-cat TL control), 51 (α-cat TL +Y27632), 37 (α-cat TS siRNA control), 55 (α-cat TS+TRIP6 siRNA), 40 (α-cat TL siRNA control) and 28 (α-cat TL+TRIP6 siRNA). Data are from three independent experiments. (O) Representative donor (mTFP) and acceptor (mVenus) channel images for the α-cat TS construct with and without ROCK inhibitor treatment. To the right of the cells an enlarged FRET index rainbow look-up table is displayed for the ROI identified by the yellow box; the scale for the FRET index is at the bottom. Statistical significance was determined using an unpaired two-tailed t-test (K,L) or a Kruskal–Wallis test (M,N) *P<0.05; **P<0.01; ****P<0.0001; ns, not significant. Error bars indicate mean±95% c.i.
To further investigate the possibility that TRIP6 is required to maintain tension at AJ, we employed a fluorescence resonance energy transfer (FRET)-based α-catenin tension sensor system (Acharya et al., 2017). This comprises constructs with the fluorescent proteins mTFP1 and Venus, separated by a flexible linker, inserted into α-catenin. In the tension-sensitive version of this construct (α-cat TS), the FRET pair is inserted N terminal to the F-actin binding domain, such that tension in the attached actin cytoskeleton can increase the separation between mTFP1 and Venus, thereby decreasing FRET. A tensionless version of the construct (α-cat TL) has the FRET pair inserted C terminal to the F-actin binding domain, such that it is unaffected by cytoskeletal tension. To assay the effectiveness of these constructs under our experimental conditions, we first examined the influence of a ROCK inhibitor, Y27632, which decreases cytoskeletal tension. In MCF10A cells transfected with α-catenin FRET constructs and treated with siRNAs to deplete endogenous α-catenin, Y27632 treatment increased the mean FRET index in α-cat TS-expressing cells (from 0.45 to 0.50), but did not significantly affect the FRET index in α-cat TL-expressing cells (0.50 versus 0.51) (Fig. 3M,O). We then performed FRET analysis on cells transfected with control or TRIP6 siRNAs. In α-cat TS-expressing cells this increased the mean FRET index (from 0.43 to 0.50), but in α-cat TL-expressing cells the mean FRET index was not significantly affected (0.50 versus 0.53) (Fig. 3N). These observations further support the inference that TRIP6 is required for the cytoskeletal tension experienced by α-catenin at AJ.
TRIP6 influences the organization of actin and myosin
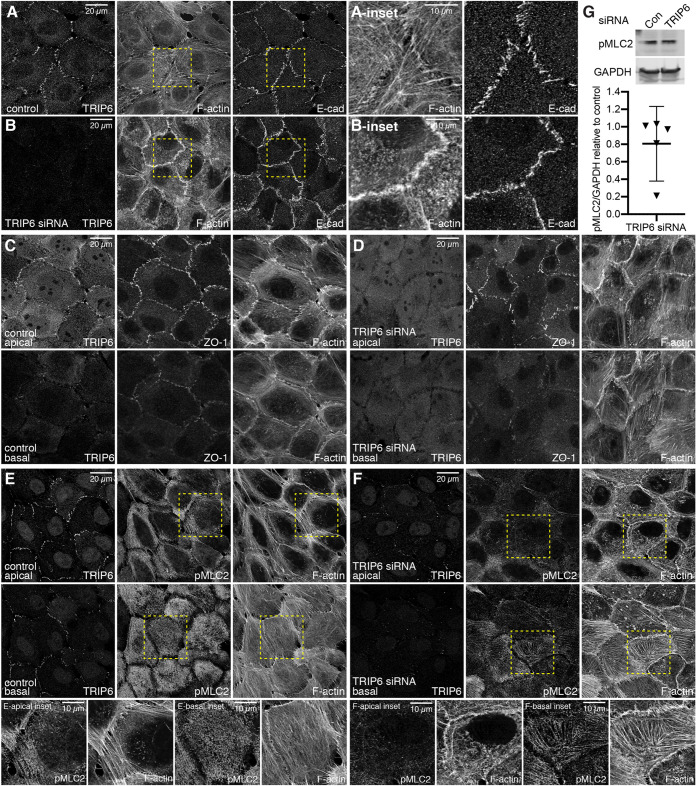
To investigate whether the requirement for TRIP6 for tension at AJ reflects a more general influence on cytoskeletal organization, we examined the distribution of F-actin by staining with fluorescently labelled phalloidin, and the distribution of myosin using antibodies against myosin IIb (MyoIIb) and phosphorylated myosin light chain (pMLC2). Overall, we observed an apparent reduction in pMLC2 and MyoIIb staining when TRIP6 was knocked down, and a reorganization of F-actin (Fig. 4; Fig. S4). In contrast, knockdown of LIMD1 was not associated with any visible changes in staining for pMLC2, MyoIIb or F-actin (Fig. S4).
Fig. 4.
TRIP6 influences the organization of actin and myosin. (A-F) MCF10A cells plated at low density and transfected with either control or TRIP6 siRNA, as indicated, and cultured for 72 h and then fixed and stained for TRIP6 (A,B,E,F, mouse Ab; C,D, rabbit Ab), F-actin and either E-cad, ZO-1 or pMLC2. Images are projections through three to seven z-sections: apical sections are upper panels in A and B and C-F, and basal sections are lower panels in C-F, and are representatives of at least three biological replicates. Insets show higher magnification of the boxed regions. (G) Western blot showing pMLC2 and loading control (GAPDH) levels in control and TRIP6 siRNA cells along with quantification of the ratio of pMLC2/GAPDH protein levels in TRIP6 siRNA-treated cells relative to that in control cells (N=5). Data from individual blots plotted using triangles. Error bars indicate mean±95% c.i.
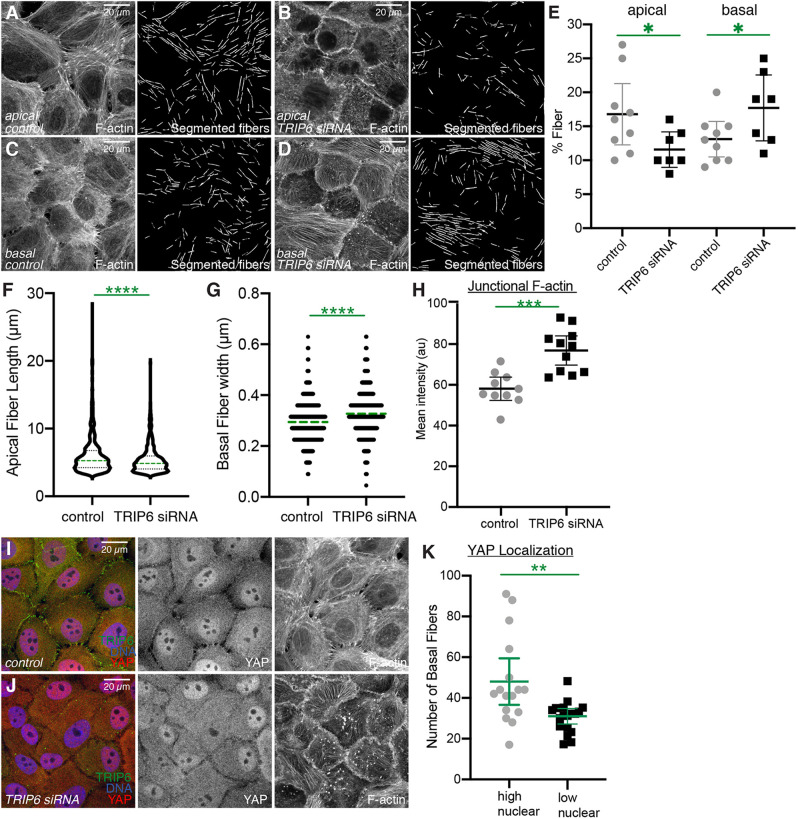
In more apical regions [e.g. overlapping junctional staining for the tight junction protein ZO-1 (also known as TJP1)], MCF10A cells cultured at low density typically had multiple long F-actin fibers extending away from the AJ (Fig. 4A,C,E). In contrast, in TRIP6 knockdown MCF10A cells fewer of these long apical F-actin fibers were observed, and instead F-actin was often concentrated in thick irregular accumulations along the AJ, and diffuse speckles of F-actin were observed within the cytoplasm (Fig. 4B,D,F, Fig. 5H). This was accompanied by an apparent reduction in apical pMLC2 and MyoIIb staining (Fig. 4E,F; Fig. S4). Consistent with previous studies on the relationship between tension at apical junctions and junction morphology (Huveneers and de Rooij, 2013; Taguchi et al., 2011), knockdown of TRIP6 was also associated with a shift in junction morphology, from predominantly punctate to more continuous, but wavy, AJ (Fig. 4A,B).
Fig. 5.
Quantification of actin fibers in TRIP6 knockdown cells. (A-D) Examples of F-actin segmentation using FSegment on MCF10A cells plated at low density and transfected with either control or TRIP6 siRNA. F-actin staining from apical (A,B) or basal (C,D) regions, together with segmented actin fibers, are shown. (E) Comparison of the percentage of the image occupied by F-actin fibers in image panels similar to those shown in A-D. Each dot represents one confocal image, comprising several cells as in the examples. (F) Comparison of the length distribution of apical actin fibers in control versus TRIP6 RNAi images, displayed in a violin plot with the median indicated by a dashed green line. N=816 fibers in control and 496 fibers in TRIP6 RNAi. (G) Comparison of the width distribution of basal actin fibers in control versus TRIP6 RNAi images, displayed in a dot plot with the median indicated by a dashed green line. Steps between points reflect 1 pixel differences in width. N=788 fibers in control and 709 fibers in TRIP6 RNAi. (H) Quantification of mean junctional F-actin intensity [in arbitrary units (au)] in control or TRIP6 siRNA-treated cells. Each point represents the measurement from an individual confocal image. N=10 for control and 11 for TRIP6 siRNA. (I,J) Examples of YAP (red/gray) localization and F-actin (gray) staining in MCF10A cells plated at low density and transfected with either control or TRIP6 siRNA. (K) Comparison of the number of basal actin fibers in TRIP6 siRNA-treated cells and YAP localization, which is categorized into high nuclear (N/C>=1.60) or low nuclear (N/C<1.60) based on the average ratio of nuclear/cytoplasmic (N/C) YAP intensity. Each dot represents one cell. N=16 for high nuclear and 18 for low nuclear. Results of statistical comparisons performed by unpaired two-tailed t-test (E-H) and by Mann–Whitney test (K) are indicated in green. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Error bars indicate mean±95% c.i.
The observation that there was reduced junctional tension and an altered actin cytoskeleton in TRIP6 knockdown cells but not in LIMD1 knockdown cells suggests that they are not related to reductions in YAP activity. To confirm this, we also examined YAP knockdown cells. Indeed, no differences in VCL or F-actin localization were detected after siRNA-mediated knockdown of YAP (Fig. S5).
In more basal regions (e.g. basal to junctional staining for ZO-1), MCF10A cells often appeared to have a meshwork of thin F-actin filaments (Fig. 4C,E). In TRIP6 knockdown MCF10A cells, basal F-actin instead concentrated in thick F-actin fibers that run along the basal surface of the cell (Fig. 4D,F). This reorganization of F-actin was accompanied by altered pMLC2 and MyoIIb staining, from diffuse basal puncta in wild-type MCF10A cells (Fig. 4E; Fig. S4), to their concentration along basal F-actin fibers in TRIP6 knockdown cells (Fig. 4F; Fig. S4). Despite the overall appearance of reduced myosin immunostaining, no significant reduction in myosin levels was detected by western blotting (Fig. 4G; Fig. S4K), implying that TRIP6 siRNA leads to a change in pMLC2 and MyoIIb localization, rather than a decrease in total pMLC2 or MyoIIb levels within the cell.
To quantify the differences in F-actin fibers, we used a published MATLAB script for segmentation and analysis of actin fibers (Rogge et al., 2017). This confirmed the visual impression of a reduction in apical actin fibers, and an increase in basal actin fibers (Fig. 5A-E). It also revealed a relative decrease in the length of apical actin fibers, and an increase in the width of basal actin fibers (Fig. 5F,G). These observations indicate that knockdown of TRIP6 is not simply associated with a reduction in cytoskeletal tension. Rather, there is a reorganization of cytoskeletal tension that includes an apparent reduction in apical actin stress fibers attached to AJ, and an increase in basal actin stress fibers.
Knockdown of TRIP6 was previously reported to be associated with reduced YAP activity (Dutta et al., 2018). As many studies have linked F-actin levels and tension in the actin cytoskeleton to YAP activation (Misra and Irvine, 2018; Sun and Irvine, 2016), we wondered whether TRIP6 knockdown could reduce YAP activity even under conditions in which prominent basal actin fibers were observed. Under our low cell density culture conditions, YAP was predominantly nuclear in MCF10A cells (Fig. 5I). The overall YAP distribution was not consistently affected by TRIP6 knockdown in our experiments (Fig. 5I,J). However, there was cell-to-cell variation both in the extent of formation of basal F-actin stress fibers in TRIP6 knockdown cells, and in the YAP localization profile. When we quantified the number of basal F-actin fibers in individual TRIP6 knockdown cells, and compared this to the YAP localization profile, we observed a correlation between higher nuclear YAP localization and higher numbers of basal F-actin fibers as compared to cells with reduced nuclear YAP (Fig. 5K). These observations suggest that the expected decrease in YAP activity generated by loss of TRIP6 from apical junctions might be compensated by an increase in YAP activity generated by elevated basal actin stress fibers.
Re-localization of VCL and VASP in the absence of TRIP6
To explain the shift in F-actin stress fibers from apical to basal in TRIP6 knockdown cells, we hypothesized that there could be a competition between apical AJ and basal FA for protein(s) that are required for the maintenance or attachment of F-actin stress fibers, and present in limiting amounts within cells. According to this model, TRIP6 could recruit these proteins to AJ, but in the absence of TRIP6 they would instead be recruited to FA (Fig. 6G). To begin to investigate this possibility, we examined the distribution of proteins that can localize to AJ and FA, and can influence or interact with F-actin. One such protein is VCL. Confluent MCF10A cells lack prominent FA, and basal accumulation of VCL is difficult to detect (Fig. 6A; Fig. S6C). However, in TRIP6 knockdown cells, basal puncta of VCL are readily apparent, particularly at the ends of F-actin stress fibers (Fig. 6B; Fig. S6D). These observations imply that knockdown of TRIP6 promotes the formation of VCL-containing FA.
Fig. 6.
TRIP6 knockdown shifts localization of VCL and VASP to FAs. (A-F) MCF10A cells plated at low density and transfected with either control or TRIP6 siRNA, and cultured for 72 h and then fixed and stained for TRIP6 (rabbit Ab A,B or mouse Ab C-F), E-cad or F-actin and VCL (mouse Ab) or VASP. Images are projections through either whole cells (C-F) or three to seven apical (upper panel) and basal (lower panel) sections (A,B), and are representatives of at least three biological replicates. Insets show higher magnification of the boxed regions. (G) Model illustrating influence of TRIP6 on actin stress fibers. In the presence of TRIP6 (left), TRIP6 recruits a crucial actin regulatory protein (X), which stabilizes attachments of tensile F-actin. In the absence of TRIP6, X is instead recruited to FA, where it stabilizes basal F-actin stress fibers. (H) Quantification of junctional levels of VASP normalized to junctional E-cad levels under control and TRIP6 knockdown conditions. Each dot represents results from a confocal image stack containing several cells. N=8 for control and 11 for TRIP6 siRNA. Statistical significance was determined using an unpaired two-tailed t-test. **P<0.01. ecm, extracellular matrix. Error bars indicate mean±95% c.i.
VASP is an actin regulatory protein that can be observed at AJ in epithelial cells, but at FA and leading edge in fibroblasts (Krause et al., 2003). Prominent VASP localization to AJ was observed in wild-type MCF10A cells (Fig. 6C,E; Fig. S6E). When TRIP6 was knocked down, VASP was lost from AJ, and was instead detected in basal puncta (Fig. 6D,F,H; Fig. S6F). Western blotting did not reveal a significant change in the total levels of VCL or VASP in TRIP6 knockdown cells (Fig. S6G,H). The basal puncta of VASP were most prominent at the ends of F-actin stress fibers, presumably representing accumulation at FA, but some VASP accumulation could also be observed along basal stress fibers (Fig. 6F). Thus, both VCL and VASP re-localize from AJ to FA in TRIP6 knockdown cells, coincident with the reorganization of F-actin and myosin. The re-localization of VCL and VASP between AJ and FA contrasted with that of the FA protein paxillin (PXN), which was detected in basal puncta both in control cells and in TRIP6 knockdown cells (Fig. S6A,B). However, there was a noticeable accumulation of PXN along the ends of actin stress fibers in TRIP6 knockdown cells, consistent with the formation of strong FAs in these cells.
VCL is required for tension at AJ and FAs
Although our observations show that TRIP6 is required for normal localization of VCL to AJ, published observations have reported that TRIP6 localization to AJ is dependent upon VCL (Dutta et al., 2018). To further investigate the relationship between VCL and TRIP6, and their influence on the cytoskeleton and Hippo signaling, we compared the consequences of VCL knockdown on tension-regulated proteins at AJ and the actomyosin cytoskeleton in MCF10A cells.
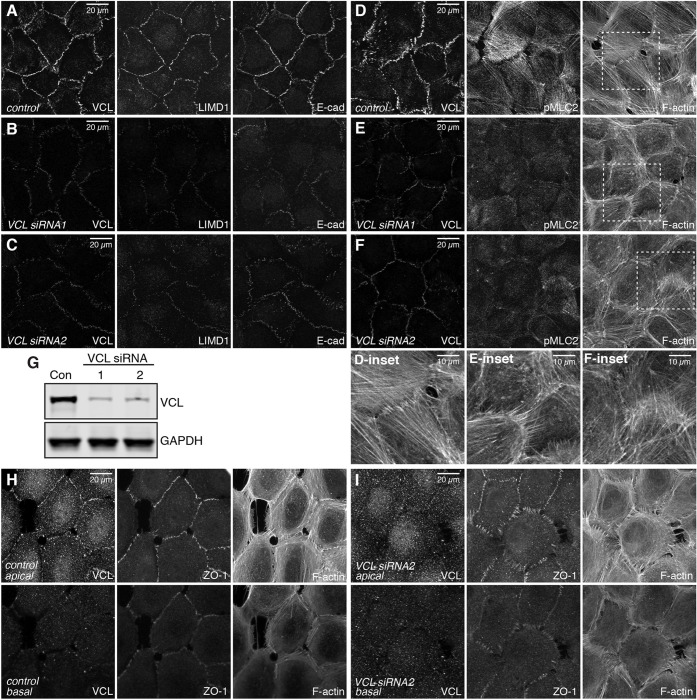
VCL siRNA reduced junctional localization of LIMD1 and resulted in a substantial reduction in pMLC2 staining, similar to TRIP6 knockdown (Fig. 7A-F). However, in other respects, the consequences of VCL knockdown differed. Strong VCL knockdown consistently reduced junctional localization of E-cad (Fig. 7A-C), whereas TRIP6 knockdown had much less effect on E-cad. VCL knockdown reduced apical F-actin stress fibers attached to AJ similar to TRIP6 knockdown (Fig. 7D-I), but did not lead to the appearance of basal F-actin stress fibers (Fig. 7H,I). Moreover, instead of thick F-actin accumulations along AJ, in apical regions VCL knockdown often resulted in the appearance of F-actin spikes (Fig. 7E,F,I). These phenotypic differences emphasize that TRIP6 and VCL have distinct roles at AJ.
Fig. 7.
VCL is required to maintain tension at AJ. (A-F) MCF10A cells plated at low-density and transfected twice with either control or VCL siRNA, and cultured for 48 h then fixed in the presence of 0.65% Triton X-100 and stained with mouse VCL, E-cad or F-actin, LIMD1 or rabbit pMLC2. Images are z-projections of confocal stacks and are representatives of at least three biological replicates. (G) Western blot showing the knockdown efficiency of the VCL siRNAs used. Con, control (negative control siRNA). GAPDH is the loading control for the blot. (H,I) MCF10A cells plated at low density, transfected with either control or VCL siRNA and stained for rabbit VCL, mouse ZO-1 and F-actin. ZO-1 staining was used to separate the images into apical (top panel) and basal sections (bottom panel). Insets show higher magnification of the boxed regions.
DISCUSSION
Previous studies ascribed similar effects on Hippo signaling to two LIM domain proteins, LIMD1 and TRIP6 (Dutta et al., 2018; Ibar et al., 2018). Our studies have now established distinct activities for these proteins, as we found that TRIP6 is required for the localization of LIMD1 to AJ, whereas LIMD1 does not affect the localization of TRIP6. The influence of TRIP6 on LIMD1 could in principle account for its reported effects on Hippo signaling, though we note our results do not exclude the possibility that TRIP6 also influences Hippo signaling through other mechanisms (Dutta et al., 2018). Our results have further identified requirements for TRIP6 in the junctional localization of VCL and VASP, and in the organization of F-actin.
Both LIMD1 and VCL are recruited to an open form of α-catenin that is generated under tension (Alégot et al., 2019; Ibar et al., 2018; Kim et al., 2015; Yao et al., 2014; Yonemura et al., 2010). The requirement for TRIP6 in the localization of LIMD1 and VCL to AJ thus implies that it is required to maintain this open form. Consistent with this, we observed that knockdown of TRIP6 results in the loss of pMLC2-associated F-actin stress fibers attached to AJ, and increases the FRET index measured for an α-catenin tension-sensor construct. These observations thus suggest that TRIP6 is required to establish or maintain attachments of tensile actin stress fibers to AJ.
Loss of TRIP6 is not only associated with a loss of apical actin stress fibers attached to AJ, but also with a gain of basal actin stress fibers attached to FA. Intriguingly, loss of E-cad in MCF10A cells has also been reported to result in the formation of thick basal actin stress fibers (Chen et al., 2014). Although it was reported that apical F-actin did not change in E-cad mutant MCF10A cells, in those experiments wild-type MCF10A cells did not exhibit the tensile apical F-actin that we observe, possibly due to higher cell density or other aspects of culture conditions. To explain the reorganization of F-actin that occurs when TRIP6 is knocked down, we suggest that there could be a competition between AJ and FA for a limiting pool of proteins needed to stabilize attachments to actin stress fibers, and that TRIP6 is needed to recruit these proteins to AJ. When TRIP6 is eliminated, these proteins are then instead able to associate with FA, at which point they promote and stabilize basal stress fibers. We identified two proteins, VCL and VASP, that are candidates to be affected by this hypothesized competition. Their localization shifts from predominantly at AJ in wild-type MCF10A cells, to predominantly at FA in TRIP6 knockdown MCF10A cells.
VCL is recruited through tension-sensitive mechanisms to AJ and FA, where it binds to F-actin, helps to stabilize association of F-actin to AJ and FA, and plays crucial roles in maintaining cell-cell and cell-matrix adhesions (Bays and DeMali, 2017). Although both VCL and TRIP6 knockdown resulted in loss of apical F-actin stress fibers, in other respects their influence on actin organization differed. VCL knockdown led to loss rather than gain of basal stress fibers. Apically, VCL knockdown led to F-actin spikes forming along cell-cell junctions. As there was a noticeable reduction of E-cad at junctions in VCL knockdown cells, the formation of F-actin spikes might be related to a recently described process in which apical actin protrusions are suggested to play a role in the repair of failing adhesive junctions (Li et al., 2020).
VASP regulates actin polymerization, and VCL and VASP have been reported to be able to interact with each other, and to function together in promoting tension-sensitive actin polymerization at AJ (Leerberg et al., 2014). VASP has also been found to be able to associate with TRIP6 and other related LIM domain proteins, including Zyxin and LPP, supporting the possibility of a direct role for TRIP6 in recruiting VASP (Hoffman et al., 2006; Petit et al., 2003; Reinhard et al., 1995). Alternatively, the influence of TRIP6 on VCL and VASP recruitment could be indirect. Indeed, as these proteins are recruited to sites of tension at AJ and FA, and they help to maintain tension at these same sites, they participate in a positive feedback loop that maintains attachments to actin stress fibers and thus their own recruitment to these sites.
Although our results indicate that TRIP6 plays an essential role in attachments of stress fibers to AJ, it was not required at FA. Studies in other cell types have yielded conflicting results on the effects of TRIP6 on FA (Lin and Lin, 2011; Willier et al., 2011). For example, in fibroblasts, overexpressed TRIP6 was reported to bind to supervillin to suppress FA maturation (Takizawa et al., 2006), and in A509 cells TRIP6 knockdown increased basal stress fibers (Guryanova et al., 2005). Conversely, in HeLa cells, TRIP6 was reported to facilitate the maturation of FAs (Bai et al., 2007), and knockdown of TRIP6 in human endothelial cells was reported to decrease the formation of actin fibers (Sanz-Rodriguez et al., 2004). In addition to TRIP6, the related Zyxin family proteins LPP and Zyxin have also been implicated in actin stress fiber organization (Smith et al., 2014), and they might contribute to the stabilization of basal stress fibers in the absence of TRIP6.
Knockdown of TRIP6 was previously reported to lead to reduced nuclear YAP localization and reduced YAP activity (Dutta et al., 2018). We did not observe an overall reduction in nuclear YAP in our experiments, but we note that Dutta et al. (2018) did not report the increase in basal actin stress fibers that we observed. Thus, the observed distinct effects on YAP might reflect differences between experimental conditions that result in different consequences for basal F-actin. Several mechanisms by which basal actin stress fibers could contribute to nuclear YAP localization have been described (Misra and Irvine, 2018), including effects on integrin signaling and on nuclear pore size (Elosegui-Artola et al., 2017; Kim and Gumbiner, 2015). The correlation between basal F-actin and nuclear YAP in TRIP6 knockdown cells suggests that loss of TRIP6 switches the predominant mechanism for cytoskeletal regulation of Hippo signaling in MCF10A cells from dependence on LATS regulation at AJ to mechanisms that depend upon FA and basal F-actin stress fibers.
MATERIALS AND METHODS
Plasmids and siRNAs
Published plasmids used include α-catenin-TL and α-catenin-TS (Acharya et al., 2017), pcDNA3.1-mTFP and pcDNA3.1-mVenus (Gates et al., 2019), mPlum-Lifeact-7 (Addgene, #54679), pEGFP-LIMD1 and pcDNA-GFP-V5 (Reddy and Irvine, 2013), and E-cad:GFP (Addgene, #67937). pEGFP-C1-hTRIP6 was generated by cloning a 1.4 kb human full-length TRIP6 cDNA into EcoRI/SalI sites of pEGFP-C1 (E. Enners, Rutgers University).
Sequences of the siRNAs (5′-> 3′) used were: human TRIP6 siRNA1, 5′-CCAAUGUUCCACUUUUGGUAUUGAT-3′ [Integrated DNA Technologies (IDT)]; human TRIP6 siRNA2, 5′-GCUCUGGAUCGAAGUUUUCACAUTG-3′ (IDT); human VCL siRNA1, 5′-CCAGUGGAUCGAUAAUGUUGAAAAA-3′ (IDT); human VCL siRNA2, 5′-GGCAAAUCAGUUACUAAGAAGAAAA-3′ (IDT); human α-catenin siRNA, 5′-CGUGAACAUGCCAACAAAUUGAUTG-3′ (IDT); and human YAP siRNA, 5′-GGUGAUACUAUCAACCAAAdTdT [Dharmacon, using sequence described in Wang et al. (2018)]. siRNAs used for LIMD1 have been described previously (Ibar et al., 2018). TRIP6 siRNA1 targets the 3′ UTR and was used for siRNA rescue experiments, as the 3′UTR is not present in the pEGFP-hTRIP6 plasmid.
Cell culture
MCF10A cells (a gift from Jay Debnath, University of California San Francisco, CA, USA) were cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 supplemented with 5% horse serum, epidermal growth factor (20 µg/ml), insulin (10 µg/ml), cholera toxin (0.1 µg/ml), hydrocortisone (0.5 µg/ml), and antibiotic-antimycotic, at 37°C at 5% CO2. Cells were used at low passage number and checked for contamination by cell morphology and mycoplasma testing. Coverslips were coated with 0.6 mg/ml of collagen for 15 min at room temperature and washed with PBS before plating cells. For low cell density experiments, cells were grown at densities of ∼35,000 cells/cm2. For Rho-activator experiments, cells were grown at high density (150,000 cells/cm2) for 48 h and were then treated with Rho-activator-II (Cytoskeleton: 1 µg/ml) for 3 h. For siRNA experiments, Lipofectamine RNAiMAX (Life Technologies) was used to deliver siRNA into MCF10A cells following the manufacturer's protocol, and cells were fixed and stained 72 h after transfection (48 h for LIMD1 siRNA transfection).
Immunostaining and imaging
Cells were either fixed with 4% paraformaldehyde (PFA) in PBS++ (PBS supplemented with 100 mM MgCl2 and 50 mM CaCl2) for 10 min at room temperature, or, when staining for VCL, LIMD1, or VASP, fixed in 1% PFA with 0.65% Triton X-100 in PBS++ for 3 min, rinsed and then fixed in 1% PFA in PBS++ for 10 min. The cells were then washed three times for 10 mins each with 200 mM glycine containing PBS, followed by permeabilization with 0.5% Triton X-100 in PBS for 20 min. After blocking with 5% bovine serum albumin (BSA) in PBS for 1 h, cells were incubated with primary antibody diluted in a 5% BSA in PBS solution overnight at 4°C (except pMLC2 staining was generally performed for two days for optimal signal). After washing with PBS, cells were incubated with Alexa Fluor 488- (Life Technologies), Cy3- or Alexa Fluor 647-conjugated (Jackson ImmunoResearch) secondary antibodies for 1 h and washed four times with PBS. Cell nuclei were counterstained with Hoechst 33342 (1 µg/ml; Invitrogen) and mounted with mounting medium (Dako). Antibodies used for immunostaining include mouse anti-Yap (1:100; Santa Cruz Biotechnology, sc-101199), rabbit anti-LATS1 (1:600; Cell Signaling Technology, 3477), mouse anti-VCL (1:2000; Sigma-Aldrich, V9131), rabbit anti-VCL (1:100; EMD Millipore, MAB3574), rabbit anti-VASP (1:100; Cell Signaling Technology, 3132S), mouse anti-ZO-1 (1:1000; Life Technology, 33-9100), mouse anti-pMLC2 (S19) (1:200; Cell Signaling Technology, 3675), rabbit anti-pMLC2 (T18/S19) (1:50; Cell Signaling Technology, 3674), rat anti-E-cadherin (1:500; Life Technology, 13-1900), rabbit anti-LIMD1 (1:500; Bethyl, A303-182A), rabbit anti-PXN (1:250; Abcam, ab32084), rabbit anti-Myosin IIB (1:200; Cell Signaling Technology, 3404S), rabbit anti-TRIP6 (1:50; Abcam, ab70747) and mouse anti-TRIP6 (1:100; Santa Cruz Biotechnology, sc-365122). F-actin was stained with Alexa Fluor 647-conjugated phalloidin (1:50; Life Technologies). For E-cad, VASP, ZO-1, pMCL2, MyoIIb and PXN antibodies we performed separate single channel stains to rule out crossover in signals (Fig. S7); other antibodies used were validated by siRNA as described in the text. Images were acquired using LAS X software on a Leica TCS SP8 confocal microscope system using an HC PL APO 63×/1.40 objective.
FRET analysis
Cells were cultured at low density (30,000/cm2) in Nunc Lab-Tek II Chambered Coverglass and imaged live at 37°C on a Leica TCS SP8 confocal microscope. α-catenin knockdown MCF10A cells were transfected with α-catenin-TL or TS constructs and live imaging was performed 24 h after transfection. For ROCK inhibitor treatment, cells were changed to serum-free DMEM/F12(1:1) and treated with either 10 µM Y-27632 (Cytoskeleton, CN06) or an equal volume of water as control for 3 h before imaging. For TRIP6 knockdown experiments, cells were co-transfected with α-catenin and TRIP6 siRNAs and then co-transfected with mPlum-Lifeact and either α-catenin-TS or TL constructs 24 h before imaging. Cells were illuminated using laser lines at 458 nm (donor and FRET channels) or 518 nm (acceptor channel), and images collected at 462-500 nm (donor channel) or 521-565 nm (acceptor and FRET channels). In TRIP6 knockdown experiments, mPlum-Lifeact was used to confirm TRIP6 knockdown. For analysis, acceptor and FRET channel images were registered with respect to the donor channel using the RiFRET plug-in of ImageJ (Roszik et al., 2009). FRET analysis was performed using Volocity software (Perkin-Elmer), with donor and acceptor bleed-through constants calculated using pcDNA3.1-mTFP- or pcDNA3.1-mVenus-expressing cells as described by Menaesse et al. (2020). A region of interest (ROI) in the background of the image field was used for background subtraction. The FRET index (Xia and Lu, 2001) was calculated using FRET index (nF/donor)=(FRET−(Acceptor×A)−(Donor×B))/Donor where A and B refer to the acceptor and donor bleed-through constants, respectively. Measurements were made in ROIs along junctions of the cells, ignoring zero intensity values. Each individual FRET index data point plotted represents the average FRET index value for all the pixels within the ROIs marked in multiple cells and junctions within an image field.
Immunostaining quantitation
For quantification of junctional protein levels, confocal stacks were segmented and measured using Volocity (Perkin Elmer) software, with E-cad signal to define apical junctions. The mean intensities of protein levels overlapping junctions were then calculated and normalized to the mean intensity of E-cad. Line scans were performed using the Plot Profile function of ImageJ.
For analysis of junctional F-actin changes between control and TRIP6 knockdown experiments, the E-cad channel was used to create a mask of AJ using ImageJ and this was used to identify the F-actin signal at the junction. Mean threshold was applied to set the background values to ‘NaN’ and then the average intensity of F-actin in the image field was determined.
Actin segmentation and quantitation
Actin stress fibers were quantified using the FSegment MATLAB script developed by Rogge et al. (2017). This script uses a trace algorithm to trace linear structures of actin and suppresses non-linear fluorescence. Segmented linear structures are then analyzed with respect to fluorescence values for the actin fibers. The application allows calculation of fiber width, fiber length, total number of actin filaments, proportion of actin in the form of stress fibers compared to total actin (% fiber actin), and total filament length. For YAP localization versus basal F-actin correlation studies in the TRIP6 siRNA-treated cells, individual cells were cropped using the ‘FreeHandCrop’ feature in the Preprocessing GUI and the actin stress fibers analysis for each cell was then compared to the YAP localization within the cell. For YAP localization measurements, the average YAP intensity was quantified in the nucleus and in the cytoplasm using Volocity, and the ratio of nuclear to cytoplasmic YAP intensity (N/C) was determined for all the cells. An N/C ratio of 1.6 was set as the cutoff as this correlated well with the visual impression of YAP localization as strong nuclear (N/C ratio above 1.6) or weak to non-nuclear (N/C ratio below 1.6). For comparison of actin between control and TRIP6 siRNA-treated cells in the apical and basal sections, the entire image field was segmented and analyzed. Individual z-sections representative of the apical and basal sections of the whole image field were selected for these analyses.
Co-immunoprecipitation
HEK293T cells were cultured in 10 cm plates for 24 h and transfected with either pcDNA-GFP-V5, pEGFP-TRIP6 or pEGFP-LIMD1 using Lipofectamine 3000. After 48 h, cells were harvested and lysed with lysis buffer [10% glycerol, 20 mM Tris-HCl (pH 7.0), 137 mM NaCl, 2 mM EDTA and 1% NP-40] supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma-Aldrich, P7426) and protease inhibitor cocktail (Roche). The lysate was then incubated with GFP-Trap_MA beads (Antibodies-Online) for 1 h at 4°C. Detergent concentration of the lysate was adjusted to 0.2% during incubation with the beads to avoid non-specific binding. The beads were then washed and resuspended in 2× Laemmli sample buffer, supplemented with PMSF and protease inhibitor cocktail. The co-immunoprecipitation supernatant and input samples were then analyzed by immunoblotting.
Immunoblotting
MCF10A cells were lysed in 2× Laemmli sample buffer (Bio-Rad) supplemented with protease inhibitor cocktail (Roche) and 1 mM PMSF (Sigma-Aldrich, P7426). Protein samples were loaded to 4%-15% gradient gels (Bio-Rad). Antibodies for immunoblotting included rabbit anti-vinculin (1:5000; EMD Millipore, MAB3574), mouse anti-phospho-myosin light chain (S19) (1:500; Cell Signaling Technology, 3675), mouse anti-TRIP6 (1:1000; Santa Cruz Biotechnology, sc-365122), rabbit anti-VASP (1:1000; Cell Signaling Technology, 3132S), rabbit anti-LIMD1 (1:1000; Bethyl, A303-182A), rabbit anti-LATS1 (1:1000; Cell Signaling Technology, 3477), rabbit anti-YAP (1:1000; Cell Signaling Technology, 14074S), rabbit anti-GFP (1:1000; Cell Signaling Technology, 2555S), mouse anti-GFP (1:1000; Cell Signaling Technology, 2955S), mouse anti-ECAD (1:2000; BD Biosciences, 610181) and rabbit-myosin light chain (1:1000; Cell Signaling Technology, 3672S). As loading control, mouse anti-α-tubulin (1:10,000; Sigma-Aldrich, T6199), rabbit anti-GAPDH (1:10,000; Santa Cruz Biotechnology, sc-25778) or mouse anti-GAPDH (1:10,000; Novus Biologicals, NBP2-27103) were used. Blots were stained with Li-COR fluorescent-conjugated secondary antibodies at 1:20,000 dilution, and were visualized and quantified using an Odyssey Imaging System (Li-COR Biosciences).
Statistical analysis
Statistical comparisons were performed using Excel and GraphPad Prism software. For ratio comparisons, analysis was conducted on the log transform of the ratio. For data with a normal distribution, we used unpaired two-tailed Student's t-tests (pairwise comparisons) or one-way ANOVA (multiple sample comparisons). In cases in which Shapiro–Wilk and Kolmogorov–Smirnov tests did not indicate a normal distribution (Fig. 3M, Fig. 5K) we used non-parametric tests (Mann–Whitney for pairwise comparison, and Kruskal–Wallis for multiple sample comparisons).
Supplementary Material
Acknowledgements
We thank Jay Debnath for MCF10A cells, Alpha Yap, Michael Davidson, Edward Enners, Nada Boustany and Brent Hoffman for plasmids, and Dan McCollum and Nada Boustany for helpful discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.V., K.D.I.; Methodology: S.V., C.I.; Investigation: S.V., C.I.; Resources: K.D.I.; Writing - original draft: S.V., K.D.I.; Writing - review & editing: S.V., C.I., K.D.I.; Supervision: K.D.I.; Funding acquisition: K.D.I.
Funding
This work was supported by the National Institutes of Health (GM131748 to K.D.I.) and a Rutgers University Busch pre-doctoral fellowship to S.V. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.247866.supplemental
Peer review history
The peer review history is available at https://dev.biologists.org/lookup/doi/10.1242/jcs.247866.reviewer-comments.pdf
References
- Acharya, B. R., Wu, S. K., Lieu, Z. Z., Parton, R. G., Grill, S. W., Bershadsky, A. D., Gomez, G. A. and Yap, A. S. (2017). Mammalian Diaphanous 1 mediates a pathway for E-cadherin to stabilize epithelial barriers through junctional contractility. Cell Rep. 18, 2854-2867. 10.1016/j.celrep.2017.02.078 [DOI] [PubMed] [Google Scholar]
- Alégot, H., Markosian, C., Rauskolb, C., Yang, J., Kirichenko, E., Wang, Y.-C. and Irvine, K. D. (2019). Recruitment of Jub by α-catenin promotes Yki activity and Drosophila wing growth. J. Cell Sci. 132, jcs222018. 10.1242/jcs.222018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, C.-Y., Ohsugi, M., Abe, Y. and Yamamoto, T. (2007). ZRP-1 controls Rho GTPase-mediated actin reorganization by localizing at cell-matrix and cell-cell adhesions. J. Cell Sci. 120, 2828-2837. 10.1242/jcs.03477 [DOI] [PubMed] [Google Scholar]
- Bays, J. L. and DeMali, K. A. (2017). Vinculin in cell–cell and cell–matrix adhesions. Cell. Mol. Life Sci. 74, 2999-3009. 10.1007/s00018-017-2511-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A., Beetham, H., Black, M. A., Priya, R., Telford, B. J., Guest, J., Wiggins, G. A. R., Godwin, T. D., Yap, A. S. and Guilford, P. J. (2014). E-cadherin loss alters cytoskeletal organization and adhesion in non-malignant breast cells but is insufficient to induce an epithelial-mesenchymal transition. BMC Cancer 14, 552. 10.1186/1471-2407-14-552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur, M., Feng, Y., Jagannathan, R., Seppa, M. J., Skeath, J. B. and Longmore, G. D. (2010). Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 20, 657-662. 10.1016/j.cub.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, S., Mana-Capelli, S., Paramasivam, M., Dasgupta, I., Cirka, H., Billiar, K. and McCollum, D. (2018). TRIP6 inhibits Hippo signaling in response to tension at adherens junctions. EMBO Rep. 19, 337-350. 10.15252/embr.201744777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola, A., Andreu, I., Beedle, A. E. M., Lezamiz, A., Uroz, M., Kosmalska, A. J., Oria, R., Kechagia, J. Z., Rico-Lastres, P., Le Roux, A.-L.et al. (2017). Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171, 1397-1410.e14. 10.1016/j.cell.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Gates, E. M., LaCroix, A. S., Rothenberg, K. E. and Hoffman, B. D. (2019). Improving quality, reproducibility, and usability of FRET-based tension sensors. Cytometry. Cytom. A 95, 201-213. 10.1002/cyto.a.23688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryanova, O. A., Sablina, A. A., Chumakov, P. M. and Frolova, E. I. (2005). Downregulation of TRIP6 gene expression induces actin cytoskeleton rearrangements in human carcinoma cell lines. Mol. Biol. 39, 792-795. 10.1007/s11008-005-0095-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, L. M., Jensen, C. C., Kloeker, S., Albert Wang, C. L., Yoshigi, M. and Beckerle, M. C. (2006). Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J. Cell Biol. 172, 771-782. 10.1083/jcb.200512115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers, S. and de Rooij, J. (2013). Mechanosensitive systems at the cadherin-F-actin interface. J. Cell Sci. 126, 403-413. 10.1242/jcs.109447 [DOI] [PubMed] [Google Scholar]
- Ibar, C., Kirichenko, E., Keepers, B., Enners, E., Fleisch, K. and Irvine, K. D. (2018). Tension-dependent regulation of mammalian Hippo signaling through LIMD1. J. Cell Sci. 131, jcs214700. 10.1242/jcs.214700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan, R., Schimizzi, G. V., Zhang, K., Loza, A. J., Yabuta, N., Nojima, H. and Longmore, G. D. (2016). AJUBA LIM proteins limit Hippo activity in proliferating cells by sequestering the Hippo core kinase complex in the cytosol. Mol. Cell. Biol. 36, 2526-2542. 10.1128/MCB.00136-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, N.-G. and Gumbiner, B. M. (2015). Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J. Cell Biol. 210, 503-515. 10.1083/jcb.201501025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.-J., Zheng, S., Sun, J., Muhamed, I., Wu, J., Lei, L., Kong, X., Leckband, D. E. and Wang, Y. (2015). Dynamic visualization of α-catenin reveals rapid, reversible conformation switching between tension states. Curr. Biol. 25, 218-224. 10.1016/j.cub.2014.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, M., Dent, E. W., Bear, J. E., Loureiro, J. J. and Gertler, F. B. (2003). Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell Dev. Biol. 19, 541-564. 10.1146/annurev.cellbio.19.050103.103356 [DOI] [PubMed] [Google Scholar]
- Leerberg, J. M., Gomez, G. A., Verma, S., Moussa, E. J., Wu, S. K., Priya, R., Hoffman, B. D., Grashoff, C., Schwartz, M. A. and Yap, A. S. (2014). Tension-sensitive actin assembly supports contractility at the epithelial zonula adherens. Curr. Biol. 24, 1689-1699. 10.1016/j.cub.2014.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. X. H., Tang, V. W. and Brieher, W. M. (2020). Actin protrusions push at apical junctions to maintain E-cadherin adhesion. Proc. Natl. Acad. Sci. USA 117, 432-438. 10.1073/pnas.1908654117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, V. T. G. and Lin, F.-T. (2011). TRIP6: An adaptor protein that regulates cell motility, antiapoptotic signaling and transcriptional activity. Cell. Signal. 23, 1691-1697. 10.1016/j.cellsig.2011.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie, H., Pratt, S. J., Betson, M., Epple, H., Kittler, J. T., Meek, L., Moss, S. J., Troyanovsky, S., Attwell, D., Longmore, G. D.et al. (2003). The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with α-catenin. J. Biol. Chem. 278, 1220-1228. 10.1074/jbc.M205391200 [DOI] [PubMed] [Google Scholar]
- Menaesse, A., Sumetsky, D., Emanuely, N., Stein, J. L., Gates, E. M., Hoffman, B. D. and Boustany, N. N. (2020). Simplified instrument calibration for wide-field fluorescence resonance energy transfer (FRET) measured by the sensitized emission method. Cytometry A, 10.1002/cyto.a.24194 [DOI] [PubMed] [Google Scholar]
- Misra, J. R. and Irvine, K. D. (2018). The Hippo signaling network and its biological functions. Annu. Rev. Genet. 52, 65-87. 10.1146/annurev-genet-120417-031621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, M. M. R., Meulemans, S. M. P., and Van, and de Ven, W. J. M. (2003). The focal adhesion and nuclear targeting capacity of the LIM-containing lipoma-preferred partner (LPP) protein. J. Biol. Chem. 278:2157-2168. 10.1074/jbc.M206106200 [DOI] [PubMed] [Google Scholar]
- Rauskolb, C., Sun, S., Sun, G., Pan, Y. and Irvine, K. D. (2014). Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell 158, 143-156. 10.1016/j.cell.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb, C., Cervantes, E., Madere, F. and Irvine, K. D. (2019). Organization and function of tension-dependent complexes at adherens junctions. J. Cell Sci. 132, jcs224063. 10.1242/jcs.224063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzell, W., Bustillo, M. E. and Zallen, J. A. (2018). The force-sensitive protein Ajuba regulates cell adhesion during epithelial morphogenesis. J. Cell Biol. 217, 3715-3730. 10.1083/jcb.201801171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, B. V. V. G. and Irvine, K. D. (2013). Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev. Cell 24, 459-471. 10.1016/j.devcel.2013.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard, M., Jouvenal, K., Tripier, D. and Walter, U. (1995). Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phosphoprotein). Proc. Natl. Acad. Sci. USA 92, 7956-7960. 10.1073/pnas.92.17.7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge, H., Artelt, N., Endlich, N. and Endlich, K. (2017). Automated segmentation and quantification of actin stress fibres undergoing experimentally induced changes. J. Microsc. 268, 129-140. 10.1111/jmi.12593 [DOI] [PubMed] [Google Scholar]
- Roszik, J., Lisboa, D., Szöllosi, J. and Vereb, G. (2009). Evaluation of intensity-based ratiometric FRET in image cytometry - approaches and a software solution. Cytometry A. 75A, 761-767. 10.1002/cyto.a.20747 [DOI] [PubMed]
- Sanz-Rodriguez, F., Guerrero-Esteo, M., Botella, L.-M., Banville, D., Vary, C. P. H. and Bernabéu, C. (2004). Endoglin regulates cytoskeletal organization through binding to ZRP-1, a member of the Lim family of proteins. J. Biol. Chem. 279, 32858-32868. 10.1074/jbc.M400843200 [DOI] [PubMed] [Google Scholar]
- Schmidt, G., Sehr, P., Wilm, M., Selzer, J., Mann, M. and Aktories, K. (1997). Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature 387, 725-729. 10.1038/42735 [DOI] [PubMed] [Google Scholar]
- Smith, M. A., Hoffman, L. M. and Beckerle, M. C. (2014). LIM proteins in actin cytoskeleton mechanoresponse. Trends Cell Biol. 24, 575-583. 10.1016/j.tcb.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S. and Irvine, K. D. (2016). Cellular organization and cytoskeletal regulation of the hippo signaling network. Trends Cell Biol. 26, 694-704. 10.1016/j.tcb.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi, K., Ishiuchi, T. and Takeichi, M. (2011). Mechanosensitive EPLIN-dependent remodeling of adherens junctions regulates epithelial reshaping. J. Cell Biol. 194, 643-656. 10.1083/jcb.201104124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, N., Smith, T. C., Nebl, T., Crowley, J. L., Palmieri, S. J., Lifshitz, L. M., Ehrhardt, A. G., Hoffman, L. M., Beckerle, M. C. and Luna, E. J. (2006). Supervillin modulation of focal adhesions involving TRIP6/ZRP-1. J. Cell Biol. 174, 447-458. 10.1083/jcb.200512051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Wu, B. and Zhong, Z. (2018). Downregulation of YAP inhibits proliferation, invasion and increases cisplatin sensitivity in human hepatocellular carcinoma cells. Oncol. Lett. 16, 585-593. 10.3892/ol.2018.8633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willier, S., Butt, E., Richter, G. H. S., Burdach, S. and Grunewald, T. G. P. (2011). Defining the role of TRIP6 in cell physiology and cancer. Biol. Cell 103, 573-591. 10.1042/BC20110077 [DOI] [PubMed] [Google Scholar]
- Xia, Z. and Liu, Y. (2001). Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J.81, 2395-2402. 10.1016/S0006-3495(01)75886-9 [DOI]
- Yao, M., Qiu, W., Liu, R., Efremov, A. K., Cong, P., Seddiki, R., Payre, M., Lim, C. T., Ladoux, B., Mège, R.-M.. et al. (2014). Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun. 5, 4525. 10.1038/ncomms5525 [DOI] [PubMed] [Google Scholar]
- Yi, J. and Beckerle, M. C. (1998). The human TRIP6gene encodes a LIM domain protein and maps to chromosome 7q22, a region associated with tumorigenesis. Genomics 49, 314-316. 10.1006/geno.1998.5248 [DOI] [PubMed] [Google Scholar]
- Yonemura, S., Wada, Y., Watanabe, T., Nagafuchi, A. and Shibata, M. (2010). α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533-542. 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- Zanconato, F., Cordenonsi, M. and Piccolo, S. (2016). YAP/TAZ at the roots of cancer. Cancer Cell 29, 783-803. 10.1016/j.ccell.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.