Graphical abstract
Keywords: SARS-CoV E SLiMs, Human PALS1, Molecular dynamics, Surface Plasmon resonance
Abstract
Short Linear Motifs (SLiMs) are functional protein microdomains that typically mediate interactions between a short linear region in one protein and a globular domain in another. Surface Plasmon Resonance assays have been performed to determine the binding affinity between PDZ domain of wild type human PALS1 protein and tetradecapeptides representing the SLiMs sequences of SARS-CoV-1 and SARS-CoV-2 E proteins (E-SLiMs). SARS-CoV-2 E-SLiM binds to the human target protein with a higher affinity compared to SARS-CoV-1, showing a difference significantly greater than previously reported using the F318W mutant of PALS1 protein and shorter target peptides. Moreover, molecular dynamics simulations have provided clear evidence of the structural determinants driving this binding process. Specifically, the Arginine 69 residue in the SARS-CoV-2 E-SLiM is the key residue able to both enhance the specific polar interaction with negatively charged pockets of the PALS1 PDZ domain and reduce significantly the mobility of the viral peptide. These experimental and computational data are reinforced by the comparison of the interaction between the PALS1 PDZ domain with the natural ligand CRB1, as well as the corresponding E-SLiMs of other coronavirus members such as MERS and OCF43. Our results provide a model at the molecular level of the strategies used to mimic the endogenous SLiM peptide in the binding of the tight junctions of the host cell, explaining one of the possible reasons of the severity of the infection and pulmonary inflammation by SARS-CoV-2.
1. Introduction
Since the first report on the scientific literature of the Severe and Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) at Wuhan at the end of 2019 which triggered the current pandemic [1], the scientific community started an intense and focused study onto the mechanism driving the transmission and the virulence of this new pathogen [2], [3], [4], [5]. In particular due to the genomic similarity with SARS-CoV-1 appeared in 2002 and 2003, a special attention was reserved to the comparison of the two viruses in term of their ability to infect the host human cells of the respiratory tract [6], [7], [8]. After more than one year, some elements emerged from this common effort as expected due to genomic similarity [5]. As a first clear evidence, both the virus SARS-CoV-1 and SARS-CoV-2 viruses enter in the human cells utilizing the angiotensin-converting enzyme 2 receptor (ACE2) [9], [10]. Accordingly, the spike proteins (S) of the two viruses were analysed and their efficiency in formation of the active complex with ACE2 receptor was deeply investigated. A large set of experimental and computational studies were focused onto the clarification of the structural details responsible for any difference between host-virus interaction, as a possible explanation for the pandemic event all over the globe caused by SARS-CoV-2, but not by SARS-CoV-1 [11], [12], [13], [14], [15], [16], [17]. These studies were also stimulated by the possible consequences, in term of a structure-based drug design of small molecules able to interfere with the process of virus entering in the host cells, to be used in clinic practice as specific antiviral compounds [18], [19].
A second set of structural studies focused their attention onto the main viral enzymes. On one hand there is the protease (Mpro, also called 3CLpro) because of its essential role in processing the polyproteins that are translated from the viral RNA. Still in the case of this topic, a strong stimulus for researchers was the idea to find a possible antiviral compound to target this enzyme, thus helping the clinicians in their dramatic effort to cope with the multifaceted syndrome that SARS-CoV 2 is producing in patients [20], [21], [22], [23]. On the other hand, there is the RNA dependent RNA Polymerase enzyme (RdRP) due to its critical role in the mechanism of viral genome production, that has been also intensively studied as a potential target for selective antiviral compounds [24], [25], [26], [27]. In both cases, structural differences and functional properties of these enzymes, among different members of the Coronaviridae family, have been compared to investigate a possible better catalytic efficiency of these key enzymes as a cause of the virus spread [28].
The last protein target that was also studied from a structural point of view, is the Envelope protein (E). The E protein is directly involved in the mechanism of virus infection due to the presence in its primary structure of Short Linear Motifs (also known as SLiMs) that are responsible of protein–protein interaction with the tight junction of the host cell, via the formation of a complex with the PDZ domain of the human PALS1 protein [29], [30]. This protein has been suggested playing a central role in the virulence of SARS-CoV-1 as demonstrated by Castaño-Rodriguez and coworkers that clearly showed in mice models that a virus lacking of the PDZ binding motif in the E protein, drastically reduced its replication and virulence [31].
In a previous paper [32], we provided a first model to explain a strengthened binding of SARS-CoV-2 E protein with the tight junction-associated PALS1, with respect to the SARS-CoV-1 by in silico analysis and docking studies. This work was performed using a SLiM octapeptide for both SARS-CoVs which represents the minimal unit identified for protein–protein interaction. At the same time, a 3-fold higher affinity for the SARS-CoV-2 peptide with respect to SARS-CoV-1 was experimentally measured by Toto et al, using a mutant of the human PALS1 protein (F318W) through equilibrium and ultra-fast kinetic binding measurements monitored by FRET [33], thus giving experimental support to our in silico model. The F318W mutant was selected for the FRET measurements due to the higher intrinsic fluorescence of the variant expressing tryptophan. Nevertheless, the topological position 318 of PALS1 in our model, was considered interacting with both the target viral peptides. Thus, in the present study, we compare the binding affinities to the SARS-CoV peptides of both the wild type and the F318W mutant PALS1 by using Surface Plasmon Resonance (SPR). We also elongated the interacting viral peptides to 14 residues each, in the experimental determination as well as in molecular dynamics simulations. This choice was a compromise: in fact, on one hand computational studies identify the length of a SLiM sequence in the range of 3–12 residues, therefore the length of 14 residues should be sufficient to take into consideration all the main structural determinants driving the binding process mediated by this protein motifs [34]. On the other hand, the 3D structure of the human PDZ domain of PALS1 in complex with its endogenous targeting peptide CRB1 was made of 17 residues, but only 14 were reported in the model, due to high mobility of the last ones [35]. Therefore, in the present study, we set the length of all peptides considered as potential target of the human PDZ PALS1 to 14 residues, as reported in the Supplementary Fig. 1.
2. Results and discussion
2.1. Surface Plasmon resonance (SPR) measurements of binding affinity
N-terminal biotinylated version of the three C-terminal tetradecapeptides, the endogenous PALS1 ligand CRB1 (MWNLMPPPAMERLI) and the envelope E protein derived peptides of SARS-CoV-1 (KNLNSSEGVPDLLV) and SARS-CoV-2 (VKNLNSSRVPDLLV) were immobilized on different streptavidin chips and used as ligands in a SPR assay. As analytes, PDZ domains of PALS1, both wild-type and F318W variant, were used. The PALS1 mutant was previously used by Toto et al., to measure the Kd values of the interaction with a shorter version of the same SARS-CoV-2 and SARS-CoV-1 envelope E protein peptides [33]. These authors, using as a probe the intrinsic fluorescence of the Trp introduced in the topological position 318 of the PALS1 protein, obtained a Kd = 40 ± 10 and a Kd = 130 ± 10 μM respectively, for SARS-CoV-2 and SARS-CoV-1 E-SLiMs peptides.
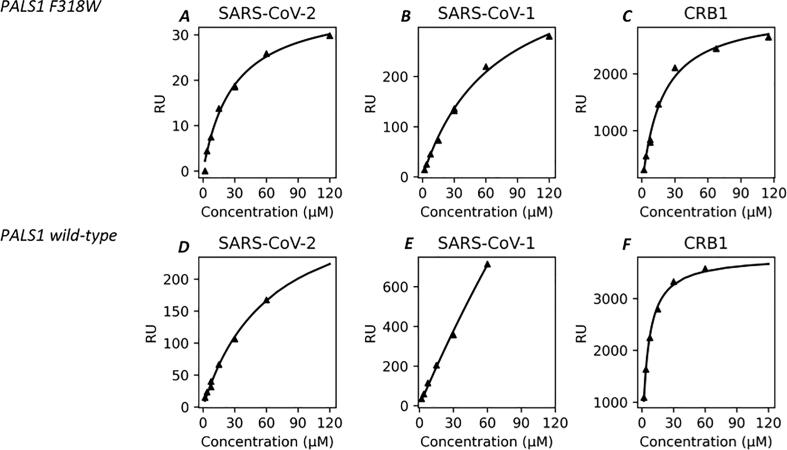
According to SPR measurements, the affinities for PALS1 F318W mutant of the longer viral tetradecapeptides used in this assay, are in good agreement with what previously reported for the shorter ones with a different experimental technique. Specifically, the Kd value measured for SARS-CoV-2 was Kd = 29 ± 4 μM while the value for SARS-CoV-1 was Kd = 69 ± 7 μM (see Table 1 and Scatchard plot in Fig. 1A and 1B). In both cases the Kd values were slightly lower than the values previously measured, possibly due to the longer peptides used that may have a larger interaction surface with the PALS1 target mutant protein, thus enhancing the binding affinity. As shown in Fig. 1C and reported in Table 1, the endogenous CRB1 peptide presented a higher affinity with a lower Kd value that is 18 ± 2 μM.
Table 1.
Kd values obtained by the SPR measurements.
| SARS-CoV-2 | SARS-CoV-1 | CRB1 | |
|---|---|---|---|
| PALS 1 F318 W | 29 ± 4 μM | 69 ± 7 μM | 18 ± 2 μM |
| PALS 1 Wild type | 63 ± 10 μM | > 500 μM | 5.11 ± 0.33 μM |
Fig. 1.
Scatchard plots of the interactions between immobilized tetradecapeptides and the analytes, PALS1 F318W and PALS1 wild-type at T = 25 °C. Triangles represent experimental data points and solid lines are the fit. Data point were obtained at the following concentration [PALS1_F318W] = 120, 60, 15, 7.5, 3.75 and 1.875 μM and [PALS1_wt] = 60, 30, 15, 7.5, 3.75 and 1.875 μM. Panel A, binding isotherm of PALS1 F318 W and SARS-CoV-2 E peptide. B, binding isotherm of PALS1 F318 W and SARS-CoV-1 E peptide, C, binding isotherm of of PALS1 F318W variant and CRB1, D, binding isotherm of PALS1 wild type and SARS-CoV-2 E peptide E, binding isotherm of PALS1 F318W variant and SARS-CoV-1 E peptide, F, binding isotherm of PALS1 wild type and CRB1 peptide.
Next, we measured the affinities of these same peptides to PALS1 wild-type protein domain. The Kd trend among the three peptides remains unchanged, having always the endogenous peptide CRB1 the lowest Kd = 5.11 ± 0.33 μM (see Table 1 and Fig. 1 E) which is comparable to that previously measured by Ivanova, et al., that is Kd = 9.2 ± 1.4 μM [35]. Moreover, SARS-CoV-2 peptide has still a lower Kd value with respect to SARS-CoV-1 even if, using the wild-type protein as a target in the SPR assay, the difference in binding affinity between the two viral peptides drastically increased. Indeed, the SARS-CoV-2 peptide Kd value was = 63 ± 10 μM (see Table 1 and Scatchard plot in Fig. 1D) while the SARS-CoV-1 peptide Kd value can be only considered having a lower limit of 500 μM. In fact, its binding isotherm maintains linear-like behaviour in the PALS1 concentration range measured (see Fig. 1E) therefore a larger experimental error is associated to this measure (see Table 1). This very weak affinity could seem hardly sufficient to be detected by co-immunoprecipitation (Co-IP), yet it was initially identified using this same technique [30]. However, it has to be noted that Co-IP in vitro experiments probed the interaction between the full-length Envelope protein and PALS1 PDZ domain and not just the E-SLiM peptide as in the present study. Accordingly, a further contribution of the entire Envelope could be sufficient to allow detection of this protein complex using Co-IP. This fact may be expected considering that, using the endogenous binding protein Crb-CT, the larger complex PALS1-PDZ-SH3-GK but not the isolated PDZ domain, binds it with very high affinity [36]. On the other hand, Teoh and coworkers clearly demonstrate that the minimal unit necessary to trigger the interaction between E protein and PDZ-PALS1 involves specifically the E-SLiM peptide [30].
Moreover, the experimental evidence that the mutant F318W has a higher affinity for both the viral peptides (but not for the endogenous peptide CRB1) with respect to wild type PALS1 protein, suggests a direct involvement of this residue into the binding mechanism of the viral peptides. As reported in our previous paper [32], where a docking of the two viral peptides is reported and compared with the crystal structure of the wild type protein in complex with CRB1, F318 is one of the key residues inside the PDZ domain, involved in the stabilization of the interaction between the last four residues of the peptides that form the Protein Binding Motif (PBM). In fact, according to the docking model, F318 interacts with the third residue in the PBM, starting from the C-terminus, which is an Arginine (R1404 see Fig. S1) for CRB1 and a Leucine (L74 and L73 see Fig. S1) for both SARS-CoVs peptides. Accordingly, its substitution with a bulkier side chain, as the Trp residue, may have an impact onto the binding affinity.
2.2. Molecular dynamics simulations
In order to better understand the differences of the affinities measured between the viral peptides and the wild-type and mutated PALS1 proteins, molecular dynamics was used to investigate the conformational variability of the two proteins. At first, two 500 ns long molecular dynamic simulations of both PALS1 wild-type and F318W mutant alone were performed. To point out the difference between these two MD simulations we have reported in Fig. 2, panels A and B the chi1-chi2 side-chain torsion angle combinations for residue 318, along the whole trajectory. In this graph the open circle colour depends on the distance between center of mass (c.o.m.) of the side chain selected (both F or W) and c.o.m. of residue L321 that is placed in the bottom-center of the binding groove of PALS1. As shown in Fig. 2 panel A, F318 in PALS1 wild-type protein is a very flexible residue, with many possible orientations that are highly distributed. Moreover, considering the distance of 7 Å from the residue L321, discriminant between open and closed conformation, the different orientations of the angles for Phe are equally distributed between these two states, therefore all the clusters in panel A contain both green and purple circles equally represented.
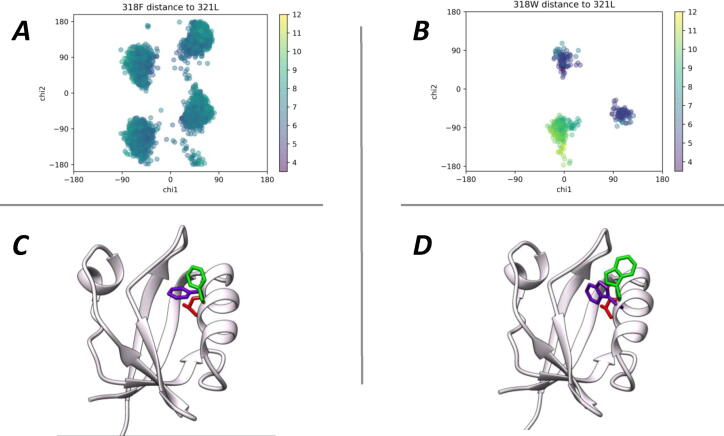
Fig. 2.
Molecular Dynamics simulation of the free PALS1 wild type and F318W variant. Panels A and B show the chi1-chi2 side-chain torsion angle combinations for residue 318, obtained by 500 ns long molecular dynamics simulations of the two PALS1 variants, where dot colour depends on the distance between center of mass (c.o.m.) of the side chain and c.o.m. of L321 (in red) placed in the bottom-center of the binding groove of PALS1. A, Wild type protein. B, F318W protein. C the 3D model of PALS1 wild type, showing two different conformation of F318 that are equally populated during the simulation. In purple the closed position of F318 in green the open one, D, the 3D model of PALS1 F318W variant showing the two different conformation of W318, keeping the same color code of panel C. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
On the contrary, a Tryptophan residue in the same position has a more restrained and defined dynamic behaviour, as W318 conformations are less disperse (see Fig. 2 panel B). In fact, its conformations distribute in just three different and clearly marked clusters that according to the relative distance between the c.o.m of W318 and L321 define an open and a closed conformation. In this case, one cluster is formed exclusively by a single set of green dots representing the conformations where PALS1 groove is clearly open (with a distance to L321 > 7 Å); while the other two clusters are made only of purple dots, outlining the conformations that lead to a closed state of the protein (with a distance to L321 < 7 Å). The sum of the purple dots, representing the closed conformation, is much more represented (about 80%) along the whole trajectory, thus indicating a preferential position for this residue in this conformation. In Fig. 2, panels C and D, we show the protein structures of PALS1, with the two alternative positions (open in green, closed in purple), for the residues in the topological position 318. These positions where selected according to a clustering of the corresponding conformations of Panel A and B, selecting as open state for Phe, the position that this residue has in the crystal structure bound to CRB1, while for the closed one, the position that is closer to the protein groove. On the other hand, the lower mobility of Trp in its most populated conformation seems consistent with the fact that this residue can slip into the hydrophobic pocket stabilizing PALS1 in a closed state, potentially hindering the binding pocket available for all the peptides. This model suggests a lower affinity of PALS1 F318W for peptides binding in the internal groove, if the affinity change is simply driven by a reduction in the on-rate of formation of the active complex, and could explain the three-fold higher value of CRB1 Kd for PALS1 F318W compared to the wild type protein. Nevertheless, this model would be in contrast with the experimental data showing a higher affinity of the mutant of PALS1 for both the viral peptides. In the absence of a kinetic determination of the kon and koff rate constants in the SPR measurements, the higher affinity of the PALS1 mutant with respect to the wild type protein, for the viral peptides, but not for CRB1, suggests that the Trp318 residue is able to better interact with these viral peptides stabilizing them in the bound state.
In order to verify this hypothesis a second set of MD simulations was performed. Starting from the reported 3D structure of the complex wild-type PALS1 and CRB1 peptide, three different computational models were prepared (see Materials and Methods section for detailed information on the molecular docking procedures): a) PALS1 F318W in complex with SARS-CoV-2 E-SLiM, b) PALS1 F318W in complex with SARS-CoV-1 E-SLiM and c) PALS1 F318W in complex with CRB1 SLiM. At least two MD runs of 500 ns each were done for any model; the simulations were analysed considering only the last 300 ns of each simulation and concatenating them. Results in term of the interaction established along this trajectory are reported in Fig. 3 panels A-C for the corresponding models. Noticeably, the complex PALS1 F318W - SARS-CoV-1 E-SLiM was run three times, as the peptide was not stable along the entire simulation in at least two over three cases. Therefore, for this system, data refer only to the last 300 ns of the more stable MD simulation. Panels D-F of the same picture show the most representative structures obtained after a clustering of the MD runs (see pie chart at the bottom of the picture) as purple peptides, while in white it is reported the starting configuration obtained after the initial docking procedure.
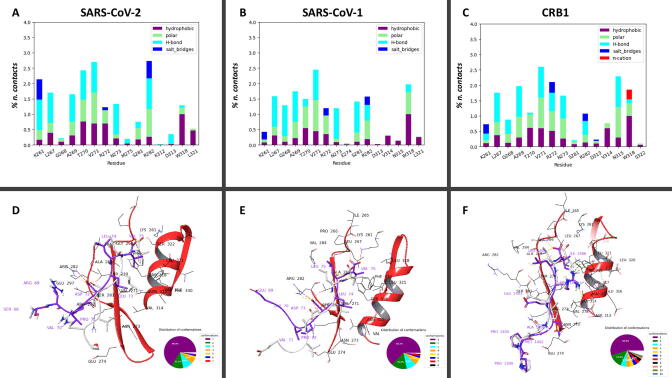
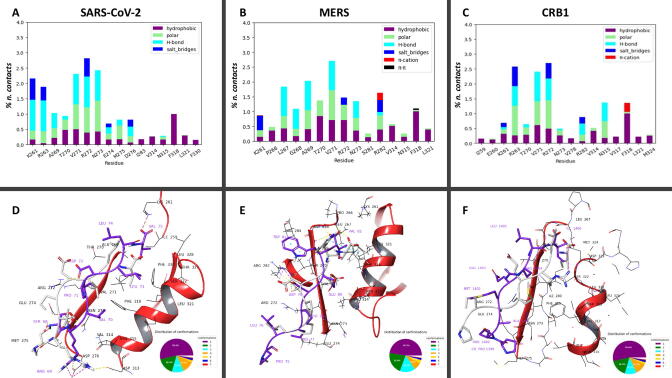
Fig. 3.
A, B, C Interaction histograms of SARS-CoV-2, SARS-CoV-1 and CRB1 E-SLiM peptides against PALS1 F318W mutant. Results include hydrophobic and polar contacts, h-bonds, salt bridges and π-cation as derived analyzing the last 300 ns of two 500 ns long molecular dynamics for each complex. D, E, F) Most representative conformations of SARS-CoV-2, SARS-CoV-1 and CRB1 are reported in purple, where only the last 8 residues are shown, in complex with PALS1 F318W. The structures superimposed in white, represent the starting conformations coinciding with the structure obtained via docking procedure for SARS-CoV-2 and SARS-CoV-1, while the minimized crystallographic conformation for CRB1 is reported (PDB code: 4UU5, after mutation F318 → W318). The pie charts below every structural representation show the distribution of peptide conformations obtained through clustering. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As expected, the interaction of the W318 was clearly evident in all the three systems analysed being the large bulky side chain able to interact with all the three peptides. Moreover, looking at the other interaction that are formed, it is also evident that the two viral peptides share a similar interaction pattern, with a significant reinforcement of the polar interaction of SARS-CoV-2 E-SLiM with K261 and R282 with respect to SARS-CoV-1 E-SLiM. On the contrary the endogenous CRB1 SLiM shows an important difference in the interaction pattern as due to the arginine residue R1404, that is able to form a strong interaction with the protein couple asparagine N315 and W318. This special condition is due to the fact that this polar residue protrudes in the direction of the two aminoacids as shown in Fig. 3 panel F.
Finally, to clarify the structural determinants driving the binding mechanism that identify SARS-CoV-2 E-SLiM as a better ligand with respect to SARS-CoV-1 E-SLiM for the wild type PALS1 protein, three further different computational models were built-up starting from the 3D structure of the complex wild-type PALS1 and CRB1 SLiM. A set of 500 ns long simulations, 2 for CRB1 SLiM-PALS1 complex, and 4 for both SARS-CoVs tetradecapeptides, were made and only the last 300 ns of each simulation were analysed in term of the interaction established along the simulations. About SARS-CoV-2 MD simulations, only one was not used in the analysis due to peptide detachment from PALS1 before the end of the simulation. On the contrary, data for SARS-CoV-1 are not reported since in none of the 4 simulations, the peptide was able to remain attached to PALS1 > 150–200 ns. In order to verify if the stability of the PALS1 E-SLiM complex was characteristic only of SARS-CoV-2 E peptide, two other models were prepared, using as a ligand to the human protein the corresponding E-SLiMs tetradecapeptides of two other members of the Coronaviridae family, MERS and OC43 (see Fig. S1). The complex formed by OC43-E-SLiM and wild type PALS1 PDZ domain was highly unstable detaching from the human protein in less than 50 ns in 4 different MD runs (data not shown). On the contrary, the complex MERS-E-SLiM with wild type PALS1 protein, was highly stable in both the 500 ns MD runs, thus allowing to analyse the binding poses and report them in Fig. 4 together with SARS-CoV-2 and CRB1.
Fig. 4.
A, B, C Interaction histograms of SARS-CoV-2, MERS and CRB1 E-SLiM peptides against PALS1 wild-type. Results include hydrophobic and polar contacts, h-bonds, salt bridges, π-cation as derived analyzing the last 300 ns of two 500 ns long molecular dynamics for each complex. D, E, F) Most representative conformations of SARS-CoV-2, MERS and CRB1 are reported in purple, where only the last 8 residues are shown, in complex with PALS1 wild-type. The structures superimposed in white represent the starting conformations, coinciding with the structure obtained via docking procedure for SARS-CoV-2 and MERS, while the crystallographic conformation is reported for CRB1 (PDB code: 4UU5). The pie charts below every structural representation show the distribution of peptide conformations obtained through clustering. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Despite sharing the same binding site, CRB1 and viral tetradecapeptides interact differently with their target. The SARS-COV-2 E-SLiM achieves to slip deeply in the groove of PALS1, indeed, the negatively charged carboxylic group of the C-terminus Valine V75 residue can reach both R263 and K261, making with them salt bridge interactions and polar contacts (as shown in Fig. 4 panel A). As a consequence, the C-terminal V75 side-chain interacts with hydrophobic residues of PALS1 domain deep in the pocket, interacting with L321, G326 and F330. Among the cited PALS1 residues, only K261 is reached by CRB1 C-terminal carboxylic group of Isoleucine I1406, as this peptide is strongly anchored in its initial position from this bulkier residue that is not able to get deeper in the groove as the corresponding Valine V75 of the SARS-COV-2 E-SLiM. The MERS-E-SLiM peptide has also a Valine C-terminal residue (V82 see Fig. S1), but it is not able to slip deeply as the other viral peptide, due to the different anchoring points to the human PALS1 protein in the following residues. Specifically, SARS-COV-2, MERS and CRB1 share a hydrophobic residue in the second position from the C-terminus (L74, W81 and L1405 for SARS-CoV-2, MERS and CRB1 respectively) while the third one is totally different. For CRB1 SLiM, there is a positively charged residue (R1404), while an other hydrophobic one (L73) is present for SARS-CoV-2 E-SLiM and a negatively charged one (E80) is present for MERS E-SLiM. As can be seen in Fig. 4, the CRB1 SLiM Arginine R1404 makes both hydrophobic and pi-cation interactions with F318 and polar contact and H-bonds with N315 of PDZ PALS1 domain. By contrast, SARS-CoV-2 E-SLiM Leucine L73 can only interact hydrophobically with F318, with negligeable interactions with N315. Also MERS E-SLiM E80 interact only hydrophobically with F318 using the aliphatic carbons of the side chain having negligeable interactions with N315. This last viral peptide though, uses the W81 to strongly interact with R282, via a pi-cation interaction thus enhancing its stability in the binding groove. Finally, considering the last residue of the PBM, CRB1 SLiM Glutamate E1403 interacts via salt bridge with R282 and R272 and has a polar interaction with T270. The arginine R282 in the human protein target, is harder to be reached by the shorter Aspartate D72 of SARS-CoV-2 E-SLiM, that achieves to interact only with R272. On the contrary, the D79 of MERS-E-SLiM is able to chelate R282 due to the combined action with W81 that fix the Arginine residue of PALS1 in the suitable position (see Fig. 4 for detailed information).
In our previous article [32], by molecular docking, we showed that SARS-CoV-2 E-SLiM Arginine R69 was able to form a salt bridge with aspartate D313. Unexpectedly, R69 does not interact with this residue significantly along the MD trajectory, but still maintains a central role in the binding mechanism. In facts, compared to the conformation obtained by molecular docking, which is the initial conformation of the simulation (white in Fig. 4 panel D), the backbone of the peptide straightens, moving away the residue from its starting point and finding a larger negative pocket in which stably lying. In fact, as shown in Fig. 4 panel D, SARS-CoV-2 E-SLiM Arginine R69 swings and finds other negatively charged residues, such as E274 and D276, as well as may form polar contacts with M275, at the edge of the binding site. Further contacts with the residues of PALS1 protein are common for all peptides and are made in an unspecific mode by peptide backbones.
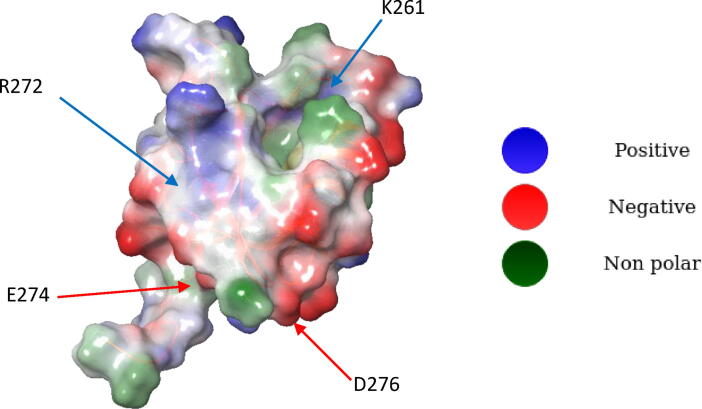
As a further consideration, SARS-CoV-2 E-SLiM finds 4 charged residues on PALS1 to anchor on it. As it is shown in Fig. 5, it constantly interacts with K261 and R272, in the C terminus part of the peptide and is able to find two negative shallow pockets, one with D276 and the other one with E274, through its Arginine R69 residue that is exclusive for SARS-CoV-2 peptide, since neither CRB1 SLiM, nor MERS E-SLiM or even SARS-CoV-1 E-SLiM, have positively charged residue in the same topological position. In particular, the endogenous peptide CRB1 has a proline residue in this position (P1400) followed by two other proline residues that strongly reduce its freedom degrees thus favouring the interaction with PALS1 PDZ partner domain. This same strategy is enhanced by MERS-E-SLiM, that has L76 in the same topological position, in the middle of four Proline residues (P74, P75, P77 and P78 see Fig. S1) that strongly stabilizes the Leucine L76 in a bound conformation. On the contrary, SARS-CoV-1 has a glycine residue (G70) in this same position, with no proximal proline residues able to reduce its mobility.
Fig. 5.
Surface of PALS1. In blue are shown side chains of positively charged residues, in red negatively charged residues and in green the non polar residues. Arrows indicate the different anchor points used by SARS-CoV-2 E SLiM. Blue arrows point in the direction of K261 and R272. Red arrows point in the direction of the negative charged pockets identified by residues D276 and E274. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
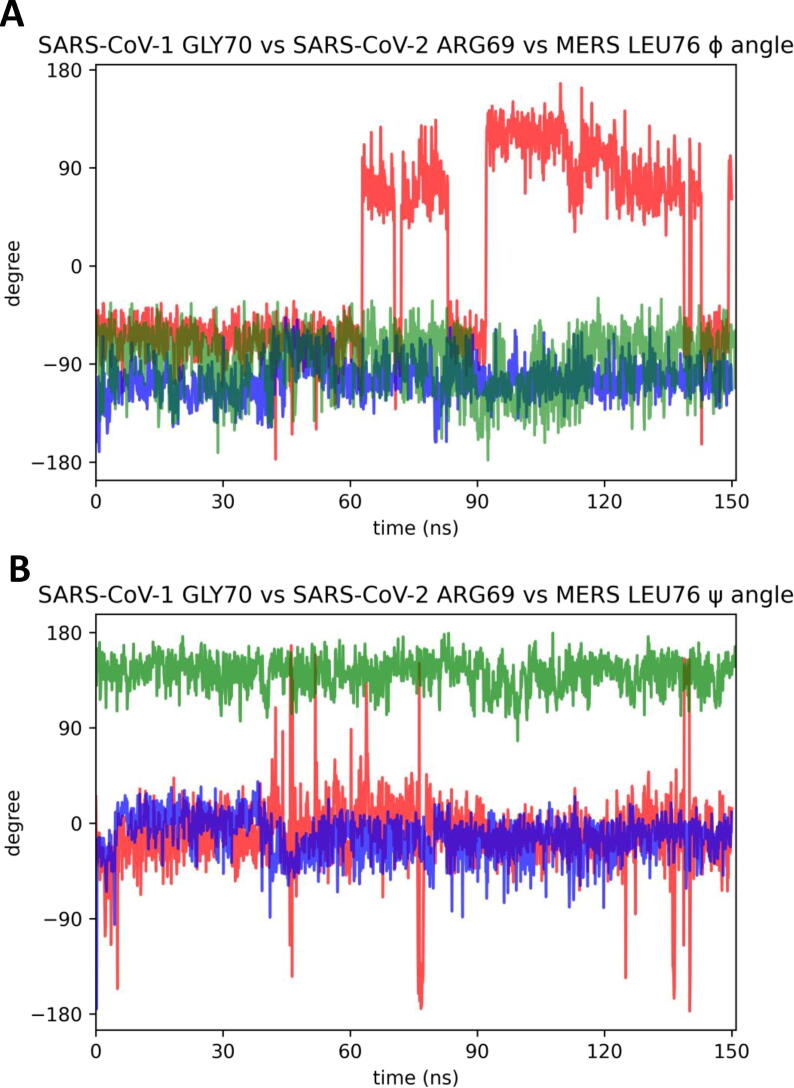
Therefore this topological position seems the key one to explain the difference in affinity between the SARS-CoVs peptides, due to the different contribution to the binding with PALS1 between R69 residue of SARS-CoV-2 and the G70 of SARS-CoV-1. In order to verify this hypothesis, we reported in Fig. 6 the result of the first 150 ns of Molecular Dynamics simulation obtained for the complex wild type PALS1 with SARS-CoV-2, SARS-CoV-1 and MERS E-SLiMs with specific reference to this topological position comparing the ψ and φ angles of the residues R69, G70 and L76 respectively. This time range was selected as SARS-CoV-1 E-SLiM still forms a stable complex with wild-type PALS1 before breaking down.
Fig. 6.
Comparison of φ/ψ angles of the residue in the same topological position of SARS-CoV-2, SARS-CoV-1 and MERS E-SLiM peptides: Variation of the angles are obtained analyzing the first 150 ns of molecular dynamic simulation of peptides-PALS1 wild-type complexes. Panel A, comparison of φ angles of SARS-CoV-2 R69 (in blue), SARS-CoV-1 G70 (in red) and MERS L76 in green. Panel B, comparison of ψ angles of same residues. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As shown in panel A and B of Fig. 6, ψ and φ angles for SARS-CoV-2 E-SLiM R69 are stable and clustered along the trajectory explored (see blue traces). The same is true for the MERS- E-SLiM L76 (green traces). On the contrary, for SARS-CoV-1 E-SLiM G70, the ψ angle (panel B), toward the C-terminal head of the peptide remains relatively stable with occasional flipping episodes, meanwhile the rotation on SARS-CoV-1 φ angle is much higher (panel A), flipping constantly and creating high instability to the N-terminal portion of the peptide, thus leading to the breakdown of the complex.
3. Conclusion
Our first aim was to compare the binding affinity of the SLiM sequences of the viral Envelope proteins of SARS-CoV-1 and SARS-CoV-2 to their protein target in the tight junction of the epithelial human cells, that is the PDZ domain of PALS1. The direct measurement of the binding affinity of both viral E-SLiMs with respect to the wild type PDZ domain of PALS1, allowed to determine a Kd value of 63 μM for SARS-CoV-2 and only a lower limit of 500 μM for SARS-CoV-1, with a difference between them close to a factor 10. Previous results underestimated this difference, measuring Kd values that were only three-fold lower for the SARS-CoV-2 E-SLiM with respect to SARS-CoV-1 (Kd value of 40 μM with respect to 130 μM).
The main reason of this discrepancy is probably the use in previous work of the F318W mutant of PALS1, instead of the wild-type protein, as we have demonstrated that the Trp substituent is actively perturbing the mechanism of interaction with the viral peptides, being less crucial for the CRB1 binding mechanism. Moreover, using smaller peptides the entropic effect due to SLiM residues farther from the binding groove is missing.
The effect of farther SLiM residues appears when we perform the MD simulations for the complex with wild type PALS1 protein. Indeed, the SARS-CoV-1 did not remain stably bound for a complete set of 500 ns simulation time, due to the significant difference that this peptide exhibits in the topological position where it has the highly mobile glycine residue (G70) that is not able to fix this peptide into the right position, thus allowing its release from PALS1. On the contrary, in SARS-CoV-2 this glycine is substituted by the positively charge residue of Arginine (R69), that anchors stably the peptide to the target human protein interacting with shallows negatively binding pockets, reducing the mobility of the whole peptide thus enhancing its binding affinity. The CRB1 endogenous ligand has a 3 proline motifs (P1398-P1400) that strongly reduce the mobility of the whole SLiM peptide, thus favouring a more stable interaction with its protein PALS1 partner. This same strategy is used by MERS-E SLiM peptide, where we observe a similar motif (PPLPP), centered to the L76 residue that corresponds to the same topological position of R69 and G70 in SARS-CoV2 and SARS-CoV-1 respectively.
In conclusion, according to the combined in silico study and SPR assay, we have been able to point out the structural determinants that allow SARS-COV-2 E-SLiM to interact with a significantly higher affinity with respect to SARS-COV-1 E-SLiM to the PALS1 wild type protein.
4. Materials and methods
4.1. Protein expression and purification
Gene encoding for the PDZ domain of PALS1 was subcloned in a pET-28b (+) plasmid vector which was used to transform Escherichia coli cells BL21 (DE3) strain. Bacterial cells were grown in LB medium, containing 30 μg/mL of kanamycin, at 37 °C until OD600 = 0.8–0.9 was reached. Protein expression was induced with 1 mM IPTG. After induction, cells were incubated at 25 °C overnight and then collected by centrifugation. Bacterial pellet was resuspended in buffer Tris-HCl 50 mM, NaCl 0.3 M, pH 7.5 with the addition of antiprotease tablet (Complete EDTA-free, Roche), then sonicated and centrifuged. The soluble fraction from bacterial lysate was loaded onto a nickel-charged HisTrap Chelating HP (GE Healthcare) column equilibrated with Tris-HCl 50 mM, NaCl 0.3 M, imidazole 10 mM, pH 7.5. Protein was then eluted with a gradient from 0 to 1 M Imidazole by using an ÄKTA-prime system. Fractions containing the protein were identified through SDS-page and collected. The imidazole present in the sample was removed by using a HiTrap Desalting column (GE Healthcare) equilibrated in Tris-HCl 50 mM, NaCl 0.3 M, pH7.5. The purity of the protein was analyzed through SDS-page.
F318W variant of the PDZ domain of PALS1 was obtained by site-directed mutagenesis using a QuickChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies), accordingly to the instructions of the manufacturer. The variant was expressed and purified following the same protocol used for the wild-type.
4.2. Surface Plasmon resonance measurements
The interactions between the peptides (ligands) and the purified PALS1 PDZ domains, both wild type and F318W mutant (analytes) were measured by surface Plasmon resonance (SPR) technique using a Biacore X100 instrument (Biacore, Uppsala, Sweden). The N-terminal biotinylated peptides were obtained from GenScript (Piscataway, NJ, USA) and were immobilized on a Sensor Chip SA, precoated with streptavidin from Biacore AB (Uppsala, Sweden). Capturing procedure on biosensor surface was performed according to manufacturer’s instructions. Running buffer was PBS, which contains 10 mM phosphate buffer pH 7.4, 0.137 M NaCl, 2.7 mM KCl. Protein domains were diluted in running buffer, and binding experiments were performed at 25 °C with a flow rate of 30 μl/min. The highest concentration obtained for the wild type protein was 60 μM while for the mutant F318W was 120 μM. The concentrations measured in the SPR assay were obtained by successive dilutions halving the concentration in each step.
The association phase was followed for 180 s, whereas the dissociation phase was followed for 300 s. Each experiment was performed using a minimum of 6 different concentration of the analyte and the intermediate concentration was repeated twice to test the reproducibility of data. The regeneration of the surface was achieved by addition of 2 M NaCl for 30 s before each new cycle start. When experimental data met quality criteria, data were analysed using Biacore X100 Evaluation Software. An affinity steady state model was applied to fit the data, as kinetic parameters were out of the range measured by the instrument but an equilibrium signal of interaction was clearly detected. Accordingly, specific Kd were determined with a confidence interval associated with a standard error value.
4.3. In silico docking of Envelope tetradecapeptides to PALS1 wild-type and preparation of the peptides-PALS1 F318W complexes
The fourteen C-terminal residues of each E protein were built, (see Fig. S1) and protonation states were assigned with PROPKA [37]. To construct the peptide-protein complexes, the tetradecapeptides were manually superimposed and aligned onto the conformations of the octapeptides already docked on PALS1, which were obtained from our previous work [32] or anew docking procedure was performed. Finally, the complexes were minimized using Prime [38].
In order to build the complexes between the peptides and PALS1 F318W mutant, the F318 residue of every peptide-protein complex was substituted with a Tryptophan, via Maestro tools, and the systems were minimized with Prime.
Moreover, in both complexes of CRB1 with PALS1 wild-type and mutant, CRB1 SME1402, a methionine sulfoxide present in the reference model (PDB code: 4UU5, [35]), was modified with a methionine.
4.4. Molecular dynamics simulations
Classical molecular dynamics simulations were performed with the GROMACS 2018.3 [39] package using the CHARMM36m force-field [40], with WYF parameter for cation-pi interactions, at the full atomistic level using a TIP3 water solvent. All the molecular dynamics sessions were performed in a water system prepared via the CHARMM-GUI server [41]. The protein–ligand systems were solvated in a cubic water box (having basis vectors lengths of 7.1 nm) under periodic boundary conditions. The total charge of the system was neutralized by randomly substituting water molecules with Na+ ions and Cl– ions, to obtain neutrality with 0.15 M salt concentration.
Following a steepest descent minimization algorithm, the system was equilibrated in canonical ensemble (NVT) conditions for 125 ps, using Nose-Hoover thermostat with position restraints for the protein-peptide complexes. Then, all restraints were removed, and molecular dynamics runs were performed under NPT conditions at 303.15 K, using Nose-Hoover thermostat, with a T-coupling constant of 1 ps, and a Parrinello-Raman barostat at 1 atm. Van der Waals interactions were modelled using 6–12 Lennard-Jones potential with a 1.2 nm cut-off. Long-range electrostatic interactions were calculated, with a cut-off for the real space term of 1.2 nm. All covalent bonds were constrained using the LINCS algorithm. The time step employed was 2 fs, and the coordinates were saved every 5 ps for analysis, which was performed using the standard GROMACS tools.
4.5. Analysis of molecular dynamics trajectories
Percentages of formation of hydrogen bonds interactions along MD trajectories have been calculated by custom procedures using the MDAnalysis python library [42]. Polar and hydrophobic interactions between peptides and receptor residues were modelled as average coordination numbers via a continuous, differentiable switching function:
with the i and j indexes running over the interacting atoms within the peptide fragment of interest and those within a chosen receptor residue, with 0 ≤ strength ≤ ni nj (where ni and nj are the total number of atoms of the peptide fragment and the receptor residue, respectively, able to make the chosen type of interaction). For hydrophobic interactions, only carbon atoms are considered, for polar interactions only oxygens and nitrogens. The same “strength” function was calculated for salt bridges, π-cation and π-π interactions. In this case ni = nj = 1 because virtual atoms are defined at the center of mass of aromatic rings or charged groups, hence 0 ≤ strength ≤ 1. For hydrophobic, salt bridges, π-cation and π-π interactions a = 6, b = 12 and r0 = 6 Å, 5 Å, 4 Å, 5.5 Å respectively, while for polar interactions a = 8, b = 12, r0 = 2.5. The chosen values of r0 account for the typical interaction distance plus thermal motion’s amplitude (e.g. ~4.5 Å + ~1.5 Å in the case of hydrophobic interactions). The Plumed package [43] was patched to the Gromacs engine to undergo this kind of analysis. These coordination numbers (one per residue of the binding site) can be calculated on single structures as well as averaged along trajectories.
Beyond the roughness of the approach used here to monitor π–π and π-cation interactions (based only on the distances between c.o.m.), a careful characterization of them would require quantum mechanics. However, it is known that classical molecular dynamics using fixed-charge force fields has been successfully applied to describe π-stacking in both parallel and T-shaped configurations and π-cation [44], [45], [46].
4.6. Artwork
3D images of peptide-receptor structures were obtained by the Chimera software [47].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Financial support by the Italian Ministry of University and Research (Linea D1 Università Cattolica del Sacro Cuore) is gratefully acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.03.014.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus I, Research T. A Novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382: 2020; 727–733 [DOI] [PMC free article] [PubMed]
- 2.Cleary S.J., Pitchford S.C., Amison R.T., Carrington R., Robaina Cabrera C.L., Magnen M., Looney M.R., Gray E., Page C.P. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br J Pharmacol. 2020;177:4851–4865. doi: 10.1111/bph.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P. (2020) SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun 11: 2020; 5885 [DOI] [PMC free article] [PubMed]
- 4.Sironi M, Hasnain SE, Rosenthal B, Phan T, Luciani F, Shaw MA, Sallum MA, Mirhashemi ME, Morand S, Gonzalez-Candelas F, Editors of Infection G, Evolution. SARS-CoV-2 and COVID-19: A genetic, epidemiological, and evolutionary perspective. Infect Genet Evol 84: 2020; 104384 [DOI] [PMC free article] [PubMed]
- 5.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies J.P., Almasy K.M., McDonald E.F., Plate L. Comparative multiplexed interactomics of SARS-CoV-2 and homologous coronavirus nonstructural proteins identifies unique and shared host-cell dependencies. ACS Infect Dis. 2020 doi: 10.1021/acsinfecdis.0c00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petersen E, Koopmans M, Go U, Hamer DH, Petrosillo N, Castelli F, Storgaard M, Al Khalili S, Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis 20: 2020; e238–e244 [DOI] [PMC free article] [PubMed]
- 8.Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir Res. 2020;21:224. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.045. 894–904 e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amin M., Sorour M.K., Kasry A. Comparing the binding interactions in the receptor binding domains of SARS-CoV-2 and SARS-CoV. J Phys Chem Lett. 2020;11:4897–4900. doi: 10.1021/acs.jpclett.0c01064. [DOI] [PubMed] [Google Scholar]
- 12.Spinello A., Saltalamacchia A., Magistrato A. Is the rigidity of SARS-CoV-2 spike receptor-binding motif the hallmark for its enhanced infectivity? Insights from all-atom simulations. J Phys Chem Lett. 2020;11:4785–4790. doi: 10.1021/acs.jpclett.0c01148. [DOI] [PubMed] [Google Scholar]
- 13.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. 281–292 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., Liu M., Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc Natl Acad Sci U S A. 2020;117:13967–13974. doi: 10.1073/pnas.2008209117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowdhury R., Boorla V.S., Maranas C.D. Computational biophysical characterization of the SARS-CoV-2 spike protein binding with the ACE2 receptor and implications for infectivity. Comput Struct Biotechnol J. 2020;18:2573–2582. doi: 10.1016/j.csbj.2020.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurini E., Marson D., Aulic S., Fermeglia M., Pricl S. Computational alanine scanning and structural analysis of the SARS-CoV-2 spike protein/angiotensin-converting enzyme 2 complex. ACS Nano. 2020;14:11821–11830. doi: 10.1021/acsnano.0c04674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalathiya U, Padariya M, Mayordomo M, Lisowska M, Nicholson J, Singh A, Baginski M, Fahraeus R, Carragher N, Ball K, Haas J, Daniels A, Hupp TR, Alfaro JA. Highly conserved homotrimer cavity formed by the SARS-CoV-2 spike glycoprotein: a novel binding site. J Clin Med 9: 2020 [DOI] [PMC free article] [PubMed]
- 19.Shekhar N, Sarma P, Prajapat M, Avti P, Kaur H, Raja A, Singh H, Bhattacharya A, Sharma S, Kumar S, Prakash A, Medhi B. In silico structure-based repositioning of approved drugs for spike glycoprotein s2 domain fusion peptide of SARS-CoV-2: rationale from molecular dynamics and binding free energy calculations. mSystems. 5: 2020 [DOI] [PMC free article] [PubMed]
- 20.Andrianov A.M., Kornoushenko Y.V., Karpenko A.D., Bosko I.P., Tuzikov A.V. Computational discovery of small drug-like compounds as potential inhibitors of SARS-CoV-2 main protease. J Biomol Struct Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1792989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Abranyi-Balogh P., Brandao-Neto J., Carbery A., Davison G., Dias A., Downes T.D., Dunnett L., Fairhead M., Firth J.D., Jones S.P., Keeley A., Keseru G.M., Klein H.F., Martin M.P., Noble M.E.M., O'Brien P., Powell A., Reddi R.N., Skyner R., Snee M., Waring M.J., Wild C., London N., von Delft F., Walsh M.A. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirza M.U., Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J Pharm Anal. 2020;10:320–328. doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su H., Su H., Zhou F., Huang Z., Ma X., Natarajan K., Zhang M., Huang Y. Molecular insights into small molecule drug discovery for SARS-CoV-2. Angew Chem Int Ed Engl. 2020 doi: 10.1002/anie.202008835. [DOI] [PubMed] [Google Scholar]
- 24.Li Q., Yi D., Lei X., Zhao J., Zhang Y., Cui X., Xiao X., Jiao T., Dong X., Zhao X., Zeng H., Liang C., Ren L., Guo F., Li X., Wang J., Cen S. Corilagin inhibits SARS-CoV-2 replication by targeting viral RNA-dependent RNA polymerase. Acta Pharm Sin B. 2021 doi: 10.1016/j.apsb.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M RK, Gideon DA, Mariadasse R, Nirusimhan V, A SR, Edward JC, Jeyaraman J, Dhayabaran VT. In silico evaluation of isatin-based derivatives with RNA-dependent RNA polymerase of the novel coronavirus SARS-CoV-2. J Biomol Struct Dyn 2021: 1–16 [DOI] [PubMed]
- 26.Naydenova K., Muir K.W., Wu L.F., Zhang Z., Coscia F., Peet M.J., Castro-Hartmann P., Qian P., Sader K., Dent K., Kimanius D., Sutherland J.D., Lowe J., Barford D., Russo C.J. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian L., Qiang T., Liang C., Ren X., Jia M., Zhang J., Li J., Wan M., YuWen X., Li H., Cao W., Liu H. RNA-dependent RNA polymerase (RdRp) inhibitors: the current landscape and repurposing for the COVID-19 pandemic. Eur J Med Chem. 2021;213 doi: 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta R., Charron J., Stenger C.L., Painter J., Steward H., Cook T.W., Faber W., Frisch A., Lind E., Bauss J., Li X., Sirpilla O., Soehnlen X., Underwood A., Hinds D., Morris M., Lamb N., Carcillo J.A., Bupp C., Uhal B.D., Rajasekaran S., Prokop J.W. SARS-CoV-2 (COVID-19) structural and evolutionary dynamicome: Insights into functional evolution and human genomics. J Biol Chem. 2020;295:11742–11753. doi: 10.1074/jbc.RA120.014873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee S., Bhattacharyya D., Bhunia A. Host-membrane interacting interface of the SARS coronavirus envelope protein: immense functional potential of C-terminal domain. Biophys Chem. 2020;266 doi: 10.1016/j.bpc.2020.106452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teoh K.T., Siu Y.L., Chan W.L., Schluter M.A., Liu C.J., Peiris J.S., Bruzzone R., Margolis B., Nal B. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol Biol Cell. 2010;21:3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castano-Rodriguez C., Honrubia J.M., Gutierrez-Alvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Baguena C., Queralt-Martin M., Kochan G., Perlman S., Aguilella V.M., Sola I., Enjuanes L. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9 doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Maio F., Lo Cascio E., Babini G., Sali M., Della Longa S., Tilocca B., Roncada P., Arcovito A., Sanguinetti M., Scambia G., Urbani A. Improved binding of SARS-CoV-2 Envelope protein to tight junction-associated PALS1 could play a key role in COVID-19 pathogenesis. Microbes Infect. 2020 doi: 10.1016/j.micinf.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toto A., Ma S., Malagrino F., Visconti L., Pagano L., Stromgaard K., Gianni S. Comparing the binding properties of peptides mimicking the Envelope protein of SARS-CoV and SARS-CoV-2 to the PDZ domain of the tight junction-associated PALS1 protein. Protein Sci. 2020;29:2038–2042. doi: 10.1002/pro.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becerra A., Bucheli V.A., Moreno P.A. Prediction of virus-host protein-protein interactions mediated by short linear motifs. BMC Bioinf. 2017;18:163. doi: 10.1186/s12859-017-1570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanova M.E., Fletcher G.C., O'Reilly N., Purkiss A.G., Thompson B.J., McDonald N.Q. Structures of the human Pals1 PDZ domain with and without ligand suggest gated access of Crb to the PDZ peptide-binding groove. Acta Crystallogr D Biol Crystallogr. 2015;71:555–564. doi: 10.1107/S139900471402776X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Wei Z., Yan Y., Wan Q., Du Q., Zhang M. Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex. Proc Natl Acad Sci U S A. 2014;111:17444–17449. doi: 10.1073/pnas.1416515111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsson M.H., Sondergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 38.Jacobson M.P., Friesner R.A., Xiang Z., Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 39.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19. [Google Scholar]
- 40.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmuller H., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., Jo S., Pande V.S., Case D.A., Brooks C.L., 3rd, MacKerell A.D., Jr., Klauda J.B., Im W. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michaud-Agrawal N., Denning E.J., Woolf T.B., Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J Comput Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tribello G.A., Bonomi M., Branduardi D., Camilloni C., Bussi PLUMED 2: new feathers for an old bird. Comput Phys Commun. 2013;185:10. [Google Scholar]
- 44.Kang M., Zhang P., Cui H., Loverde S.M. Pi-pi stacking mediated chirality in functional supramolecular filaments. Macromolecules. 2016;49:994–1001. doi: 10.1021/acs.macromol.5b02148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macias A.T., Mackerell A.D., Jr. CH/pi interactions involving aromatic amino acids: refinement of the CHARMM tryptophan force field. J Comput Chem. 2005;26:1452–1463. doi: 10.1002/jcc.20281. [DOI] [PubMed] [Google Scholar]
- 46.Khan H.M., MacKerell A.D., Jr., Reuter N. Cation-pi interactions between methylated ammonium groups and tryptophan in the CHARMM36 additive force field. J Chem Theory Comput. 2019;15:7–12. doi: 10.1021/acs.jctc.8b00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.