Abstract
Xanthomonas oryzae pv. oryzae is a bacterial pathogen that gives rise to diseases in rice all over the world. A bacteriophage infecting this bacterium was isolated from rice fields in China. Here, we report the complete genome sequence of this phage, which has a linear dsDNA genome of 309,023 bp and a G + C content of 42.43%. It contains 401 open reading frames and encodes 28 tRNAs. It belongs to the family Myoviridae and has a broad host range, making it a possible candidate for phage therapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-021-04985-4.
Xanthomonas oryzae pv. oryzae is considered the most important agent of bacterial blight of rice [1], a disease that can cause major crop destruction and threatens global food security [10]. Bacteriophages are ubiquitous in the environment, and some have the potential to control bacterial diseases [2]. Therefore, various strategies have been developed to control pathogens by utilizing either a single phage or a cocktail of phages [3]. To gain further insights into the genetic diversity of Xanthomonas phages, we previously isolated a novel phage, Xoo-sp13, a polyvalent phage with a wide spectrum of activity and potential as a biocontrol agent against X. oryzae. This phage may be useful for curing bacterial diseases and other biotechnological applications.
Phage Xoo-sp13 was isolated from a soil sample in Shandong, East China, using the PXO99A strain, which was derived from Philippine race 6 strain PXO99, as a host. Methods for isolation, purification, and the host range determination of phage Xoo-sp13 were described previously [4]. The phage was visualized by transmission electron microscopy (TEM) at 200 kV, and images were produced at the Wuhan Institute of Virology, China. Phage genomic DNA was extracted using ZnCl2 precipitation [5]. A phage sequencing library was prepared using a NEBNext Ultra II kit v3 (New England Biolabs). Whole-genome sequencing of the phage was performed by Berry Genomics Biotechnology Co., Ltd (Beijing, China) using Illumina HiSeq 2500 paired-end sequencing technology. The complete genome sequence of Xoo-sp13 was annotated using Rapid Annotation using Subsystem Technology (RAST; http://rast.nmpdr.org) [6]. All predicted ORFs were checked manually against the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/) using PSI-BLAST (E-value = 0.0001) [7]. tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/index.html) was used for the prediction of genes encoding tRNAs [8].
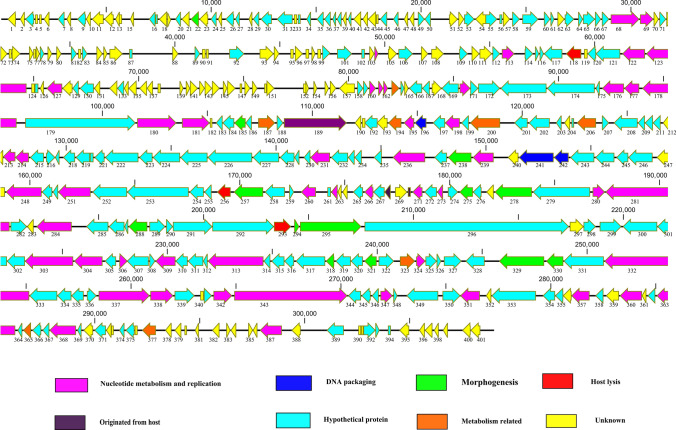
TEM results showed that Xoo-sp13 has an isometric head (60.56 ± 3 nm) and a contractile tail (121.56 ± 2 nm). Its genome sequence and morphology suggest that it belongs to the family Myoviridae, based on the current International Committee on Taxonomy of Viruses classification system (Fig. S1). A comparison of the genome sequence of Xoo-sp13 with other sequences in the GenBank Database using BLASTn showed it to be 75% identical to Xanthomonas phage XacN1 (accession no. AP018399.1). Phage XacN1 was isolated from a soil sample and infects Xanthomonas citri, a causative agent of Asian citrus canker. Xoo-sp13 had a linear dsDNA genome of 309,023 bp and a G+C content of 42.43%. Interestingly, Xoo-sp13 possesses unique characteristics due to the presence of a long terminal repeat of about 50 kb, which was detected as described previously [9]. A total of 401 ORFs and 28 tRNAs were found in the genome of Xoo-sp13 (Table S1). These include 135 genes that are unrelated to any genes from the prokaryotic or viral database and 188 that have been annotated as hypothetical proteins. Only 79 ORFs were identified as encoding proteins with known functions (Table S2), thus highlighting the novelty of this phage (Fig. 1). Phage Xoo-sp13 was tested against sixteen different strains of X. oryzae for host range analysis and found to infect 9 of them (Table S3), indicating a broad host range.
Fig. 1.
Schematic representation of the dsDNA genome of phage Xoo-sp13. Putative ORFs are presented as arrows, with predicted functions where available. Proposed modules are based on predicted functions. Turquoise, hypothetical protein; yellow, unknown; pink, nucleotide metabolism and replication; green, Morphogenesis; red, lysis; blue, DNA packaging. The map was drawn with CLC Genomics main Workbench version 3.6.1 (CLC bio, Aarhus, Denmark)
In conclusion, Xoo-sp13 is a bacteriophage with a large DNA genome and low sequence similarity to other known phages. It represents a new addition to the list of X. oryzae phages. Interestingly, most of the open reading frames in its genome are not functionally annotated. Determining the functions of these genes will be an exciting subject of study for understanding the biology of this novel phage.
Nucleotide sequence accession number
The complete genome sequence of phage Xoo-sp13 with annotations was submitted to the GenBank database under the accession number MN047793.
Supplementary Information
Below is the link to the electronic supplementary material.
Transmission electron microscopy of Xoo-sp13. Phages were stained with 2 % uranyl acetate. Morphology was observed under transmission electron microscope at institute of virology Wuhan, China (TIFF 5795 KB)
Funding
This research was supported by a grant from the National Key Research and Development Program of China (2018YFA0903000), the National Natural Science Foundation of China (81672001), and the China MoST Emergency Project on COVID-19 (2020YFC0840800).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Amina Nazir, Zhaoxia Dong and Jin Liu contributed equally.
Contributor Information
Hong Qing, Email: donghaipeng@mail.hzau.edu.cn.
Donghai Peng, Email: hqing@bit.edu.cn.
Yigang Tong, Email: tong.yigang@gmail.com.
References
- 1.Liu W, Liu J, Triplett L, Leach JE, Wang G-L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol. 2014;52:213–241. doi: 10.1146/annurev-phyto-102313-045926. [DOI] [PubMed] [Google Scholar]
- 2.Xi H, Dai J, Tong Y, Cheng M, Zhao F, Fan H, Li X, Cai R, Ji Y, Sun C. The characteristics and genome analysis of vB_AviM_AVP, the first phage infecting Aerococcus viridans. Viruses. 2019;11(2):104. doi: 10.3390/v11020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim MS, Hong SS, Park K, Myung H. Genomic analysis of bacteriophage PBECO4 infecting Escherichia coli O157: H7. Adv Virol. 2013;158(11):2399–2403. doi: 10.1007/s00705-013-1718-3. [DOI] [PubMed] [Google Scholar]
- 4.Dong Z, Xing S, Liu J, Tang X, Ruan L, Sun M, Tong Y, Peng D. Isolation and characterization of a novel phage Xoo-sp2 that infects Xanthomonas oryzae pv. oryzae. J Gen Virol. 2018;99(10):1453–1462. doi: 10.1099/jgv.0.001133. [DOI] [PubMed] [Google Scholar]
- 5.MrA S. An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. Nucleic Acids Res. 1991;19(19):5442. doi: 10.1093/nar/19.19.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. The RAST Server: rapid annotations using subsystems technology. BMC Genom. 2008;9(1):75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephen FA. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33(suppl_2):W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Wang Y, Tong Y, et al. Analyzing genome termini of bacteriophage through high-throughput sequencing. In: Clokie Martha RJ, et al., editors. Bacteriophages: Methods and Protocols. Springer; 2018. pp. 139–163. [DOI] [PubMed] [Google Scholar]
- 10.Nazir A, Dong Z, Liu J, Zhang X, Tahir RA, Ashraf N, Qing H, Peng D, Tong Y. Sequence analysis of a jumbo bacteriophage, Xoo-sp14, that infects Xanthomonas oryzae pv. oryzae. Microbiol Resour Announc. 2020;9:e01072–e1120. doi: 10.1128/MRA.01072-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transmission electron microscopy of Xoo-sp13. Phages were stained with 2 % uranyl acetate. Morphology was observed under transmission electron microscope at institute of virology Wuhan, China (TIFF 5795 KB)