Abstract
Genetic polymorphisms in select genes, including APOE (apolipoprotein E), COMT (Catechol-O-Methyltransferase), MDR1 (multi-drug resistance 1), BDNF (brain derived neurotrophic factor), and GST (glutathione-S-transferase), have been associated with vulnerability to cognitive impairment. In this study, we evaluated the relationship of these genetic variants to measures of brain health in patients with breast cancer, including neurocognitive testing and functional connectome analysis. Women with breast cancer (n = 83) and female healthy controls (n = 53) were evaluated. They underwent resting-state functional MRI scans and neurocognitive testing. Polymerase chain reaction (PCR) was performed on saliva samples to identify single nucleotide polymorphisms (SNPs) in candidate genes: APOE, COMT, MDR1, BDNF, and GST. Breast cancer patients treated with chemotherapy had slower processing speed (p = 0.04) and poorer reported executive function (p < 0.0001) than healthy controls. Those chemotherapy-treated patients that were APOE e4 carriers had significantly slower processing speed. A greater number of risk-related alleles was associated with poorer connectivity in the regions of the left cuneus and left calcarine. While breast cancer patients that are APOE e4 carriers may have a select vulnerability to processing speed impairments, other risk-related alleles were not found to influence cognitive test performance in this population. Conversely, regions of impaired functional connectivity appeared to be related to risk-related genetic polymorphisms in breast cancer patients. This suggests that a cancer patient’s SNPs in candidate genes may influence the risk of neurotoxicity. Further study evaluating the impact of genotype on biomarkers of brain health in cancer survivors is warranted.
Subject terms: Cancer, Neuroscience, Psychology, Neurology, Oncology
Introduction
With an increasing cancer incidence and decreasing death rate1, the public health impact of cancer survivorship is of central importance. In addition to the direct sequelae of a malignancy itself, cancer has an established impact on cognitive function2,3. Cancer patients have a 40% greater likelihood of reporting memory complaints than those without cancer, affecting those with active cancer and those in remission4. There are often multiple contributors to cancer related cognitive impairment (CRCI), with surgery, chemotherapy, radiation, psychosocial stress, and the cancer itself all having demonstrated contributions5. The biologic underpinnings of CRCI are likely complex, with both preclinical work and human neuroimaging studies have played central roles in elucidating some of these underlying mechanisms6. Mechanisms, including oxidative stress, mitotic inhibition of neural and glial progenitor cells, and induction of neural apoptosis have supportive contributory data7, and accelerated cortical ageing is now a central biologic construct in CRCI8.
More recently, the concept of germline genetic differences imparting increased vulnerability to CRCI has been explored, with genes associated with neurodegenerative diseases and enhanced toxicity from neurologic insults being implicated9–12. Carriers of the epsilon 4 allele of apolipoprotein E (APOE), associated with impaired neuronal membrane repair and synaptic plasticity13, has been associated with increased incidence of cognitive impairment after chemotherapy exposure9,14. Similarly, polymorphisms of other genes associated with neuronal and glial health and resilience, including COMT (Catechol-O-Methyltransferase), MDR1 (multi-drug resistance 1), BDNF (brain derived neurotrophic factor), and GST (glutathione-S-transferase), also have a proposed role in augmenting neurotoxicity11,15–17.
Neuroimaging studies have demonstrated changes in brain structure and function in cancer patients, lending insight into the pathophysiology of cognitive impairment in these patients6. Connectome analysis is the mathematical representation on the brain’s functional and structural networks. It provides insight into brain circuitry, including the integration and segregation of networks18–21. Functional connectome organization in this cohort of chemotherapy-treated breast cancer survivors compared to healthy female controls was previously evaluated, with decreased clustering as well as alterations of nodal degree in frontal, striatal and temporal areas being observed22. Here we expand upon those results by including a chemotherapy-naïve breast cancer comparison and exploring genetic contributions to chemotherapy-related connectome injury and cognitive deficit. We also focused on evaluation of regional effects using network-based analysis which represents an improvement over our previous approach.
Methods
Participants
For this retrospective study, we used all available cases from our laboratory for breast cancer survivors who had neuroimaging, neuropsychological and genetic data. This included 83 women age 41–74 years with a history of primary breast cancer (stage I-IIIA) who had completed their primary treatment (surgery, chemotherapy and/or locoregional radiation therapy) more than 6 months (mean = 5.5 + /- 5.2 years; range = 0.7–28 years) prior to study entry and were currently without evidence of active cancer or gross neuropathology. They were compared with 53 healthy female controls (Table 1). In the breast cancer group, 42 women had a history of chemotherapy treatment while the remaining 41 were chemotherapy naïve. Individual chemotherapy treatment protocols included doxorubicin/cyclophosphamde/paclitaxel or docetaxel = 16, cyclophosphamide/methotrexate/5-fluorouracil = 4, doxorubicin/cyclophosphamide = 11, cyclophosphamide/paclitaxel or docetaxel = 9, doxorubicin/cyclophosphamide/5-fluorouracil = 1 and epirubicin/ cyclophosphamide/paclitaxel = 1. Additional treatment information is provided in Table 1. The Stanford University Institutional Review Board approved this study which was conducted according to the principles expressed in the Declaration of Helsinki. All participants provided written informed consent.
Table 1.
Demographic, genotype and clinical data show as mean (standard deviation) unless otherwise noted.
| Chemotherapy N = 42 |
Chemotherapy naive N = 41 |
Healthy control N = 53 |
|
|---|---|---|---|
| Age | 55 (7) | 59 (7)* | 55 (9) |
| Age range | 43–73 | 41–74 | 41–71 |
| Education (yr) | 16 (3) | 17 (2) | 17 (3) |
| Breast radiation | 73% | 65% | |
| Endocrine therapy (tamoxifen) | 43% | 58% | |
| Disease stage at diagnosis (0, I, II, III) | 0%, 26%, 55%, 19% | 37%*, 50%*, 13%*, 0%* | |
| Time since primary treatment (yr) | 4.8 (4.8) | 6.5 (5.6) | |
| Postmenopausal | 89% | 82% | 44%* |
| Clinical assessment of depression score | 51 (11)* | 44 (10) | 44 (9) |
| APOE e4 | 31% | 24% | 23% |
| BDNF Met | 43% | 28% | 36% |
| MDR1 T | 81% | 78% | 83% |
| GST Null | 38% | 42% | 51% |
| COMT Val | 88% | 91% | 95% |
*Denotes significant (p < 0.05) difference.
Breast cancer survivors were excluded for history of relapse or prior anti-cancer treatment. All participants were excluded for diagnosed psychiatric, neurologic or comorbid medical conditions that are known to affect cognitive function as well as pregnancy, MRI contraindications or major sensory deficits (e.g. blindness)23. Breast cancer and comparison participants were recruited via the Army of Women (http://www.armyofwomen.org/), community flyer postings and advertisements. Breast cancer participants were also recruited via local support groups and physician referrals from the Stanford Women’s Cancer Center.
Neurocognitive function testing
Subtests from the Wechsler Adult Intelligence Scales Fourth Edition24 were used to measure processing speed (Symbol Search) and working memory (Digit Span). Executive function was assessed using the Neuropsychological Assessment Battery Categories test25 and verbal fluency was measured using the Delis-Kaplan Executive Function System Letter Fluency subtest26. Verbal memory was assessed using the Hopkins Verbal Learning Test Revised Version27, which assesses both immediate and delayed recall. We also administered the Behavioral Rating Inventory of Executive Function (BRIEF), a patient reported measure of executive function28 and the Clinical Assessment of Depression (CAD) which measures depression, anxiety and fatigue29. Test scores were converted to T scores (mean = 50 + /- 10) based on the published normative data for each test. Some participants had missing data for certain assessments.
MRI acquisitions
A subset of participants underwent MRI scanning which was performed on a GE Discovery MR750 3.0 T whole body scanner (GE Medical Systems, Milwaukee, WI). Functional MRI (fMRI) data were acquired while participants rested in the scanner with their eyes closed using a T2* weighted gradient echo spiral pulse sequence: relaxation time = 2000 ms, echo time = 30 ms, flip angle = 80° and 1 interleave, field of view = 220, matrix = 64 × 64, in-plane resolution = 3.125. Number of data frames collected was 216, thus total scan time was 7:12. An automated high-order shimming method based on spiral acquisitions was employed to reduce field heterogeneity30. To coregister and normalize functional images with a standardized template, a high-resolution, 3 dimension inversion-recovery prepared fast spoiled gradient echo anatomical scan was acquired: relaxation time: minimum, echo time: minimum, flip: 11 degrees, inversion time: 300 ms, bandwidth: ± 31.25 kHz, field of view: 24 cm, phase field of view: 0.75, slice thickness: 1.5 mm, 125 slices, 256 × 256 at 1 excitation, scan time: 4:26. Two task based fMRI scans and a diffusion weighted scan were also acquired during the MRI session that are utilized in other analyses not reported here. However, the resting state scan was acquired prior to any other scans to reduce the effects of specific cognitive processes on the resting state networks.
Functional MRI preprocessing
Image preprocessing was performed using Statistical Parametric Mapping 8 (Wellcome Trust Centre, London, UK). Briefly, images were realigned to correct for head motion, segmented and coregistered with the anatomic MRI, normalized to a standard space (MNI) template and smoothed with an 8 mm FWHM kernel. We defined 90 cortical and subcortical regions of interest (ROIs) using the Automated Anatomical Labeling atlas31 which were used to extract regional time series data. This was performed using the CONN Toolbox32. First, data were band pass filtered to 0.008 Hz—0.09 Hz. Then, the CompCor method33 was used to reduce physiological and other non-neuronal artifacts. This method involves extracting signal from white matter and cerebrospinal fluid regions using principal component analysis and then regressing these signals out of the total fMRI signal. Finally, temporal correlations between all possible pairs of regions were computed based on the corrected fMRI signal and normalized to z-scores, resulting in a 90 × 90 functional connectivity matrix for each participant. Negative functional edges were zeroed given evidence that properties of negative correlation networks are different than those of positive correlation networks34,35.
Genotyping
Saliva samples were obtained from all participants using the Oragene DNA OG-250 collection kit (DNA Genotek, Kanata, Ontario). Genotyping was accomplished by polymerase chain reaction (PCR) fragment length polymorphism analysis with restricted fragment length polymorphisms for APOE, COMT, MDR1, BDNF, GST.
Statistical analysis
Neurocognitive function
Test scores were compared between groups using analysis of covariance controlling for age. Significant omnibus tests were supplemented with Tukey’s honestly significant difference test.
Connectome
Global connectome properties including clustering, path length, and small-worldness were calculated using graph theoretical analysis as described previously36–39. Briefly, a network is defined as a set nodes (regions) and their connections (edges). The clustering coefficient of a node is the proportion of a nodes neighbors that are also neighbors with each other and is a measure of network segregation. Path length describes the minimum number of edges that separate pairs of nodes and is an indication of network integration. Small-worldness is an organization characteristic of brain networks and other large-scale complex biological networks that differentiates them from random networks. Small-worldness was calculated as [CC/CCR]/[PL/PLR] where CC = clustering coefficient and PL = characteristic path length and CCR and PLR are the mean clustering coefficient and characteristic path length of 100 random benchmark networks40.
Thresholding connectomes is necessary for removing false positive edges and facilitating between group comparisons but can remove potentially valid information regarding differences in network topology41,42. Therefore, we compared connectome properties across multiple densities using the area under the curve (AUC)43,44. Connectomes were first corrected for age via linear regression conducted at every ROI45. We then measured connectome properties at each density from minimum connection density to the last density associated with a small-world organization46,47 up to a maximum density of 0.548 and calculated the AUC across this entire range. AUCs were compared pairwise between groups using nonparametric permutation testing using 5000 iterations and two-tailed p values as in our previous studies22,49. Specifically, AUCs from randomized groups with the same number of participants as the original groups were calculated to create a permutation distribution of between-group differences. The actual between group difference was then placed in the corresponding permutation distribution and a two-tailed p value was calculated based on its percentile position45.
Regional connectome differences were assessed using the Network-Based Statistic50 with age as a covariate. Unlike our previous approach which examined connectivity for individual nodes, this method identifies connected substructures, or components, within the larger network50. Permutation testing with 5000 permutations was then used to determine group differences in components controlling for multiple comparisons using false discovery rate (FDR)50.
Neurocognitive test scores and connectome properties that showed between group differences were further compared between genotypes (e.g. presence vs. absence of APOE e4, COMT Val, BDNF Met, MDR1 T and GST null variants) within each participant group using t-tests or Wilcoxon rank tests, as appropriate, based on data visualization.
Results
Neurocognitive function
As shown in Table 2, the chemotherapy group demonstrated lower processing speed as measured by the Symbol Search test (F = 3.2, p = 0.04). They also rated themselves as having significantly more difficulties with executive functions than the other two groups (F = 20.4, p < 0.0001). Although it differed between groups, CAD score was not significantly associated with any objective cognitive scores but was related to BRIEF score (r = 0.62, p < 0.0001). However, the group comparison remained significant even after adding CAD as a covariate (F = 12.0, p < 0.0001).
Table 2.
Neurocognitive function data shown as mean (standard deviation).
| N | Chemotherapy | N | Chemotherapy naive | N | Healthy control | p value | |
|---|---|---|---|---|---|---|---|
| Verbal fluency | 42 | 55 (11) | 41 | 58 (11) | 53 | 60 (12) | 0.09 |
| HVLT-total | 42 | 52 (8.8) | 41 | 53 (7.6) | 41 | 53 (10) | 0.79 |
| HVLT-delayed | 42 | 53 (7.5) | 41 | 53 (9.1) | 41 | 52 (9.8) | 0.99 |
| NAB categories | 42 | 52 (9.8) | 41 | 54 (7.5) | 39 | 53 (9.4) | 0.66 |
| Symbol search | 29 | 59 (8.3) | 29 | 64 (9.3) | 35 | 60 (9.6) | 0.04 |
| Digit span | 41 | 53 (8.3) | 40 | 55 (9.2) | 49 | 55 (9.7) | 0.55 |
| BRIEF | 42 | 61 (11) | 41 | 50 (9.7) | 52 | 48 (8.4) | < .0001 |
Connectome properties
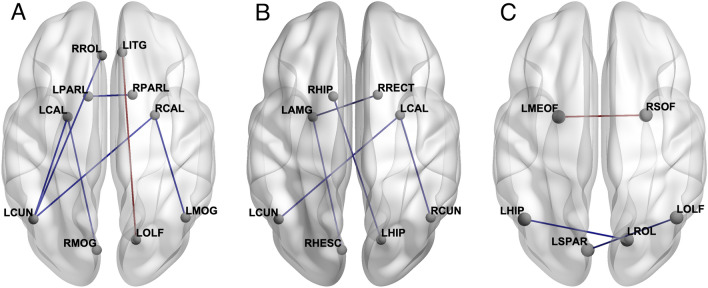
The chemotherapy group demonstrated significantly lower clustering compared to the chemotherapy naïve group (p = 0.02). Additionally, they showed significantly lower characteristic path length (p = 0.04). The chemotherapy group showed hypo- and hyper-connectivity between several regions (Fig. 1).
Figure 1.
The chemotherapy group demonstrated significantly (p < 0.05, FDR corrected) hypo-connected (blue lines) and hyper-connected (red lines) edges compared to the chemotherapy naïve group (A) and hypo-connected edges compared to healthy controls (B). The chemotherapy naïve group showed both hypo-connected (blue lines) and hyper-connected (red lines) edges compared to healthy control (C). LAMG left amygdala, LCAL left calcarine, LCUN left cuneus, LHIP left hippocampus, LITG left inferior temporal gyrus, LMEOF left medial orbital frontal, LMOG left middle occipital gyrus, LOLF left olfactory, LPARL left parietal lobule, LROL left rolandic operculum, LSPAR left superior parietal lobe, RCAL right calcarine, RCUN right cuneus, RHESC right Heschl’s gyrus, RHIP right hippocampus, RMOG right middle occipital gyrus, RPARL right parietal lobule, RRECT right rectus gyrus, RROL right rolandic operculum, RSOF right superior orbital frontal. Figure created using BrainNet Viewer51.
Compared to healthy controls, the chemotherapy group demonstrated significantly lower clustering (p = 0.005) and characteristic path length (p = 0.02). Regionally, there were multiple areas of hypo-connectivity in the chemotherapy group (Fig. 1).
There were no differences in global connectome metrics between healthy controls and chemotherapy naïve survivors. However, there were some regions of hyper- and hypo-connectivity in the chemotherapy naïve group (Fig. 1). CAD score was not associated with connectome metrics (p > 0.52).
Genotype and neurocognitive function
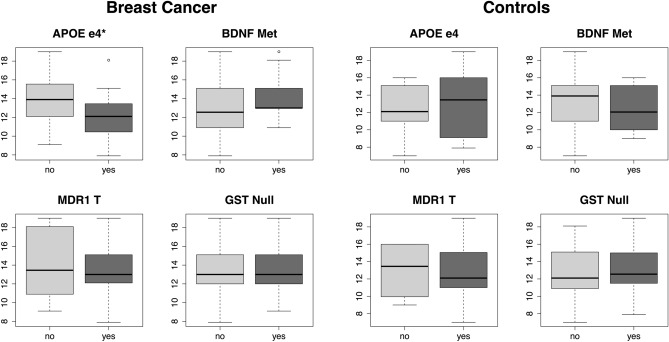
Symbol Search (processing speed) performance was significantly lower in chemotherapy-treated survivors who had the APOE e4 allele (Fig. 2). No other comparisons were significant.
Figure 2.
Processing speed performance by genotype. Symbol Search test score differed significantly only between APOE e4 groups. * p < 0.05.
Genotype and connectome function
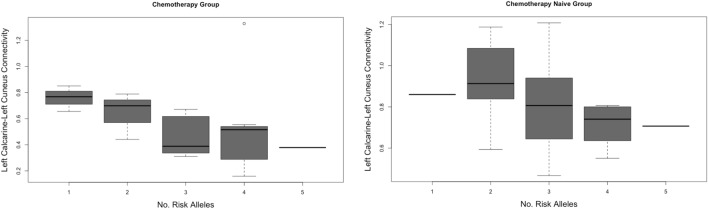
There were no differences between genotype groups and global connectome properties. Regionally, connectivity between the left calcarine and left cuneus, which was lower in the chemotherapy group compared to both control groups, was significantly lower in chemotherapy-treated survivors who had the BDNF Met allele (W = 84, p = 0.04) as well as those who possessed the MDR1 T allele (W = 49, p = 0.02). Additionally, the connectivity between right and left paracentral lobule, which was lower in the chemotherapy group compared to chemotherapy naïve group, was significantly lower in chemotherapy-treated survivors who had the MDR1 T allele (W = 3, p = 0.01). Further, as shown in Fig. 3, lower left calcarine to left cuneus connectivity was significantly correlated with higher number of risk-related alleles in the chemotherapy group (r = −0.51, p = 0.02) and in the chemotherapy naïve group (r = −0.47, p = 0.01), but not in healthy controls (p = 0.20). In healthy controls who carried the APOE e4 allele, connectivity between left olfactory and left superior parietal lobe was higher (W = 30, p < 0.0001) while connectivity between left hippocampus and left rolandic operculum was lower (W = 199, p = 0.004). No other genotype comparisons or correlations were significant. There was not enough variance to conduct comparisons for the COMT genotype.
Figure 3.
Correlation between left calcarine-left cuneus connectivity and number of risk-related alleles in the breast cancer groups.
Discussion
This innovative study evaluates the relationship of genetic polymorphisms associated with neural ageing and vulnerability to neurologic insults on cerebral connectivity. This is the first study to our knowledge where both cognitive function and the functional connectome, a demonstrated biomarker of CRCI18,22,52, have been evaluated in relation to these genetic signatures. With roles identified in neuronal repair and survival, dendritic and axonal growth, long term potentiation, CNS concentrations of cytotoxic therapies16,53–57, the genes evaluated in this study have an association with brain ageing, a proposed concept behind CRCI8,58. The functional connectome, representing key contributions of genetic, epigenetic, and environmental variables59–61, has been shown to be a viable biomarker of neurologic impact of cancer18,52,62. In this study, we identified that patients harboring those genetic signatures associated with accelerate cognitive ageing have associated alterations in brain functional connectivity, supporting genetic relationship to cancer-related neurotoxicity.
Of all cognitive domains evaluated on clinical testing, only processing speed had a significant interrelationship with genotype, with APOE4 carriers showing impairment. The lack of association with other cognitive functions may be secondary to the low rates of cognitive impairment identified in this population as a whole. A lack of sensitivity of formal neuropsychological evaluation in corroborating subjective cognitive complaints has previously been identified63. The disparity between self-reported and objectively defined cognitive dysfunction is further supported in this study, with patients rating themselves as having significant cognitive issues illustrated by different BRIEF scores between the groups, despite the lack of executive dysfunction by clinical testing. The adult children of Alzheimer’s patients carrying the APOE4 allele Alzheimer’s have been found to have slower processing speed than those that do not harbor this allele, and have lower white matter volumes64. Processing speed is a commonly affected domain in CRCI, and the APOE4 allele could render this domain particularly vulnerable to dysfunction58.
A distinct global connectome organization was demonstrated in the chemotherapy group, including lower clustering, shorter characteristic path length, and hypo- and hyper-connectivity between multiple regions. These distinct alterations in large-scale brain networks have been highlighted previously in chemotherapy-treated patients22, highlighting the general impairments in information processing and network integration that can occur in cancer survivors. Conversely, no global differences were observed in connectome properties between genotype groups. There are several potential explanations for this finding. The selected at-risk alleles may not impact on global connectivity, instead leading to more focused regions of aberrant cerebral connectivity with relative preservation of global brain function. Furthermore, differences in the allele frequency in our test group may have led to these distinct results. While variable amongst ethnicities65, the prevalence of APOE4 carriers in the general population is estimated at 14% globally66, a smaller proportion than in our study population. Similarly, the MDR1 T allele has been identified in 53%67, but was higher in our evaluated population. As such, these could alter our identification of abnormalities.
The genetic polymorphisms evaluated in this study have may also predispose to cancer itself, adding further complexity to the interrelationship with brain health. APOE has been found to mediate tissue repair, immune response and regulation, and cell growth and differentiation68, biologic processes that influence individual susceptibility to malignancy. A meta-analysis evaluating the APOE gene in breast cancer patients identified one and a half times increased risk of breast cancer among Asians carrying the e4 protein isoform of APOE, supporting this allele as a low-penetrant risk factor for development of breast cancer69. As the COMT gene is an estrogen-metabolizing enzyme, polymorphisms have been postulated to impact breast cancer risk, with catechol estrogens having proposed oncogenic influence. Despite this theoretical relationship, population-based studies have failed to demonstrate a compelling relationship between COMT polymorphisms and breast cancer carcinogenesis70,71. As the MDR1 gene encodes the ATP-dependent cellular efflux pump p-glycoprotein, protecting cells from xenobiotics and metabolites, it has also had a proposed role in carcinogenesis. Multiple cancer types have been associated with the MDR1 C3435T polymorphism in a meta-analysis of case–control studies72. These findings suggest complex interrelationships between these genetic polymorphisms and carcinogenesis and may render individuals vulnerable to both cancer the neurotoxicity of its therapies. These relationships warrant further exploration.
In contrast to global connectivity, regional changes were found to be related to at-risk allele status. Regional connectivity involving left calcarine and left cuneus as well as right and left paracentral lobule was significantly lower in chemotherapy-treated patients treated with BDNF Met allele or MDR1 T allele. Further, lower left calcarine and left cuneus connectivity was also associated with a higher number of at-risk alleles in the breast cancer population, independent of chemotherapy exposure, but not in healthy controls. These regions have had implicated vulnerability in other imaging studies of CRCI. In a study of hippocampal-cortical functional connectivity, increased functional connectivity was demonstrated between the hippocampus and left cuneus in breast cancer survivors, as opposed to healthy controls73. We previously observed significant hyper-activation in left (and right) cuneus among chemotherapy-treated breast cancer survivors compared to healthy controls during a verbal memory task74. Increased perfusion in the left cuneus and left calcarine has been demonstrated in patients with breast cancer exposed to chemotherapy compared to both healthy and chemotherapy-naïve controls, a finding also correlated with dysfunction changes in executive control network scores75. Our analysis suggests dysfunction in these regions may be influenced by the presence of at-risk alleles, a finding that warrants further exploration.
Another unique finding in our analysis were the differences in connectivity between left olfactory and left superior parietal lobe as well as left hippocampus and left rolandic operculum for healthy controls with the APOE e4 allele. The olfactory system and hippocampi are of central interest in neurobiology given their roles in neurogenesis. Neurogenesis extends well-beyond the embryonic and post-natal system in select regions of the brain. The subventricular zone generates interneurons for the olfactory bulb, and the olfactory neurons generate new excitatory sensory neurons that send their axons to the olfactory bulb76. Similarly, adult neurogenesis also occurs in the subgranular zone of the hippocampal dentate gyrus77. Alterations of connectivity in these regions among individuals harboring the APOE e4 allele may be associated with increased vulnerability to age-related cognitive decline. For example, APOE e4 is a risk factor for Alzheimer’s disease, where neurogenesis is known to be dysfunctional78. However, post hoc analysis did not indicate significant differences in cognitive performance between healthy controls based on APOE e4 genotype in this sample. It is also unclear why this finding was specific to healthy controls but may suggest an altered relationship between genotype and neurogenesis in patients with breast cancer that requires further study.
The potential for genetic polymorphisms to modulate the impact of neurologic insults has been identified in several diseases79,80, but is not defined in cancer. Recent systematic review has found variable correlations between genetic polymorphisms and CRCI81, precluding the conclusive definition of a relationship. Connectomics provides a novel contribution to elucidating this relationship. The data presented in this study supports at-risk alleles impacting vulnerability to CRCI, with the numbers of alleles expressed in an individual predicting poorer cerebral connectivity. Limitations of this study include the small number of patients studied and the differential frequencies of at-risk alleles compared to the normal population. The latency between initial breast cancer therapy and neurocognitive testing and imaging was greater than 5 years, significantly longer than most studies of CRCI, which generally evaluate patients within a year of treatment completion82–84. This prolonged interval may have influenced our identification of cognitive impairment on formal testing and our findings on connectome analysis. However, many studies have identified cognitive deficit in cross-sectional cohorts of breast cancer survivors who are decades off-therapy85,86. Therefore, many factors may contribute such as choice of neuropsychological tests and/or small sample. The focused study of breast cancer patients also suggests these findings should be validated in different oncology patient groups to determine their generalizability. Furthermore, a predictive relationship between these genetic polymorphisms and CRCI or connectome alterations cannot be definitively drawn given the lack of baseline neurocognitive and neuroimaging data. Larger prospective study in this area will allow for more definitive analysis of the influence of genetic polymorphisms on brain network topology and the development of CRCI. Ultimately, a comprehensive understanding of at-risk alleles to the development of cancer-related neurotoxicity may assist in identifying high-risk patients, allowing for appropriate counseling and risk-reduction strategies to be enacted where feasible.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee (Stanford Institutional Review Board, #14623) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Author contributions
R.A.H., V.R., and S.R.K. were involved in the concept and design of the study, V.R. and S.K. performed the analysis, R.A.H. and S.R.K. wrote the main manuscript text. All authors reviewed the manuscript.
Funding
This study was supported by the National Institutes of Health DP2OD004445, R01CA226080 and R01CA172145 grants.
Data availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Brezden CB, Phillips KA, Abdolell M, Bunston T, Tannock IF. Cognitive function in breast cancer patients receiving adjuvant chemotherapy. J. Clin. Oncol. 2000;18(14):2695–2701. doi: 10.1200/JCO.2000.18.14.2695. [DOI] [PubMed] [Google Scholar]
- 3.Schagen SB, Muller MJ, Boogerd W, Rosenbrand RM, van Rhijn D, Rodenhuis S, van Dam FS. Late effects of adjuvant chemotherapy on cognitive function: a follow-up study in breast cancer patients. Ann. Oncol. 2002;13(9):1387–1397. doi: 10.1093/annonc/mdf241. [DOI] [PubMed] [Google Scholar]
- 4.Janelsins MC, Kesler SR, Ahles TA, Morrow GR. Prevalence, mechanisms, and management of cancer-related cognitive impairment. Int. Rev. Psychiatry (Abingdon, England) 2014;26(1):102–113. doi: 10.3109/09540261.2013.864260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison RA, Wefel JS. Neurocognitive function in adult cancer patients. Neurol. Clin. 2018;36(3):653–674. doi: 10.1016/j.ncl.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Miao H, Li J, Hu S, He X, Partridge SC, Ren J, Bian Y, Yu Y, Qiu B. Long-term cognitive impairment of breast cancer patients after chemotherapy: a functional MRI study. Eur. J. Radiol. 2016;85(6):1053–1057. doi: 10.1016/j.ejrad.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Vichaya EG, Chiu GS, Krukowski K, Lacourt TE, Kavelaars A, Dantzer R, Heijnen CJ, Walker AK. Mechanisms of chemotherapy-induced behavioral toxicities. Front Neurosci. 2015;9:131. doi: 10.3389/fnins.2015.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henneghan A, Rao V, Harrison RA, Karuturi M, Blayney DW, Palesh O, Kesler SR. Cortical brain age from pre-treatment to post-chemotherapy in patients with breast cancer. Neurotoxicol. Res. 2020;37:788–799. doi: 10.1007/s12640-019-00158-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speidell AP, Demby T, Lee Y, Rodriguez O, Albanese C, Mandelblatt J, Rebeck GW. Development of a human APOE knock-in mouse model for study of cognitive function after cancer chemotherapy. Neurotox. Res. 2019;35(2):291–303. doi: 10.1007/s12640-018-9954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng H, Li W, Gan C, Zhang B, Jia Q, Wang K. The COMT (rs165599) gene polymorphism contributes to chemotherapy-induced cognitive impairment in breast cancer patients. Am. J. Transl. Res. 2016;8(11):5087–5097. [PMC free article] [PubMed] [Google Scholar]
- 11.Ng T, Lee YY, Chae J-W, Yeo AHL, Shwe M, Gan YX, Ng RCH, Chu PPY, Khor CC, Ho HK, Chan A. Evaluation of plasma brain-derived neurotrophic factor levels and self-perceived cognitive impairment post-chemotherapy: a longitudinal study. BMC Cancer. 2017;17(1):867–867. doi: 10.1186/s12885-017-3861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng T, Teo SMJ, Yeo HLA, Shwe M, Gan YX, Cheung YT, Tan YP, Yong WS, Madhukumar P, Loh KW-J, Koo S-L, Jain A, Lee GE, Wong M, Dent RA, Yap YS, Ng RCH, Khor CC, Ho HK, Chan A. Evaluation of brain-derived neurotrophic factor (BDNF) genetic polymorphism (rs6265) on chemotherapy-associated cognitive impairment in early-stage breast cancer (ESBC) patients. J. Clin. Oncol. 2015;33(15_suppl):9616. doi: 10.1200/jco.2015.33.15_suppl.9616. [DOI] [Google Scholar]
- 13.White F, Nicoll JA, Roses AD, Horsburgh K. Impaired neuronal plasticity in transgenic mice expressing human apolipoprotein E4 compared to E3 in a model of entorhinal cortex lesion. Neurobiol. Dis. 2001;8(4):611–625. doi: 10.1006/nbdi.2001.0401. [DOI] [PubMed] [Google Scholar]
- 14.Ahles TA, Saykin AJ, Noll WW, Furstenberg CT, Guerin S, Cole B, Mott LA. The relationship of APOE genotype to neuropsychological performance in long-term cancer survivors treated with standard dose chemotherapy. Psychooncology. 2003;12(6):612–619. doi: 10.1002/pon.742. [DOI] [PubMed] [Google Scholar]
- 15.Small BJ, Rawson KS, Walsh E, Jim HS, Hughes TF, Iser L, Andrykowski MA, Jacobsen PB. Catechol-O-methyltransferase genotype modulates cancer treatment-related cognitive deficits in breast cancer survivors. Cancer. 2011;117(7):1369–1376. doi: 10.1002/cncr.25685. [DOI] [PubMed] [Google Scholar]
- 16.Kerb R. Implications of genetic polymorphisms in drug transporters for pharmacotherapy. Cancer Lett. 2006;234(1):4–33. doi: 10.1016/j.canlet.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 17.Barahmani N, Carpentieri S, Li XN, Wang T, Cao Y, Howe L, Kilburn L, Chintagumpala M, Lau C, Okcu MF. Glutathione S-transferase M1 and T1 polymorphisms may predict adverse effects after therapy in children with medulloblastoma. Neuro Oncol. 2009;11(3):292–300. doi: 10.1215/15228517-2008-089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kesler SR, Adams M, Packer M, Rao V, Henneghan AM, Blayney DW, Palesh O. Disrupted brain network functional dynamics and hyper-correlation of structural and functional connectome topology in patients with breast cancer prior to treatment. Brain Behav. 2017;7(3):e00643–e00643. doi: 10.1002/brb3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menning S, de Ruiter MB, Veltman DJ, Koppelmans V, Kirschbaum C, Boogerd W, Reneman L, Schagen SB. Multimodal MRI and cognitive function in patients with breast cancer prior to adjuvant treatment–the role of fatigue. Neuroimage Clin. 2015;7:547–554. doi: 10.1016/j.nicl.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Chen H, Lv Y, Chao HH, Gong L, Li C-SR, Cheng H. Diminished gray matter density mediates chemotherapy dosage-related cognitive impairment in breast cancer patients. Sci. Rep. 2018;8(1):13801–13801. doi: 10.1038/s41598-018-32257-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen C-Y, Chen VC-H, Yeh D-C, Huang S-L, Zhang X-R, Chai J-W, Huang Y-H, Chou M-C, Weng J-C. Association of functional dorsal attention network alterations with breast cancer and chemotherapy. Sci. Rep. 2019;9(1):104. doi: 10.1038/s41598-018-36380-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruno J, Hosseini SM, Kesler S. Altered resting state functional brain network topology in chemotherapy-treated breast cancer survivors. Neurobiol. Dis. 2012;48(3):329–338. doi: 10.1016/j.nbd.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kesler SR, Watson CL, Blayney DW. Brain network alterations and vulnerability to simulated neurodegeneration in breast cancer. Neurobiol. Aging. 2015;36(8):2429–2442. doi: 10.1016/j.neurobiolaging.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wechsler, D., Wechsler Adult Intelligence Scale Fourth Edition. 2008, San Antonio, TX: The Psychological Corporation.
- 25.Stern RAW. Neuropsychological Assessment Battery. Lutz, FL: Psychological Assessment Resources, Inc; 2003. [Google Scholar]
- 26.Delis D, Kaplan E, Kramer J. Delis-Kaplan Executive Function System. Psychological Corporation; 2001. [Google Scholar]
- 27.Benedict RHB, Schretlen D, Groninger L, Brandt J. Hopkins verbal learning test revised: normative data and analysis of inter-form and test-retest reliability. Clin. Neuropsychol. 1998;12(1):43–55. doi: 10.1076/clin.12.1.43.1726. [DOI] [Google Scholar]
- 28.Roth RM, Isquith PK, Gioia G. Behavioral Rating Inventory of Executive Function—Adult Version. Lutz, FL: Psychological Assessment Resources; 2005. [Google Scholar]
- 29.Aghakhani, A., and E.K. Chan (2007) Test Reviews: Bracken, B. A., & Howell, K. (2004). Clinical assessment of depression. Odessa, FL: Psychological Assessment Resources. J. Psychoeduc. Assessment 2007. 25(4), 416–422.
- 30.Glover GH, Lai S. Self-navigated spiral fMRI: interleaved versus single-shot. Magn. Reson. Med. 1998;39(3):361–368. doi: 10.1002/mrm.1910390305. [DOI] [PubMed] [Google Scholar]
- 31.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 32.Whitfield-Gabrieli S, Nieto-Castanon A. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2012;2(3):125–141. doi: 10.1089/brain.2012.0073. [DOI] [PubMed] [Google Scholar]
- 33.Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37(1):90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hosseini SM, Kesler SR. Comparing connectivity pattern and small-world organization between structural correlation and resting-state networks in healthy adults. Neuroimage. 2013;78:402–414. doi: 10.1016/j.neuroimage.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz AJ, McGonigle J. Negative edges and soft thresholding in complex network analysis of resting state functional connectivity data. Neuroimage. 2011;55(3):1132–1146. doi: 10.1016/j.neuroimage.2010.12.047. [DOI] [PubMed] [Google Scholar]
- 36.Bassett DS, Bullmore E. Small-world brain networks. Neuroscientist. 2006;12(6):512–523. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- 37.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10(3):186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 38.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52(3):1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Sporns O. The human connectome: a complex network. Ann. N. Y. Acad. Sci. 2011;1224:109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- 40.Zalesky A, Fornito A, Bullmore E. On the use of correlation as a measure of network connectivity. Neuroimage. 2012;60(4):2096–2106. doi: 10.1016/j.neuroimage.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 41.van Wijk BCM, Stam CJ, Daffertshofer A. Comparing brain networks of different size and connectivity density using graph theory. PLoS ONE. 2010;5(10):e13701. doi: 10.1371/journal.pone.0013701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fornito A, Zalesky A, Breakspear M. Graph analysis of the human connectome: promise, progress, and pitfalls. Neuroimage. 2013;80:426–444. doi: 10.1016/j.neuroimage.2013.04.087. [DOI] [PubMed] [Google Scholar]
- 43.Bassett DS, Meyer-Lindenberg A, Achard S, Duke T, Bullmore E. Adaptive reconfiguration of fractal small-world human brain functional networks. Proc. Natl. Acad. Sci. USA. 2006;103(51):19518–19523. doi: 10.1073/pnas.0606005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bassett DS, Nelson BG, Mueller BA, Camchong J, Lim KO. Altered resting state complexity in schizophrenia. Neuroimage. 2012;59(3):2196–2207. doi: 10.1016/j.neuroimage.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosseini SM, Hoeft F, Kesler SR. GAT: a graph-theoretical analysis toolbox for analyzing between-group differences in large-scale structural and functional brain networks. PLoS ONE. 2012;7:e40709. doi: 10.1371/journal.pone.0040709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A. Hierarchical organization of human cortical networks in health and schizophrenia. J. Neurosci. 2008;28(37):9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Humphries MD, Gurney K. Network 'small-world-ness': a quantitative method for determining canonical network equivalence. PLoS ONE. 2008;3(4):e0002051. doi: 10.1371/journal.pone.0002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaiser M, Hilgetag CC. Nonoptimal component placement, but short processing paths, due to long-distance projections in neural systems. PLoS Comput. Biol. 2006;2(7):e95. doi: 10.1371/journal.pcbi.0020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kesler SR, Ogg R, Reddick WE, Phillips N, Scoggins M, Glass JO, Cheung YT, Pui CH, Robison LL, Hudson MM, Krull KR. Brain network connectivity and executive function in long-term survivors of childhood acute lymphoblastic leukemia. Brain Connect. 2018;8(6):333–342. doi: 10.1089/brain.2017.0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zalesky A, Fornito A, Bullmore ET. Network-based statistic: identifying differences in brain networks. Neuroimage. 2010;53(4):1197–1207. doi: 10.1016/j.neuroimage.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 51.Xia M, Wang J, He Y. BrainNet Viewer: a network visualization tool for human brain connectomics. PLoS ONE. 2013;8(7):e68910. doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kesler SR, Gugel M, Huston-Warren E, Watson C. Atypical structural connectome organization and cognitive impairment in young survivors of acute lymphoblastic leukemia. Brain Connect. 2016;6(4):273–282. doi: 10.1089/brain.2015.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lima Giacobbo B, Doorduin J, Klein HC, Dierckx RAJO, Bromberg E, de Vries EFJ. Brain-derived neurotrophic factor in brain disorders: focus on neuroinflammation. Mol. Neurobiol. 2019;56(5):3295–3312. doi: 10.1007/s12035-018-1283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savitz J, Solms M, Ramesar R. The molecular genetics of cognition: dopamine, COMT and BDNF. Genes Brain Behav. 2006;5(4):311–328. doi: 10.1111/j.1601-183X.2005.00163.x. [DOI] [PubMed] [Google Scholar]
- 55.Korwek KM, Trotter JH, Ladu MJ, Sullivan PM, Weeber EJ. ApoE isoform-dependent changes in hippocampal synaptic function. Mol. Neurodegen. 2009;4:21–21. doi: 10.1186/1750-1326-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horch HW. Local effects of BDNF on dendritic growth. Rev. Neurosci. 2004;15(2):117–129. doi: 10.1515/REVNEURO.2004.15.2.117. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y.X., Y. Ma, H. Zhang, W.P. Zhang, and X.Y. Yang, Genetic polymorphism in MDR1 C3435T is a determinant of methotrexate cerebrospinal fluid concentrations in Chinese children with acute lymphoblastic leukemia.Int. J. Clin. Pharmacol. Ther. 2020. [DOI] [PubMed]
- 58.Ahles TA, Root JC, Ryan EL. Cancer and cancer treatment-associated cognitive change: an update on the state of the science. J. Clin. Oncol. 2012;30(30):3675–3686. doi: 10.1200/JCO.2012.43.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ingalhalikar M, Smith A, Parker D, Satterthwaite TD, Elliott MA, Ruparel K, Hakonarson H, Gur RE, Gur RC, Verma R. Sex differences in the structural connectome of the human brain. Proc. Natl. Acad. Sci. USA. 2014;111(2):823–828. doi: 10.1073/pnas.1316909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krishnadas R, Kim J, McLean J, Batty GD, McLean JS, Millar K, Packard CJ, Cavanagh J. The envirome and the connectome: exploring the structural noise in the human brain associated with socioeconomic deprivation. Front Hum. Neurosci. 2013;7:722. doi: 10.3389/fnhum.2013.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun J, Tong S, Yang GY. Reorganization of brain networks in aging and age-related diseases. Aging Dis. 2012;3(2):181–193. [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng Y, Cheng ASK, Song T, Sheng X, Zhang Y, Liu X, Chan CCH. Subjective cognitive impairment and brain structural networks in Chinese gynaecological cancer survivors compared with age-matched controls: a cross-sectional study. BMC Cancer. 2017;17(1):796. doi: 10.1186/s12885-017-3793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bray VJ, Dhillon HM, Vardy JL. Systematic review of self-reported cognitive function in cancer patients following chemotherapy treatment. J. Cancer Surviv. 2018;12(4):537–559. doi: 10.1007/s11764-018-0692-x. [DOI] [PubMed] [Google Scholar]
- 64.Ready RE, Baran B, Chaudhry M, Schatz K, Gordon J, Spencer RM. Apolipoprotein E-e4, processing speed, and white matter volume in a genetically enriched sample of midlife adults. Am. J. Alzheimers Dis. Other Demen. 2011;26(6):463–468. doi: 10.1177/1533317511421921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kern S, Mehlig K, Kern J, Zetterberg H, Thelle D, Skoog I, Lissner L, Blennow K, Borjesson-Hanson A. The distribution of apolipoprotein E genotype over the adult lifespan and in relation to country of birth. Am. J. Epidemiol. 2015;181(3):214–217. doi: 10.1093/aje/kwu442. [DOI] [PubMed] [Google Scholar]
- 66.ALZGENE. Meta-analysis of all published AD association studies (case-control only) APOE_E2/3/4. 2010 March 15, 2020]; Available from: http://www.alzgene.org/meta.asp? geneID=83.
- 67.Baldissera VD, de Mattos AA, Coral GP, de Araujo FB, Marroni CA, de Mello Brandao AB, Ott Fontes PR, Schmidt Cerski CT, Hartmann AA, Kretzmann Filho NA. Evaluation of the C3435T polymorphism in the MDR1 gene in patients with hepatocellular carcinoma. Ann. Hepatol. 2012;11(6):899–906. doi: 10.1016/S1665-2681(19)31416-4. [DOI] [PubMed] [Google Scholar]
- 68.Dose J, Huebbe P, Nebel A, Rimbach G. APOE genotype and stress response—a mini review. Lipids Health Dis. 2016;15:121. doi: 10.1186/s12944-016-0288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saadat M. Apolipoprotein E (APOE) polymorphisms and susceptibility to breast cancer: a meta-analysis. Cancer Res. Treat. 2012;44(2):121–126. doi: 10.4143/crt.2012.44.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peterson NB, Trentham-Dietz A, Garcia-Closas M, Newcomb PA, Titus-Ernstoff L, Huang Y, Chanock SJ, Haines JL, Egan KM. Association of COMT haplotypes and breast cancer risk in caucasian women. Anticancer Res. 2010;30(1):217–220. [PMC free article] [PubMed] [Google Scholar]
- 71.Bergman-Jungestrom M, Wingren S. Catechol-O-Methyltransferase (COMT) gene polymorphism and breast cancer risk in young women. Br. J. Cancer. 2001;85(6):859–862. doi: 10.1054/bjoc.2001.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheng X, Zhang L, Tong N, Luo D, Wang M, Xu M, Zhang Z. MDR1 C3435T polymorphism and cancer risk: a meta-analysis based on 39 case-control studies. Mol. Biol. Rep. 2012;39(7):7237–7249. doi: 10.1007/s11033-012-1554-7. [DOI] [PubMed] [Google Scholar]
- 73.Apple AC, Schroeder MP, Ryals AJ, Wagner LI, Cella D, Shih PA, Reilly J, Penedo FJ, Voss JL, Wang L. Hippocampal functional connectivity is related to self-reported cognitive concerns in breast cancer patients undergoing adjuvant therapy. Neuroimage Clin. 2018;20:110–118. doi: 10.1016/j.nicl.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kesler SR, Bennett FC, Mahaffey ML, Spiegel D. Regional brain activation during verbal declarative memory in metastatic breast cancer. Clin. Cancer Res. 2009;15(21):6665–6673. doi: 10.1158/1078-0432.CCR-09-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen X, He X, Tao L, Cheng H, Li J, Zhang J, Qiu B, Yu Y, Wang K. The attention network changes in breast cancer patients receiving neoadjuvant chemotherapy: evidence from an arterial spin labeling perfusion study. Sci. Rep. 2017;7:42684. doi: 10.1038/srep42684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brann JH, Firestein SJ. A lifetime of neurogenesis in the olfactory system. Front Neurosci. 2014;8:182. doi: 10.3389/fnins.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Toda T, Parylak SL, Linker SB, Gage FH. The role of adult hippocampal neurogenesis in brain health and disease. Mol. Psychiatry. 2019;24(1):67–87. doi: 10.1038/s41380-018-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Demars M, Hu YS, Gadadhar A, Lazarov O. Impaired neurogenesis is an early event in the etiology of familial Alzheimer's disease in transgenic mice. J. Neurosci. Res. 2010;88(10):2103–2117. doi: 10.1002/jnr.22387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rajan KB, Aggarwal NT, Schneider JA, Wilson RS, Everson-Rose SA, Evans DA. Role of APOE ε4 Allele and incident stroke on cognitive decline and mortality. Alzheimer Dis. Assoc. Disord. 2016;30(4):318–323. doi: 10.1097/WAD.0000000000000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Houlden H, Greenwood R. Apolipoprotein E4 and traumatic brain injury. J. Neurol. Neurosurg. Psychiatry. 2006;77(10):1106–1107. doi: 10.1136/jnnp.2006.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buskbjerg CDR, Amidi A, Demontis D, Nissen ER, Zachariae R. Genetic risk factors for cancer-related cognitive impairment: a systematic review. Acta Oncol. 2019;58(5):537–547. doi: 10.1080/0284186X.2019.1578410. [DOI] [PubMed] [Google Scholar]
- 82.Bender CM, Sereika SM, Berga SL, Vogel VG, Brufsky AM, Paraska KK, Ryan CM. Cognitive impairment associated with adjuvant therapy in breast cancer. Psychooncology. 2006;15(5):422–430. doi: 10.1002/pon.964. [DOI] [PubMed] [Google Scholar]
- 83.Hermelink K, Untch M, Lux MP, Kreienberg R, Beck T, Bauerfeind I, Münzel K. Cognitive function during neoadjuvant chemotherapy for breast cancer: results of a prospective, multicenter, longitudinal study. Cancer. 2007;109(9):1905–1913. doi: 10.1002/cncr.22610. [DOI] [PubMed] [Google Scholar]
- 84.Stewart A, Collins B, Mackenzie J, Tomiak E, Verma S, Bielajew C. The cognitive effects of adjuvant chemotherapy in early stage breast cancer: a prospective study. Psychooncology. 2008;17(2):122–130. doi: 10.1002/pon.1210. [DOI] [PubMed] [Google Scholar]
- 85.van der Willik KD, Koppelmans V, Hauptmann M, Compter A, Ikram MA, Schagen SB. Inflammation markers and cognitive performance in breast cancer survivors 20 years after completion of chemotherapy: a cohort study. Breast Cancer Res. 2018;20(1):135. doi: 10.1186/s13058-018-1062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koppelmans V, Breteler MM, Boogerd W, Seynaeve C, Gundy C, Schagen SB. Neuropsychological performance in survivors of breast cancer more than 20 years after adjuvant chemotherapy. J. Clin. Oncol. 2012;30(10):1080–1086. doi: 10.1200/JCO.2011.37.0189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.