Abstract
Exocytosis is a vesicle fusion process driven by soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs). A classic exocytic pathway is insulin-stimulated translocation of the glucose transporter type 4 (GLUT4) from intracellular vesicles to the plasma membrane in adipocytes and skeletal muscles. The GLUT4 exocytic pathway plays a central role in maintaining blood glucose homeostasis and is compromised in insulin resistance and type 2 diabetes. A candidate regulator of GLUT4 exocytosis is tomosyn, a soluble protein expressed in adipocytes. Tomosyn directly binds to GLUT4 exocytic SNAREs in vitro but its role in GLUT4 exocytosis was unknown. In this work, we used CRISPR-Cas9 genome editing to delete the two tomosyn-encoding genes in adipocytes. We observed that both basal and insulin-stimulated GLUT4 exocytosis was markedly elevated in the double knockout (DKO) cells. By contrast, adipocyte differentiation and insulin signaling remained intact in the DKO adipocytes. In a reconstituted liposome fusion assay, tomosyn inhibited all the SNARE complexes underlying GLUT4 exocytosis. The inhibitory activity of tomosyn was relieved by NSF and α-SNAP, which act in concert to remove tomosyn from GLUT4 exocytic SNAREs. Together, these studies revealed an inhibitory role for tomosyn in insulin-stimulated GLUT4 exocytosis in adipocytes. We suggest that tomosyn-arrested SNAREs represent a reservoir of fusion capacity that could be harnessed to treat patients with insulin resistance and type 2 diabetes.
Keywords: exocytosis, membrane fusion, membrane protein, membrane trafficking, vesicle fusion, SNARE, glucose transporter type 4 (GLUT4), tomosyn, insulin signaling
INTRODUCTION
Exocytosis is a vesicle fusion event delivering membrane proteins to the cell surface and soluble molecules to the extracellular space (1). A classic exocytic pathway is insulin-stimulated exocytosis of the glucose transporter GLUT4, which plays a central role in maintaining blood glucose homeostasis (2-5). Under the basal condition, GLUT4 is largely sequestered in intracellular storage vesicles in adipocytes and skeletal muscles (6-10). Insulin stimulation activates a cascade of signaling events that ultimately triggers the fusion of GLUT4 storage vesicles (GSVs) with the plasma membrane, delivering GLUT4 to the cell surface to facilitate glucose uptake (2, 11-16). Impairment of insulin-stimulated GLUT4 exocytosis is a hallmark of insulin resistance and type 2 diabetes (6, 11, 17, 18).
Exocytosis, like other intracellular vesicle fusion pathways, is driven by SNARE complexes assembled from the vesicle-anchored v-SNARE and target membrane-rooted t-SNAREs (1, 19-21). SNARE complexes zipper progressively toward membranes, forcing the vesicle and target membrane into close proximity to fuse (19, 22-26). In GLUT4 exocytosis, the primary v-SNARE is VAMP2/synaptobrevin whereas VAMP3/cellubrevin and VAMP8/endobrevin play redundant or compensatory roles (27-32). The t-SNAREs of GLUT4 exocytosis are syntaxin-4 and SNAP-23 (18, 33-35).
Besides SNAREs, an exocytic pathway also requires SNARE-binding regulatory factors to achieve spatial and temporal precision of the vesicle fusion reaction (36, 37). A candidate regulator of GLUT4 exocytosis is tomosyn, a soluble 120-130 kDa protein that negatively regulates SNARE-dependent membrane fusion (38-46). Tomosyn possesses two distinct domains – a large N-terminal domain containing WD40 repeats and a small C-terminal domain harboring a v-SNARE-like motif (46-49). Both of the N- and C-terminal domains of tomosyn are involved in SNARE association (50, 51). Tomosyn is expressed in insulin-responsive tissues including adipocytes (52, 53), and its overexpression in adipocytes reduces GLUT4 exocytosis (52). In vitro, tomosyn directly binds to GLUT4 exocytic t-SNAREs and prevents the formation of SNARE complexes (50, 54). These data suggest that tomosyn has the potential to negatively regulate GLUT4 vesicle fusion. However, direct loss-of-function evidence for a role of tomosyn in the GLUT4 exocytic pathway was still lacking.
In this work, we used CRISPR-Cas9 genome editing to delete the two tomosyn-encoding genes (Tomosyn-1 and Tomosyn-2) in adipocytes. We observed that both basal and insulin-stimulated GLUT4 exocytosis was markedly elevated in Tomosyn-1/2 double knockout (DKO) cells. Adipocyte differentiation and insulin signaling remained intact in the mutant cells. We then characterized the molecular mechanism of tomosyn in GLUT4 vesicle fusion using a reconstituted liposome fusion system. We observed that tomosyn bound to GLUT4 exocytic t-SNAREs and arrested the fusion reaction. Tomosyn inhibited all the SNARE complexes involved in GLUT4 exocytosis. Tomosyn was removed from GLUT4 exocytic t-SNAREs by NSF and α-SNAP, relieving its inhibitory function in the fusion reaction. Together, these studies uncovered an inhibitory role of tomosyn in GLUT4 exocytosis in adipocytes.
RESULTS
DKO of Tomosyn-1 and Tomosyn-2 using CRISPR-Cas9 genome editing
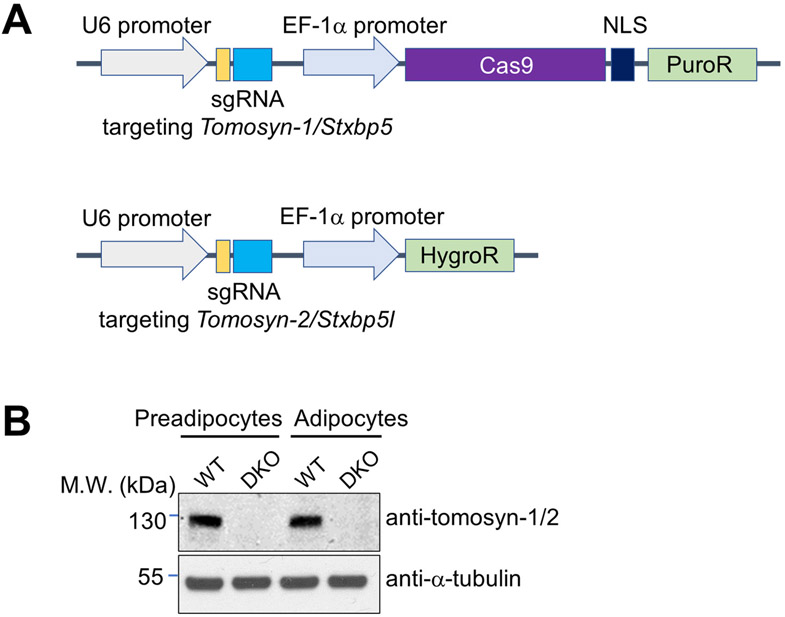
The mouse genome possesses two tomosyn-encoding genes – Tomosyn-1 (Stxbp5) and Tomosyn-2 (Stxbp5l). The sequences of tomosyn-1 and tomosyn-2 proteins are highly similar and both of them are expressed in mouse adipocytes (53, 55). To investigate the functional role of tomosyn in insulin-stimulated GLUT4 exocytosis, we deleted both Tomosyn-1 and Tomosyn-2 genes in mouse preadipocytes using CRISPR-Cas9 genome editing (Fig. 1A). Wild-type (WT) and Tomosyn-1/2 DKO preadipocytes were then differentiated into mature adipocytes. We observed that tomosyn protein expression was abolished in Tomosyn-1/2 DKO preadipocytes and mature adipocytes (Figs. 1B and S1).
Figure 1. Deletion of Tomosyn-1 and Tomosyn-2 using CRISPR-Cas9 genome editing.
A) Diagrams of CRISPR constructs used to generate Tomosyn-1/2 DKO cells. NLS: nuclear localization signal. B) Representative immunoblots showing the expression of the indicated proteins in WT or Tomosyn-1/2 DKO preadipocytes and adipocytes. The anti-tomosyn antibodies recognize both tomosyn-1 and tomosyn-2. A full anti-tomosyn blot is shown in Figure S1A. Tomosyn mRNA levels were not significantly altered in the DKO cells (Figure S1B), indicating that the loss of tomosyn protein expression was due to protein degradation.
Intact differentiation and insulin signaling in Tomosyn-1/2 DKO adipocytes
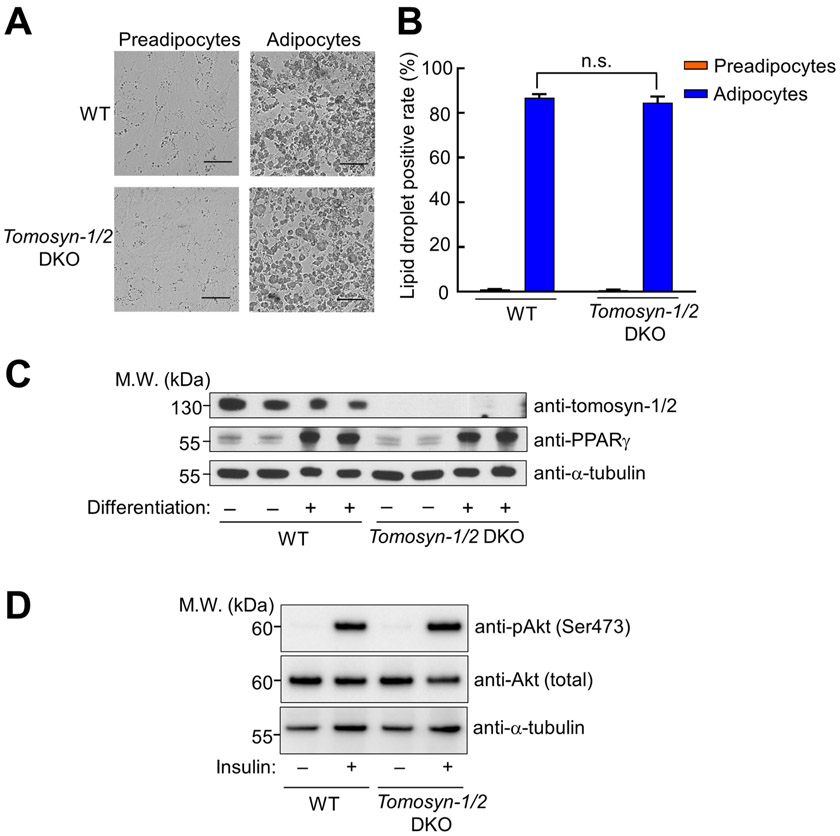
The overall morphology of Tomosyn-1/2 DKO adipocytes was indistinguishable from that of WT adipocytes (Fig. 2A). A characteristic feature of mature adipocytes is the presence of large lipid droplets (Fig. 2A) (11, 56). Using microscopy and flow cytometry, we observed comparable lipid droplets in WT and Tomosyn-1/2 DKO adipocytes (Fig. 2A-B). The expression of peroxisome proliferator-activated receptor gamma (PPARγ), a central regulator of adipocyte differentiation (57), was also comparable in WT and Tomosyn-1/2 DKO cells (Fig. 2C). These results indicate that tomosyn is dispensable for adipocyte differentiation.
Figure 2. Normal adipocyte differentiation and insulin signaling in Tomosyn-1/2 DKO cells.
(A) Representative images of preadipocytes and adipocytes captured using a 20x objective on an Olympus IX81 Microscope. Scale bars: 100 μm. (B) Quantification of lipid droplet-positive cells. Cells were stained with Nile red and the fluorescence of Nile red was measured using flow cytometry. Data are presented as mean ± SD. n = 3. P values were calculated using Student’s t-test. n.s., P>0.05. (C) Representative immunoblots showing the expression of the indicated proteins in preadipocytes (before differentiation) and adipocytes (after differentiation). Two independent samples were prepared for each cell line. (D) Representative immunoblots showing the indicated proteins in WT and Tomosyn-1/2 DKO adipocytes.
Next, we examined whether insulin signaling is impacted in Tomosyn-1/2 DKO cells. Insulin stimulation triggered the phosphorylation and activation of Akt/PKB, a key kinase in the insulin signaling pathway controlling GLUT4 trafficking and other cellular responses (Fig. 2D) (53, 58, 59). We found that insulin-stimulated Akt phosphorylation was similar between WT and Tomosyn-1/2 DKO adipocytes (Fig. 2D). Thus, tomosyn does not regulate insulin signaling.
GLUT4 exocytosis is elevated in Tomosyn-1/2 DKO adipocytes
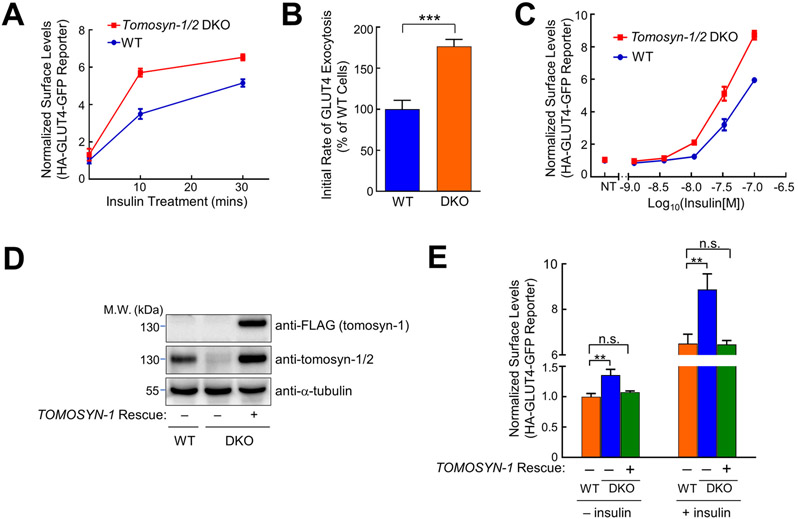
To determine the role of tomosyn in GLUT4 exocytosis, we used flow cytometry to measure the translocation of an HA-GLUT4-GFP reporter stably expressed in adipocytes (60, 61). This GLUT4 reporter accurately recapitulates insulin-regulated GLUT4 trafficking in live cells (60-62). Insulin stimulation strongly promoted the translocation of the GLUT4 reporter to the cell surface (Figs. 3A). We observed that insulin-stimulated GLUT4 translocation was markedly increased in Tomosyn-1/2 DKO adipocytes (Fig. 3A). Kinetic analysis revealed that the elevated translocation of the GLUT4 reporter was due to its accelerated exocytosis in the DKO cells (Fig. 3B). We also measured the responses of the adipocytes to various concentrations of insulin. Elevated GLUT4 translocation was observed in the DKO cells at all the insulin concentrations (Fig. 3C). The basal surface level of the GLUT4 reporter (without insulin stimulation) was also increased in the DKO adipocytes (Fig. 3A and C). We then used confocal microscopy to visualize GLUT4 reporters. We observed that surface levels of the GLUT4 reporter were substantially higher in Tomosyn-1/2 DKO adipocytes under both unstimulated and insulin-stimulated conditions (Fig. S2). These results are consistent with the flow cytometry data and demonstrate that tomosyn negatively regulates both basal and insulin-stimulated GLUT4 exocytosis in adipocytes.
Figure 3. GLUT4 exocytosis is elevated in Tomosyn-1/2 DKO adipocytes.
(A) Normalized surface levels of the GLUT4 reporter in WT and Tomosyn-1/2 DKO adipocytes. After treatment with 100 nM insulin for the indicated periods, surface levels of the GLUT4 reporter were measured using flow cytometry and normalized to untreated WT samples. Data are presented as mean ± SD. n = 3. (B) Initial exocytosis rates of the GLUT4 reporter in WT and Tomosyn-1/2 DKO adipocytes. Initial exocytosis rates were calculated based on increases in surface levels of the GLUT4 reporter during the first 10 minutes of insulin treatment shown in A. Datasets are normalized to untreated WT samples. Data are presented as mean ± SD. n = 3. P values were calculated using Student’s t-test. *** P<0.001. (C) Dose responses of insulin-stimulated translocation of the GLUT4 reporter in WT and Tomosyn-1/2 DKO adipocytes. Cells were treated with the indicated concentrations of insulin for 30 minutes before surface levels of the GLUT4 reporter were measured by flow cytometry. Datasets are normalized to untreated WT samples. Data are presented as mean ± SD. n = 3. (D) Representative immunoblots showing the expression levels of the indicated proteins in preadipocytes. The rescue gene encodes a 3xFLAG-tagged tomosyn-1 protein. (E) Normalized surface levels of the GLUT4 reporter in WT and Tomosyn-1/2 DKO adipocytes. In rescue samples, the human TOMOSYN-1 gene was expressed in Tomosyn-1/2 DKO adipocytes. Datasets are normalized to untreated WT samples. Data are presented as mean ± SD. n = 3. P values were calculated using Two-way ANOVA. ** P<0.01. n.s., P>0.05.
To rule out off-target effects, we expressed the human TOMOSYN-1 gene in the Tomosyn-1/2 DKO cells (Fig. 3D). The human TOMOSYN-1 gene, which was not targeted by the mouse Tomosyn-1/2 gRNAs, restored tomosyn expression (Fig. 3D). Using flow cytometry and confocal microcopy, we observed that the human TOMOSYN-1 gene fully rescued the phenotype of the DKO cells such that both basal and insulin-stimulated GLUT4 translocation returned to WT levels (Figs. 3E and S2). Thus, the accelerated GLUT4 exocytosis observed in the DKO adipocytes was caused by a loss of tomosyn expression. Since insulin signaling was intact in the DKO cells (Fig. 2D), the elevated GLUT4 exocytosis was not due to abnormal insulin signaling, consistent with a direct role of tomosyn in SNARE-mediated vesicle fusion. Together, these genetic studies established an inhibitory role of tomosyn in insulin-stimulated GLUT4 exocytosis.
Tomosyn inhibits all SNARE complexes involved in GLUT4 exocytosis
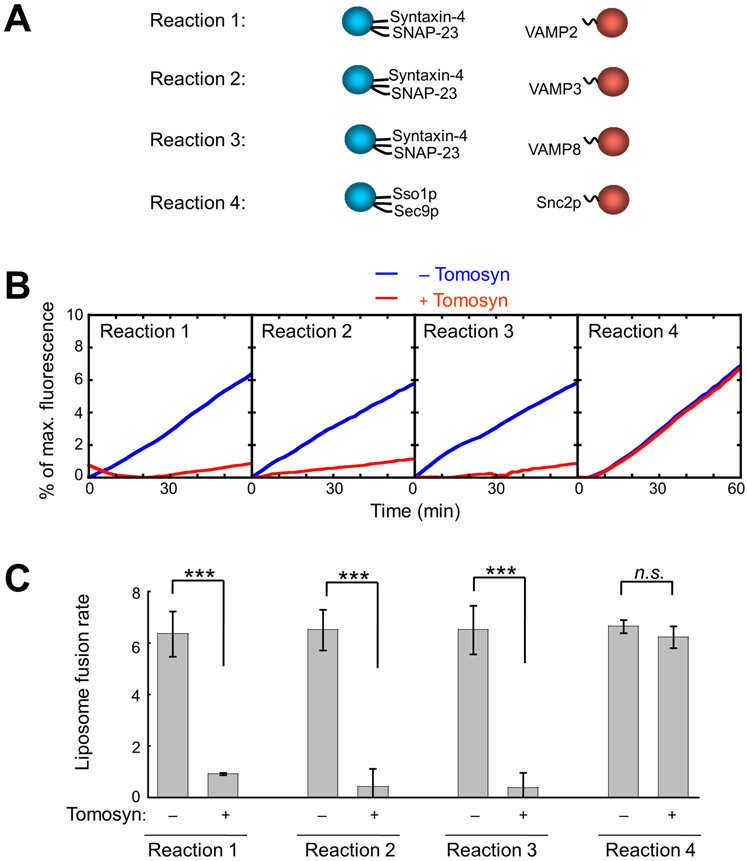
Tomosyn binds to GLUT4 exocytic SNAREs (Fig. S3) (50, 52), but it remains incompletely understood how it regulates SNARE-mediated GLUT4 vesicle fusion. To address this question, we reconstituted tomosyn into a liposome fusion reaction mediated by GLUT4 exocytic SNAREs – syntaxin-4, SNAP-23 and VAMP2 (Fig. 4A). A major advantage of this defined system is that regulatory factors can be individually added and perturbed. As such, their effects on membrane fusion kinetics can be precisely determined without the complication of other factors naturally present in the cell. Previous biochemical studies of tomosyn were mainly performed in solution using truncated fragments (48, 63). While these studies yielded important initial insights, it is critical to examine full-length (FL) tomosyn because both the N-terminal and C-terminal regions are involved in its function (50, 51, 64, 65).
Figure 4. Tomosyn inhibits all SNARE complexes involved in GLUT4 vesicle fusion.
(A) Illustrations of the v- and t-SNARE pairs in liposome fusion reactions. Each fusion reaction contained 5 μM t-SNAREs, 1.5 μM v-SNARE, and 100 mg/mL Ficoll 70 as the crowding agent. (B) Liposome fusion reactions depicted in A were carried out in the absence or presence of 5 μM recombinant FL tomosyn-1. Lipid mixing of the liposomes was measured using a FRET-based assay. (C) Lipid-mixing rates of the liposome fusion reactions shown in B. Data are presented as mean ± SD. n = 3. P values were calculated using Student’s t-test. *** P<0.001. n.s., P>0.05.
Using FL tomosyn proteins prepared in an insect cell expression system (50), we first determined how tomosyn regulates the SNARE complexes involved in GLUT4 exocytosis. Tomosyn bound to GLUT4 exocytic t-SNAREs (syntaxin-4 and SNAP-23) and prevented their pairing with the v-SNARE VAMP2, arresting the fusion reaction at an intermediate stage (Fig. 4) (50). By contrast, tomosyn had no effect on a liposome fusion reaction driven by yeast exocytic SNAREs – Sso1p, Sec9p and Snc2p (Fig. 4), suggesting that tomosyn inhibits membrane fusion through specific interactions with SNAREs. We also tested how tomosyn regulates the fusion of preincubated SNARE liposomes. Low-temperature preincubation allows v- and t-SNAREs to form partially assembled trans-SNARE complexes between membrane bilayers (66). Tomosyn strongly inhibited the fusion of the preincubated SNARE liposomes (Fig. S4), suggesting that it can also arrest membrane fusion when SNARE complexes are partially assembled. Using a liposome co-flotation assay, we observed that fully assembled SNARE complexes remained intact when tomosyn was added (Fig. S5). Thus, tomosyn is unable to dissociate VAMP2 from t-SNAREs when the SNARE complex is fully assembled.
The v-SNAREs VAMP3 and VAMP8 also participate in GLUT4 exocytosis (30), but it was unclear whether tomosyn inhibits VAMP3- or VAMP8-mediated fusion reactions. It is critical to examine these v-SNAREs because a v-SNARE may directly relieve the inhibitory activity of a SNARE regulator (67). Liposomes bearing VAMP3 or VAMP8 fused with t-SNARE liposomes as efficiently as VAMP2 liposomes (Fig. 4B-C). We observed that these liposome fusion reactions were also strongly inhibited by tomosyn (Fig. 4B-C). Thus, the inhibitory activity of tomosyn cannot be overcome by any of the v-SNAREs involved in GLUT4 exocytosis. Together, these data demonstrate that tomosyn inhibits all the three SNARE complexes mediating GLUT4 vesicle fusion.
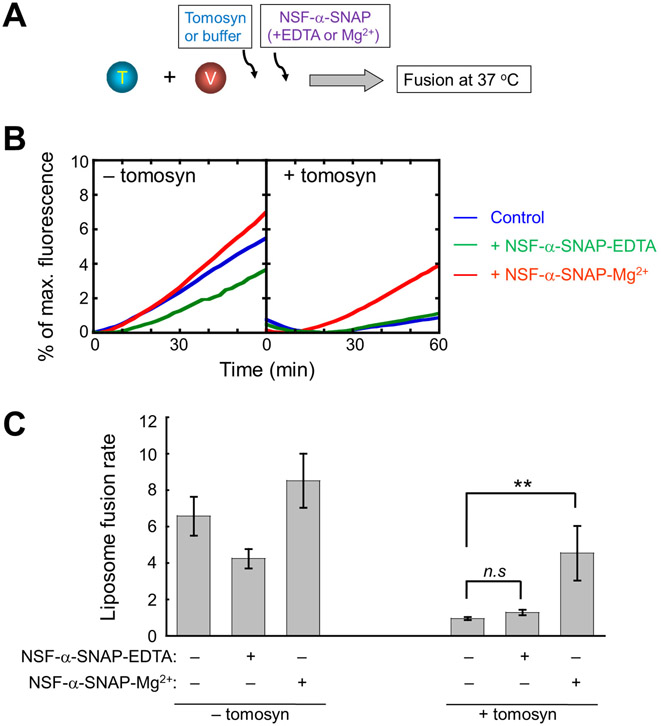
NSF and α-SNAP relieve the inhibitory activity of tomosyn in GLUT4 vesicle fusion
We then examined how the tomosyn-SNARE interaction is regulated by NSF and α-SNAP, a pair of SNARE-binding factors recycling SNARE complexes (68-70). Although NSF and α-SNAP are known to dissociate v-SNAREs and v-SNARE-like motifs from t-SNAREs (48, 63), it was unclear whether FL tomosyn could be dissociated from GLUT4 exocytic t-SNAREs. We observed that, while tomosyn strongly inhibited SNARE-mediated liposome fusion, the fusion was restored when NSF and α-SNAP were added in the presence of ATP and Mg2+ (Fig. 5A-C). In the absence of tomosyn, NSF and α-SNAP had limited effects on SNARE-mediated liposome fusion (Fig. 5B-C). Thus, NSF and α-SNAP are able to relieve the inhibitory activity of FL tomosyn in GLUT4 vesicle fusion.
Figure 5. NSF and α-SNAP relieve the inhibitory activity of tomosyn in GLUT4 vesicle fusion.
(A) Diagram illustrating the reconstituted liposome fusion reactions. The t-SNARE liposomes containing syntaxin-4 and SNAP-23 were directed to fuse with VAMP2-bearing liposomes in the absence or presence of 5 μM tomosyn-1. Each fusion reaction contained 5 μM t-SNAREs, 1.5 μM v-SNARE, and 100 mg/mL Ficoll 70 as the crowding agent. To test the activities of NSF and α-SNAP, the following components were added to a fusion reaction: 0.5 μM NSF, 1 μM α-SNAP, 2.5 mM ATP, and 5 mM MgCl2 or EDTA. (B) Lipid mixing of the fusion reactions was measured using a FRET-based assay. Control: liposome fusion reactions without NSF or α-SNAP. (C) Lipid-mixing rates of the liposome fusion reactions shown in B. Data are presented as mean ± SD. n = 3. P values were calculated using Student’s t-test. ** P<0.01. n.s., P>0.05.
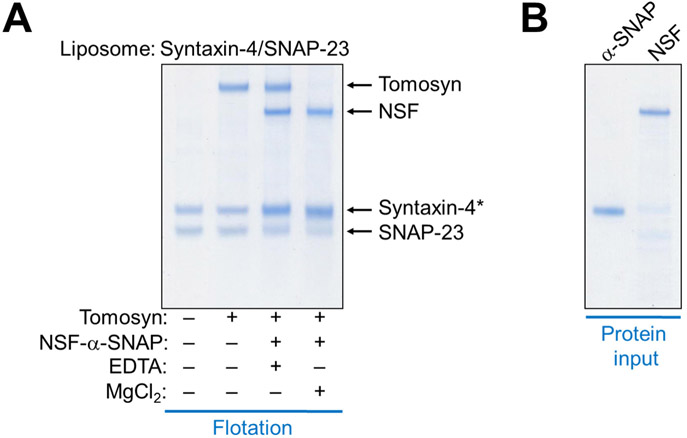
Tomosyn is dissociated from GLUT4 exocytic t-SNAREs by NSF and α-SNAP
To further characterize how NSF and α-SNAP modulate tomosyn function, we examined tomosyn-SNARE interactions in a liposome co-flotation assay. We observed that FL tomosyn bound stoichiometrically to GLUT4 exocytic t-SNAREs (syntaxin-4 and SNAP-23) anchored to liposomes (Fig. 6A). Tomosyn was fully dissociated from the t-SNAREs when NSF and α-SNAP were introduced in the presence of ATP and Mg2+ (Fig. 6A-B). Tomosyn dissociation was accompanied by stoichiometric binding of NSF and α-SNAP to GLUT4 exocytic t-SNAREs (Fig. 6A). These data demonstrate that NSF and α-SNAP directly dissociate tomosyn from GLUT4 exocytic t-SNAREs, correlating with their ability to relieve the inhibitory activity of tomosyn in the fusion reaction (Fig. 5).
Figure 6. NSF and α-SNAP disassociate tomosyn from GLUT4 exocytic t-SNAREs.
(A) Liposomes containing GLUT4 exocytic t-SNAREs (syntaxin-4 and SNAP-23) were incubated with or without 5 μM tomosyn-1 for one hour at 4 °C to form the t-SNARE/tomosyn-1 complex. Liposomes bearing the t-SNARE/tomosyn-1 complex were then incubated with or without 0.5 μM NSF, 1 μM α-SNAP, 2.5 mM ATP, and 5 mM MgCl2/EDTA at 37 °C for another hour. After flotation on a Nycodenz gradient, proteins bound to the liposomes were resolved on SDS-PAGE and stained with coomassie blue. Asterisk: α-SNAP co-migrated with syntaxin-4 on SDS-PAGE but its binding to t-SNARE liposomes was evident. (B) Coomassie blue-stained SDS-PAGE gel showing recombinant NSF and α-SNAP proteins used in this study.
DISCUSSION
In this work, we provide direct genetic evidence for an inhibitory role of tomosyn in GLUT4 exocytosis. Both basal and insulin-stimulated GLUT4 exocytosis is substantially elevated in the absence of tomosyn. Tomosyn inhibits GLUT4 exocytosis by binding to GLUT4 exocytic t-SNAREs and preventing their pairing with the v-SNARE. Tomosyn inhibits all the three SNARE complexes underlying GLUT4 vesicle fusion and none of the v-SNAREs – VAMP2, VAMP3 and VAMP8 – can overcome the inhibitory activity of tomosyn. These v-SNAREs drive comparable levels of membrane fusion and are equally sensitive to tomosyn inhibition. However, it is difficult to estimate the abilities of the v-SNAREs to support GLUT4 exocytosis in the cell because they may behave differently in interacting with other GLUT4 exocytic regulators. Our previous studies showed that tomosyn’s inhibitory activity is dominant over the stimulatory function of Munc18c/Munc18-3, a key positive regulator of GLUT4 exocytosis (50). More recently, we discovered that Munc18c uses a SNARE-like peptide to recognize the C-terminal domains of t-SNAREs (71), which overlap with the tomosyn-binding site on t-SNAREs. These observations suggest that tomosyn physically blocks the association of Munc18c with t-SNAREs.
Tomosyn-arrested t-SNAREs are not dead-end assemblies. Tomosyn can be readily removed from GLUT4 exocytic SNAREs by NSF and α-SNAP using energy from ATP hydrolysis, suggesting that the inhibitory activity of tomosyn in GLUT4 exocytosis is reversible. The overall fusion capacity of GLUT4 exocytic SNAREs is determined by the dynamic balance between the fusion-arresting activity of tomosyn and the fusion-permitting function of NSF and α-SNAP. A key question raised by this work is whether the inhibitory activity of tomosyn is modulated by insulin signaling. Tomosyn is phosphorylated in adipocytes upon insulin stimulation, leading to the speculation that insulin may promote GLUT4 exocytosis by silencing its inhibitory activity (53, 54). However, the physiological roles of the phosphorylations remain to be determined. To address this question, genetic studies would be required to determine whether and how tomosyn function in GLUT4 exocytosis is influenced by phospho-null and phospho-mimetic mutations. These genetic experiments are crucial because many insulin-induced phosphorylation events are likely non-functional (72). Once the physiological role of a phosphorylation(s) is validated, phospho-null and phospho-mimetic mutations could be introduced into recombinant tomosyn proteins to define their effects on protein-protein interactions and membrane fusion kinetics using biochemical assays described in this work.
Our data suggest that a significant portion of GLUT4 exocytic t-SNAREs are arrested by tomosyn in adipocytes and thus are unable to participate in GLUT4 vesicle fusion. Like its role in synaptic transmission (39, 73), tomosyn enables insulin-responsive tissues to fine-tune GLUT4 exocytosis such that glucose uptake can be adjusted according to physiological demands. Thus, tomosyn-arrested t-SNAREs represent a reservoir of fusion capacity that could be harnessed to enhance GLUT4 translocation. An important future direction is to determine whether tomosyn also negatively regulates GLUT4 exocytosis in patients with insulin resistance and type 2 diabetes. If this is the case, it is conceivable that tomosyn-silencing therapeutics will help unleash the full fusion capacity of GLUT4 exocytic SNAREs to improve blood glucose homeostasis in the patients.
MATERIALS AND METHODS
Cell culture and adipocyte differentiation
Mouse preadipocytes were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FB Essence (FBE, VWR, #10803-034) and penicillin/streptomycin. The preadipocyte was a spontaneously arising immortalized cell line from mouse inguinal adipose tissue (74, 75). The cells were grown to ~95% confluence before a differentiation cocktail was added at the following concentrations: 5 μg/mL insulin (Sigma, #I0516), 1 nM Triiodo-L-thyronine (T3, Sigma, #T2877), 125 μM indomethacin (Sigma, #I-7378), 5 μM dexamethasone (Sigma, #D1756), and 0.5 mM 3-isobutyl-1-methylxanthine (IBMX, Sigma, #I5879). After two days, the cells were switched to DMEM supplemented with 10% FBE, 5 μg/mL insulin, and 1 nM T3. After another two days, fresh DMEM media containing 10% FBE and 1 nM T3 were added to the cells. Differentiated adipocytes were usually analyzed six days after addition of the differentiation cocktail.
CRISPR-Cas9 Genome editing
The gRNA targeting Tomosyn-1/Stxbp5 was subcloned into the pLenti-CRISPR-V2 vector (Addgene, #52961). The gRNA targeting Tomosyn-2/Stxbp5l was subcloned into a modified version of the pLentiGuide-Puro vector (Addgene, #52963) in which the puromycin selection marker was replaced with a hygromycin selection marker (60). CRISPR plasmids were transfected into 293T cells along with pAdVAntage (Promega, #E1711), pCMV-VSVG (Addgene, #8454), and psPax2 (Addgene, #12260) as previously described (60). The 293T cell culture media containing lentiviral particles were harvested daily for four days and centrifuged at 25,000 RPM for 1.5 hours in a Beckman SW28 rotor. Viral pellets were resuspended in DMEM and used to infect preadipocytes. The cells were consecutively selected using 3.5 μg/mL puromycin (Sigma, #3101118) and 500 μg/mL hygromycin B (Thermo, #10687010).
The gRNA sequences were: TCCCGTTCAGAAGATCCTGG (Tomosyn-1) TGGATCAAAGGCCAATGCTG (Tomosyn-2).
Expression of the TOMOSYN-1 rescue gene
The human TOMOSYN-1 gene was amplified using PCR, introducing a NheI site at the 5’ end and a SalI site plus a 3xFLAG-encoding sequence at the 3’ end. The PCR product was subcloned into the SalI and NheI sites of the SHC003BSD-GFPD vector (Addgene, #133301). The resulted SHC003BSD-TOMOSYN1 construct was transfected into 293T cells to produce lentiviral particles, which were used to infect preadipocytes. Transduced preadipocytes were selected using 10 μg/mL blasticidin (Thermo Fisher Scientific, # BP2647). The human TOMOSYN-1 gene is not targeted by the Tomosyn-1/2 gRNAs used in this work. Primers used for amplification of the human TOMOSYN-1 gene were:
CGCGCTAGCCTAGCCACCATGAGGAAATTCAACATCAG (forward)
GCGGTCGACCTACTTGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCGAACTGGTACCACTTCTTAT (reverse)
Measurement of insulin-stimulated GLUT4 exocytosis
Insulin-stimulated GLUT4 exocytosis was measured using flow cytometry as previously described (60, 76, 77). Briefly, cells were washed three times with the KRH buffer (121 mM NaCl, 4.9 mM KCl, 1.2 mM MgSO4, 0.33 mM CaCl2, and 12 mM HEPES [pH 7.4]). After incubation in the KRH buffer for two hours, the cells were treated with insulin. Subsequently, the cells were rapidly chilled, and surface GLUT4 reporters were stained using anti-HA antibodies (BioLegend, #901501) and APC-conjugated secondary antibodies (eBioscience, #17-4014). The cells were dislodged using Accutase (Innovative Cell Technologies, #AT 104), and the GFP and APC fluorescence was measured on a CyAN ADP analyzer (Beckman Coulter). To calculate normalized surface levels of GLUT4 reporters, the mean APC fluorescence (surface staining) was divided by mean GFP fluorescence (total reporters), and the obtained values were normalized to those of untreated WT samples. Data from populations of ~5,000 cells were analyzed using the FlowJo software (FlowJo, LLC, v10) based on experiments run in biological triplicate.
To visualize GLUT4 translocation using confocal microscopy, preadipocytes expressing the HA-GLUT4-GFP reporter were grown and differentiated in glass-bottom plates. After starvation in the KRH buffer for two hours, adipocytes were either untreated or treated with 100 nM insulin for 30 minutes. Subsequently, the cells were fixed with 4% paraformaldehyde (TED PELLA, INC. #18505) and incubated with 2% bovine serum albumin (Thermo, #064985). Surface GLUT4 reporters were stained with anti-HA antibodies and Alexa Fluor 568-conjugated secondary antibodies (Thermo, #A-11004). Nuclei were stained with Hoechst 33342 (Sigma, #D9642). Images were captured using a 100× oil immersion objective on a Nikon A1 Laser Scanning confocal microscope.
Immunoprecipitation (IP)
Cells were lysed in an IP buffer (25 mM HEPES [pH 7.4], 138 mM NaCl, 10 mM Na3PO4, 2.7 mM KCl, 0.5% CHAPS, 1 mM DTT, and a protease inhibitor cocktail). Tomosyn and associated proteins were precipitated from cell lysates using anti-FLAG antibodies (Sigma, #F1804) and protein A agarose beads. Immunoprecipitates and whole cells were dissolved in 1× SDS protein sample buffer and resolved on 8% Bis-Tris SDS-PAGE. After transferring to PVDF membranes, proteins were detected using primary antibodies and horseradish peroxidase-conjugated secondary antibodies. The experiments were repeated three times with similar results obtained.
Immunoblotting
Cells grown in 24-well plates were lysed in an SDS protein sample buffer. Cell lysates were resolved on 8% Bis-Tris SDS-PAGE and probed using primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Primary antibodies used in immunoblotting included mouse monoclonal anti-tomosyn-1/2 antibodies (BD Biosciences, #611296), rabbit polyclonal anti-syntaxin-4 antibodies (Sigma, #S9924), mouse monoclonal anti-PPARγ antibodies (Santa Cruz Biotechnology, #sc-7273), anti-α-tubulin (DSHB, clone, #12G10), rabbit polyclonal anti-phospho-Akt (Ser473) antibodies (Cell Signaling Technology, #9271), rabbit polyclonal anti-Akt antibodies (Cell Signaling Technology, #9272), and mouse monoclonal anti-FLAG antibodies.
Real-time quantitative reverse transcription PCR (qRT-PCR)
Total RNAs were isolated using the TRI reagent (Sigma, #93289) and the ezDNAse Enzyme (Thermo, #18091150). First strand synthesis was performed using a Superscript IV kit (Thermo, #18091050). Gene expression was determined by qRT-PCR on a Bio-Rad CFX384 Real-time PCR Detection System using SsoAdvanced Universal SYBER Green Supermix (Bio-Rad, #172-5272) with gene-specific primer sets. The cycle threshold values of a candidate gene were normalized to those of Gapdh, a reference gene, and the Δcycle threshold values were calculated. The results were plotted as fold changes relative to the control sample.
The PCR primers for Tomosyn-1 were:
ACCATCGAACTTTACGGCTCA (forward)
ACATCTTTGCGGAAGTGGGAG (reverse)
The PCR primers for Gapdh were:
AGGTCGGTGTGAACGGATTTG (forward)
TGTAGACCATGTAGTTGAGGTCA (reverse)
Lipid droplet staining and analysis
Preadipocytes and adipocytes grown in clear-bottom 96-well plates were fixed by incubating with 4% paraformaldehyde for 20 minutes. The cells were washed three times with PBS and visualized on an Olympus IX81 Microscope using a 20x objective. For flow cytometry analysis of lipid droplets, cells were stained with 5 μg/mL Nile red (Sigma, #N3013) in the presence of 10 μg/mL Hoechst 33342. Subsequently, the cells were detached using Accutase and the fluorescence of Nile red and Hoechst 33342 was quantified on a CyAN ADP analyzer. Data from populations of ~5,000 cells were analyzed using the FlowJo software based on experiments run in biological triplicate. Fluorescence of unstained WT preadipocytes was used as a blank control.
Protein expression and purification
Recombinant v- and t-SNARE proteins were expressed in E. coli and purified by affinity chromatography. GLUT4 exocytic t-SNAREs consisted of untagged syntaxin-4 and His6-tagged SNAP-23 whereas yeast exocytic t-SNAREs consisted of His8-tagged Sso1p and untagged Sec9p (78, 79). The v-SNAREs VAMP2, VAMP3, VAMP8 and Snc2p were expressed as His6-SUMO-tagged proteins before the tags were proteolytically removed (80). All SNARE proteins were stored in buffer A containing 25 mM HEPES (pH 7.4), 400 mM KCl, 1% n-octyl-β-D-glucoside, 10% glycerol, and 0.5 mM Tris (2-carboxyethyl) phosphine (TCEP). GST-tagged cytoplasmic domain of VAMP2 (GST-VAMP2 CD) was purified by GST affinity chromatography, followed by dialysis against buffer B containing 25 mM HEPES (pH 7.4), 150 mM KCl, 10% glycerol, and 5 mM TCEP. His6-MBP-tagged human tomosyn-1 was expressed and purified from Sf9 cells as previously described (50). After purification, the His6-MBP moiety was proteolytically removed using the tobacco etch virus protease, followed by overnight dialysis against buffer B. His6-NSF-myc was prepared in a previous study (68). Recombinant α-SNAP protein was expressed and purified using a His6-SUMO tagging system as previously described for Munc18-1 (78).
Reconstitution of proteoliposomes
All lipids were obtained from Avanti Polar Lipids Inc. To prepare t-SNARE liposomes, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) and cholesterol were mixed in a molar ratio of 60:20:10:10. To prepare v-SNARE liposomes, POPC, POPE, POPS, cholesterol, (N-(7-nitro-2,1,3-benzoxadiazole-4-yl)-1,2-dipalmitoyl phosphatidylethanolamine (NBD-DPPE) and N-(Lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl phosphatidylethanolamine (rhodamine-DPPE) were mixed at a molar ratio of 60:17:10:10:1.5:1.5. SNARE proteoliposomes were formed by detergent dilution and isolated on a Nycodenz density gradient flotation (78). Complete detergent removal was achieved by overnight dialysis of the samples in Novagen dialysis tubes against the reconstitution buffer (25 mM HEPES [pH 7.4], 100 mM KCl, 10% glycerol, and 1 mM DTT). The protein: lipid ratio was 1:200 for v-SNARE liposomes, and 1:500 for t-SNARE liposomes.
Liposome lipid-mixing assay
A standard liposome fusion reaction contained 45 μL unlabeled t-SNARE liposomes and 5 μL v-SNARE liposomes. Liposome fusion was measured using a fluorescence dequenching assay (78). Increase in NBD-fluorescence at 538 nm (excitation 460 nm) was measured every two minutes in a BioTek Synergy HT microplate reader. At the end of the reaction, 10% CHAPSO was added to the samples to obtain values of maximum fluorescence. The rate of a liposome fusion reaction is presented as the fluorescence change during the 60-minute reaction normalized to maximum fluorescence. Full accounting of statistical significance was included for each figure based on at least three independent experiments.
Liposome co-flotation assay
The interactions of soluble factors with liposomes were measured using a liposome co-flotation assay (60, 71, 78, 81). Soluble proteins were incubated with liposomes at 4 °C with gentle agitation. After one hour, an equal volume of 80% Nycodenz (w/v) in reconstitution buffer was added and the mixture was transferred to 5 x 41 mm centrifuge tubes. The samples were overlaid with 200 μL each of 35% and 30% Nycodenz, followed by 20 μL reconstitution buffer on the top. The samples were centrifuged at 52,000 rpm for four hours in a Beckman SW55 rotor. Liposome samples were collected from the 0/30% Nycodenz interface (2 x 20 μL) and analyzed by SDS-PAGE.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Gus Lienhard, Shingo Kajimura, Wei Guo and David James for advice or reagents, and Yan Ouyang for technical assistance. This work was supported by National Natural Science Foundation of China grants 31871425 and 91854117 (H.Y.), a Fellowship from the Postdoctoral Overseas Training Program at Beijing University of Chinese Medicine (S.W.), National Institutes of Health (NIH) grants DK124431, GM126960, AG061829 (JS), an American Diabetes Association Basic Science Award (J.S.), an NIH institutional predoctoral training grant GM088759 (L.C.), and a Fellowship from the Undergraduate Research Opportunities Program at University of Colorado Boulder (J.M.).
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science (New York, NY) 2009;323(5913):474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant NJ, Govers R, James DE. Regulated transport of the glucose transporter GLUT4. Nature reviews Molecular cell biology 2002;3(4):267–277. [DOI] [PubMed] [Google Scholar]
- 3.Klip A, McGraw TE, James DE. Thirty sweet years of GLUT4. J Biol Chem 2019;294(30):11369–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavan BE, Lienhard GE. Insulin signalling and the stimulation of glucose transport. Biochemical Society transactions 1994;22(3):676–680. [DOI] [PubMed] [Google Scholar]
- 5.Kioumourtzoglou D, Pryor PR, Gould GW, Bryant NJ. Alternative routes to the cell surface underpin insulin-regulated membrane trafficking of GLUT4. Journal of cell science 2015;128(14):2423–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogan JS, Kandror KV. Biogenesis and regulation of insulin-responsive vesicles containing GLUT4. Current opinion in cell biology 2010;22(4):506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Rubin BR, Orme CM, Karpikov A, Yu C, Bogan JS, Toomre DK. Dual-mode of insulin action controls GLUT4 vesicle exocytosis. The Journal of cell biology 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jewell JL, Oh E, Thurmond DC. Exocytosis mechanisms underlying insulin release and glucose uptake: conserved roles for Munc18c and syntaxin 4. Am J Physiol Regul Integr Comp Physiol 2010;298(3):R517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vassilopoulos S, Esk C, Hoshino S, Funke BH, Chen CY, Plocik AM, Wright WE, Kucherlapati R, Brodsky FM. A role for the CHC22 clathrin heavy-chain isoform in human glucose metabolism. Science (New York, NY) 2009;324(5931):1192–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogan JS, Hendon N, McKee AE, Tsao TS, Lodish HF. Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature 2003;425(6959):727–733. [DOI] [PubMed] [Google Scholar]
- 11.Leto D, Saltiel AR. Regulation of glucose transport by insulin: traffic control of GLUT4. Nature reviews Molecular cell biology 2012;13(6):383–396. [DOI] [PubMed] [Google Scholar]
- 12.Hou JC, Pessin JE. Ins (endocytosis) and outs (exocytosis) of GLUT4 trafficking. Current opinion in cell biology 2007;19(4):466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, Lifshitz LM, Jones C, Bellve KD, Standley C, Fonseca S, Corvera S, Fogarty KE, Czech MP. Insulin stimulates membrane fusion and GLUT4 accumulation in clathrin coats on adipocyte plasma membranes. Mol Cell Biol 2007;27(9):3456–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonescu CN, McGraw TE, Klip A. Reciprocal regulation of endocytosis and metabolism. Cold Spring Harbor perspectives in biology 2014;6(7):a016964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kandror KV, Pilch PF. The Sugar Is sIRVed: Sorting Glut4 and Its Fellow Travelers. Traffic 2011;12(6):665–671. [DOI] [PubMed] [Google Scholar]
- 16.Shi J, Kandror KV. Sortilin is essential and sufficient for the formation of Glut4 storage vesicles in 3T3-L1 adipocytes. Developmental cell 2005;9(1):99–108. [DOI] [PubMed] [Google Scholar]
- 17.Kewalramani G, Bilan PJ, Klip A. Muscle insulin resistance: assault by lipids, cytokines and local macrophages. Curr Opin Clin Nutr Metab Care 2010;13(4):382–390. [DOI] [PubMed] [Google Scholar]
- 18.Aslamy A, Thurmond DC. Exocytosis proteins as novel targets for diabetes prevention and/or remediation? American journal of physiology Regulatory, integrative and comparative physiology 2017;312(5):R739–R752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature 1993;362(6418):318–324. [DOI] [PubMed] [Google Scholar]
- 20.Rizo J, Sudhof TC. The Membrane Fusion Enigma: SNAREs, Sec1/Munc18 Proteins, and Their Accomplices-Guilty as Charged? Annu Rev Cell Dev Biol 2012;28:279–308. [DOI] [PubMed] [Google Scholar]
- 21.Wickner W. Membrane Fusion: Five Lipids, Four SNAREs, Three Chaperones, Two Nucleotides, and a Rab, All Dancing in a Ring on Yeast Vacuoles. Annu Rev Cell Dev Biol 2010;26:115–136. [DOI] [PubMed] [Google Scholar]
- 22.Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, Sirinakis G, Rothman JE, Zhang Y. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science (New York, NY) 2012;337(6100):1340–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 1998;395(6700):347–353. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez JM, Stein A, Behrmann E, Riedel D, Cypionka A, Farsi Z, Walla PJ, Raunser S, Jahn R. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science (New York, NY) 2012;336(6088):1581–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ungar D, Hughson FM. SNARE protein structure and function. Annu Rev Cell Dev Biol 2003;19:493–517. [DOI] [PubMed] [Google Scholar]
- 26.Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem 2008;77:615–641. [DOI] [PubMed] [Google Scholar]
- 27.Olson AL, Knight JB, Pessin JE. Syntaxin 4, VAMP2, and/or VAMP3/cellubrevin are functional target membrane and vesicle SNAP receptors for insulin-stimulated GLUT4 translocation in adipocytes. Mol Cell Biol 1997;17(5):2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jedrychowski MP, Gartner CA, Gygi SP, Zhou L, Herz J, Kandror KV, Pilch PF. Proteomic analysis of GLUT4 storage vesicles reveals LRP1 to be an important vesicle component and target of insulin signaling. The Journal of biological chemistry 2010;285(1):104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazakerley DJ, Naghiloo S, Chaudhuri R, Koumanov F, Burchfield JG, Thomas KC, Krycer JR, Prior MJ, Parker BL, Murrow BA, Stockli J, Meoli CC, Holman GD, James DE. Proteomic Analysis of GLUT4 Storage Vesicles Reveals Tumor Suppressor Candidate 5 (TUSC5) as a Novel Regulator of Insulin Action in Adipocytes. The Journal of biological chemistry 2015;290(39):23528–23542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao P, Yang L, Lopez JA, Fan J, Burchfield JG, Bai L, Hong W, Xu T, James DE. Variations in the requirement for v-SNAREs in GLUT4 trafficking in adipocytes. Journal of cell science 2009;122(Pt 19):3472–3480. [DOI] [PubMed] [Google Scholar]
- 31.Williams D, Pessin JE. Mapping of R-SNARE function at distinct intracellular GLUT4 trafficking steps in adipocytes. The Journal of cell biology 2008;180(2):375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams D, Vicogne J, Zaitseva I, McLaughlin S, Pessin JE. Evidence that electrostatic interactions between vesicle-associated membrane protein 2 and acidic phospholipids may modulate the fusion of transport vesicles with the plasma membrane. Molecular biology of the cell 2009;20(23):4910–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie L, Zhu D, Dolai S, Liang T, Qin T, Kang Y, Xie H, Huang YC, Gaisano HY. Syntaxin-4 mediates exocytosis of pre-docked and newcomer insulin granules underlying biphasic glucose-stimulated insulin secretion in human pancreatic beta cells. Diabetologia 2015;58(6):1250–1259. [DOI] [PubMed] [Google Scholar]
- 34.Smithers NP, Hodgkinson CP, Cuttle M, Sale GJ. Insulin-triggered repositioning of munc18c on syntaxin-4 in GLUT4 signalling. The Biochemical journal 2008;410(2):255–260. [DOI] [PubMed] [Google Scholar]
- 35.Tellam JT, Macaulay SL, McIntosh S, Hewish DR, Ward CW, James DE. Characterization of Munc-18c and syntaxin-4 in 3T3-L1 adipocytes. Putative role in insulin-dependent movement of GLUT-4. J Biol Chem 1997;272(10):6179–6186. [DOI] [PubMed] [Google Scholar]
- 36.Tamm LK, Crane J, Kiessling V. Membrane fusion: a structural perspective on the interplay of lipids and proteins. Curr Opin Struct Biol 2003;13(4):453–466. [DOI] [PubMed] [Google Scholar]
- 37.Zick M, Orr A, Schwartz ML, Merz AJ, Wickner WT. Sec17 can trigger fusion of trans-SNARE paired membranes without Sec18. Proc Natl Acad Sci U S A 2015;112(18):E2290–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehman K, Rossi G, Adamo JE, Brennwald P. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. The Journal of cell biology 1999;146(1):125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuda ES, Huang BC, Fisher JM, Luo Y, Scheller RH. Tomosyn binds t-SNARE proteins via a VAMP-like coiled coil. Neuron 1998;21(3):479–480. [DOI] [PubMed] [Google Scholar]
- 40.Fujita Y, Shirataki H, Sakisaka T, Asakura T, Ohya T, Kotani H, Yokoyama S, Nishioka H, Matsuura Y, Mizoguchi A, Scheller RH, Takai Y. Tomosyn: a syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron 1998;20(5):905–915. [DOI] [PubMed] [Google Scholar]
- 41.Yizhar O, Matti U, Melamed R, Hagalili Y, Bruns D, Rettig J, Ashery U. Tomosyn inhibits priming of large dense-core vesicles in a calcium-dependent manner. Proc Natl Acad Sci U S A 2004;101(8):2578–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakisaka T, Baba T, Tanaka S, Izumi G, Yasumi M, Takai Y. Regulation of SNAREs by tomosyn and ROCK: implication in extension and retraction of neurites. The Journal of cell biology 2004;166(1):17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gracheva EO, Burdina AO, Holgado AM, Berthelot-Grosjean M, Ackley BD, Hadwiger G, Nonet ML, Weimer RM, Richmond JE. Tomosyn inhibits synaptic vesicle priming in Caenorhabditis elegans. PLoS Biol 2006;4(8):e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gladycheva SE, Lam AD, Liu J, D'Andrea-Merrins M, Yizhar O, Lentz SI, Ashery U, Ernst SA, Stuenkel EL. Receptor-mediated regulation of tomosyn-syntaxin 1A interactions in bovine adrenal chromaffin cells. J Biol Chem 2007;282(31):22887–22899. [DOI] [PubMed] [Google Scholar]
- 45.Ashery U, Bielopolski N, Barak B, Yizhar O. Friends and foes in synaptic transmission: the role of tomosyn in vesicle priming. Trends Neurosci 2009;32(5):275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhatnagar S, Oler AT, Rabaglia ME, Stapleton DS, Schueler KL, Truchan NA, Worzella SL, Stoehr JP, Clee SM, Yandell BS, Keller MP, Thurmond DC, Attie AD. Positional cloning of a type 2 diabetes quantitative trait locus; tomosyn-2, a negative regulator of insulin secretion. PLoS Genet 2011;7(10):e1002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pobbati AV, Razeto A, Boddener M, Becker S, Fasshauer D. Structural basis for the inhibitory role of tomosyn in exocytosis. J Biol Chem 2004;279(45):47192–47200. [DOI] [PubMed] [Google Scholar]
- 48.Hatsuzawa K, Lang T, Fasshauer D, Bruns D, Jahn R. The R-SNARE motif of tomosyn forms SNARE core complexes with syntaxin 1 and SNAP-25 and down-regulates exocytosis. J Biol Chem 2003;278(33):31159–31166. [DOI] [PubMed] [Google Scholar]
- 49.Williams AL, Bielopolski N, Meroz D, Lam AD, Passmore DR, Ben-Tal N, Ernst SA, Ashery U, Stuenkel EL. Structural and functional analysis of tomosyn identifies domains important in exocytotic regulation. J Biol Chem 2011;286(16):14542–14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu H, Rathore SS, Gulbranson DR, Shen J. The N- and C-terminal domains of tomosyn play distinct roles in soluble N-ethylmaleimide-sensitive factor attachment protein receptor binding and fusion regulation. J Biol Chem 2014;289(37):25571–25580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burdina AO, Klosterman SM, Shtessel L, Ahmed S, Richmond JE. In vivo analysis of conserved C. elegans tomosyn domains. PLoS One 2011;6(10):e26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Widberg CH, Bryant NJ, Girotti M, Rea S, James DE. Tomosyn interacts with the t-SNAREs syntaxin4 and SNAP23 and plays a role in insulin-stimulated GLUT4 translocation. J Biol Chem 2003;278(37):35093–35101. [DOI] [PubMed] [Google Scholar]
- 53.Humphrey SJ, Yang G, Yang P, Fazakerley DJ, Stockli J, Yang JY, James DE. Dynamic Adipocyte Phosphoproteome Reveals that Akt Directly Regulates mTORC2. Cell Metab 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagano K, Takeuchi H, Gao J, Mori Y, Otani T, Wang D, Hirata M. Tomosyn is a novel Akt substrate mediating insulin-dependent GLUT4 exocytosis. Int J Biochem Cell B 2015;62:62–71. [DOI] [PubMed] [Google Scholar]
- 55.Cheviet S, Bezzi P, Ivarsson R, Renstrom E, Viertl D, Kasas S, Catsicas S, Regazzi R. Tomosyn-1 is involved in a post-docking event required for pancreatic beta-cell exocytosis. Journal of cell science 2006;119(Pt 14):2912–2920. [DOI] [PubMed] [Google Scholar]
- 56.Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. The Journal of cell biology 1985;100(3):965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spiegelman BM. PPAR-gamma: adipogenic regulator and thiazolidinedione receptor. Diabetes 1998;47(4):507–514. [DOI] [PubMed] [Google Scholar]
- 58.Bai L, Wang Y, Fan J, Chen Y, Ji W, Qu A, Xu P, James DE, Xu T. Dissecting multiple steps of GLUT4 trafficking and identifying the sites of insulin action. Cell Metab 2007;5(1):47–57. [DOI] [PubMed] [Google Scholar]
- 59.Sano H, Eguez L, Teruel MN, Fukuda M, Chuang TD, Chavez JA, Lienhard GE, McGraw TE. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab 2007;5(4):293–303. [DOI] [PubMed] [Google Scholar]
- 60.Gulbranson DR, Davis EM, Demmitt BA, Ouyang Y, Ye Y, Yu H, Shen J. RABIF/MSS4 is a Rab-stabilizing holdase chaperone required for GLUT4 exocytosis. Proc Natl Acad Sci U S A 2017;114(39):E8224–E8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blot V, McGraw TE. Use of quantitative immunofluorescence microscopy to study intracellular trafficking: studies of the GLUT4 glucose transporter. Methods Mol Biol 2008;457:347–366. [DOI] [PubMed] [Google Scholar]
- 62.Lizunov VA, Matsumoto H, Zimmerberg J, Cushman SW, Frolov VA. Insulin stimulates the halting, tethering, and fusion of mobile GLUT4 vesicles in rat adipose cells. The Journal of cell biology 2005;169(3):481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Wang S, Li T, Zhu L, Ma C. Tomosyn guides SNARE complex formation in coordination with Munc18 and Munc13. FEBS Lett 2018;592(7):1161–1172. [DOI] [PubMed] [Google Scholar]
- 64.Yizhar O, Lipstein N, Gladycheva SE, Matti U, Ernst SA, Rettig J, Stuenkel EL, Ashery U. Multiple functional domains are involved in tomosyn regulation of exocytosis. J Neurochem 2007;103(2):604–616. [DOI] [PubMed] [Google Scholar]
- 65.Geerts CJ, Mancini R, Chen N, Koopmans FTW, Li KW, Smit AB, van Weering JRT, Verhage M, Groffen AJA. Tomosyn associates with secretory vesicles in neurons through its N- and C-terminal domains. PLoS One 2017;12(7):e0180912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective Activation of Cognate SNAREpins by Sec1/Munc18 Proteins. Cell 2007;128(1):183–195. [DOI] [PubMed] [Google Scholar]
- 67.Schollmeier Y, Krause JM, Kreye S, Malsam J, Sollner TH. Resolving the function of distinct Munc18-1/SNARE protein interaction modes in a reconstituted membrane fusion assay. The Journal of biological chemistry 2011;286(35):30582–30590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber T, Parlati F, McNew JA, Johnston RJ, Westermann B, Sollner TH, Rothman JE. SNAREpins are functionally resistant to disruption by NSF and alphaSNAP. The Journal of cell biology 2000;149(5):1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 1993;75(3):409–418. [DOI] [PubMed] [Google Scholar]
- 70.Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, Brunger AT. Mechanistic insights into the recycling machine of the SNARE complex. Nature 2015;518(7537):61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu H, Shen C, Liu Y, Menasche BL, Ouyang Y, Stowell MHB, Shen J. SNARE zippering requires activation by SNARE-like peptides in Sec1/Munc18 proteins. Proc Natl Acad Sci U S A 2018;115(36):E8421–E8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lienhard GE. Non-functional phosphorylations? Trends in biochemical sciences 2008;33(8):351–352. [DOI] [PubMed] [Google Scholar]
- 73.Chen K, Richlitzki A, Featherstone DE, Schwarzel M, Richmond JE. Tomosyn-dependent regulation of synaptic transmission is required for a late phase of associative odor memory. Proc Natl Acad Sci U S A 2011;108(45):18482–18487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang S, Crisman L, Miller J, Datta I, Gulbranson DR, Tian Y, Yin Q, Yu H, Shen J. Inducible Exoc7/Exo70 knockout reveals a critical role of the exocyst in insulin-regulated GLUT4 exocytosis. J Biol Chem 2019;294(52):19988–19996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gulbranson DR, Crisman L, Lee M, Ouyang Y, Menasche BL, Demmitt BA, Wan C, Nomura T, Ye Y, Yu H, Shen J. AAGAB Controls AP2 Adaptor Assembly in Clathrin-Mediated Endocytosis. Developmental cell 2019;50(4):436–446 e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gulbranson DR, Crisman L, Lee M, Ouyang Y, Menasche BL, Demmitt BA, Wan C, Nomura T, Ye Y, Yu H, Shen J. AAGAB Controls AP2 Adaptor Assembly in Clathrin-Mediated Endocytosis. Developmental cell 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Menasche BL, Crisman L, Gulbranson DR, Davis EM, Yu H, Shen J. Fluorescence Activated Cell Sorting (FACS) in Genome-Wide Genetic Screening of Membrane Trafficking. Curr Protoc Cell Biol 2018:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu H, Crisman L, Stowell MHB, Shen J. Functional Reconstitution of Intracellular Vesicle Fusion Using Purified SNAREs and Sec1/Munc18 (SM) Proteins. Methods Mol Biol 2019;1860:237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu H, Rathore SS, Lopez JA, Davis EM, James DE, Martin JL, Shen J. Comparative studies of Munc18c and Munc18-1 reveal conserved and divergent mechanisms of Sec1/Munc18 proteins. Proc Natl Acad Sci U S A 2013;110(35):E3271–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu H, Rathore SS, Davis EM, Ouyang Y, Shen J. Doc2b promotes GLUT4 exocytosis by activating the SNARE-mediated fusion reaction in a calcium- and membrane bending-dependent manner. Molecular biology of the cell 2013;24(8):1176–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rathore SS, Liu Y, Yu H, Wan C, Lee M, Yin Q, Stowell MHB, Shen J. Intracellular Vesicle Fusion Requires a Membrane-Destabilizing Peptide Located at the Juxtamembrane Region of the v-SNARE. Cell Rep 2019;29(13):4583–4592 e4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.