The Hippo kinase Mst1 enforces the establishment of cellular quiescence for terminal maturation and reciprocal fate decisions between iNKT1 and iNKT17 effector cells, which respectively depend upon Opa1-mediated mitochondrial dynamics and ICOS–mTORC2 signaling.
Abstract
Invariant natural killer T (iNKT) cells acquire effector functions during development by mechanisms that remain poorly understood. Here, we show that the Hippo kinases Mst1 and Mst2 act as molecular rheostats for the terminal maturation and effector differentiation programs of iNKT cells. Loss of Mst1 alone or together with Mst2 impedes iNKT cell development, associated with defective IL-15–dependent cell survival. Mechanistically, Mst1 enforces iNKT cellular and transcriptional quiescence associated with maturation and commitment to iNKT1 cells by suppressing proliferation and Opa1-related mitochondrial metabolism that are dynamically regulated during iNKT cell development. Furthermore, Mst1 shapes the reciprocal fate decisions between iNKT1 and iNKT17 effector cells, which respectively depend upon mitochondrial dynamics and ICOS–mTORC2 signaling. Collectively, these findings establish Mst1 as a crucial regulator of mitochondrial homeostasis and quiescence in iNKT cell development and effector lineage differentiation and highlight that establishment of quiescence programs underlies iNKT cell development and effector maturation.
Introduction
Invariant natural killer T (iNKT) cells are an innate-like T cell subset that promotes rapid responses to infections and influences downstream adaptive immune responses (Crosby and Kronenberg, 2018; Wang and Hogquist, 2018). iNKT cells develop in the thymus at the CD4+CD8+ double positive (DP) stage and to a lesser extent at the double negative (DN) stage (Dashtsoodol et al., 2017), where they undergo temporally regulated maturation through stages 0–3 (S0–S3) and stage-associated effector differentiation programs (Lee et al., 2013) that lead to the generation of iNKT1, iNKT2, and iNKT17 cells (Crosby and Kronenberg, 2018; Wang and Hogquist, 2018). Many studies have emphasized the upstream signals and transcriptional regulators critical for iNKT cell lineage commitment from DP cells, including TCRs and costimulatory receptors, leading to the induction of promyelocytic leukemia zinc finger (PLZF), a master transcription factor required for iNKT cell development (Crosby and Kronenberg, 2018; Wang and Hogquist, 2018). In contrast, mechanisms regulating the stage-specific maturation and differentiation of iNKT effector cells, especially the intracellular processes and signaling networks, are not fully understood.
A unique feature of iNKT cell development is the proliferative expansion that occurs after positive selection at the DP stage, followed by a reduction in proliferation (Benlagha et al., 2002, 2005) as iNKT cells mature within the thymus. In developing and mature αβ T cells, the induction of cellular proliferation requires the PI3K and mechanistic target of rapamycin (mTOR) pathways, which also shape iNKT cell development and maturation (Finlay et al., 2010; Henao-Mejia et al., 2013; Shin et al., 2014; Wei et al., 2014; Wu et al., 2014). Defective PI3K signaling has been shown to reduce metabolic gene signatures, glycolysis, and nutrient transporter expression in early developing T cells and iNKT cells (Finlay et al., 2010; Henao-Mejia et al., 2013), suggesting a role for anabolic metabolism in iNKT cell development. However, nutrient stress-induced catabolic programs also favor iNKT cell development, as evidenced by reduced iNKT cell development in mice lacking molecular regulators in the 5′ AMP-activated protein kinase and autophagy pathways (Park et al., 2014; Pei et al., 2015; Salio et al., 2014). Thus, metabolic regulation of iNKT cell development is not well understood, with limited information available about how metabolic programs support iNKT cell developmental maturation and effector lineage decisions. Furthermore, whether additional signaling pathways other than PI3K–mTOR are involved in iNKT cell metabolism and development is underexplored.
The conserved Hippo signaling pathway is emerging as a critical regulator of immune cell function (Hong et al., 2018). Mst1 and Mst2, two integral serine/threonine kinases in the mammalian Hippo pathway (Yu et al., 2015), can regulate immune cell function by promoting cellular adhesion mechanisms (Hong et al., 2018; Katagiri et al., 2009), as well as cell-intrinsic regulation of intracellular signaling (Shi et al., 2018). Further, more recent evidence suggests that the Hippo pathway orchestrates diverse cellular processes, including metabolic control. Specifically, the Hippo pathway can both influence metabolic processes and be regulated by metabolic cues to control cell growth and survival (Ardestani et al., 2018; Koo and Guan, 2018). Whether Mst1 and Mst2, along with canonical Hippo signaling, play any role in iNKT cell development is unknown.
In this study, we show that maturation and differentiation of effector iNKT cells require Mst1, and to a lesser extent Mst2, but not other canonical Hippo signaling components. We find that Mst1 deficiency disrupts iNKT cell development and survival, associated with impaired IL-15 signaling. Our unbiased transcriptional network analysis and experimental validation reveal that the dynamic regulation of proliferation and mitochondrial metabolism is associated with the establishment of quiescence programs during iNKT cell maturation and effector differentiation. Additionally, we uncover Mst signaling as a negative regulator of proliferation and mitochondrial homeostasis during the maturation of S3 iNKT cells and development of iNKT1 effector cells. Moreover, Mst1 deficiency leads to impaired iNKT1 but enhanced iNKT17 cell development. We further identify mitochondrial dynamics regulated by Opa1 and inducible costimulator (ICOS)–mTORC2 signaling as crucial regulators of iNKT1 and iNKT17 cells, respectively, thereby highlighting Mst1 function and the coordination of metabolism and immune signaling as novel processes underlying iNKT fate decisions. These findings establish the integration of cellular and metabolic quiescence with the terminal maturation and effector programming of iNKT cells, with Mst1 as a key molecular rheostat in linking metabolism and immune signaling in this process.
Results
Mst1 and Mst2 are required for iNKT cell terminal maturation
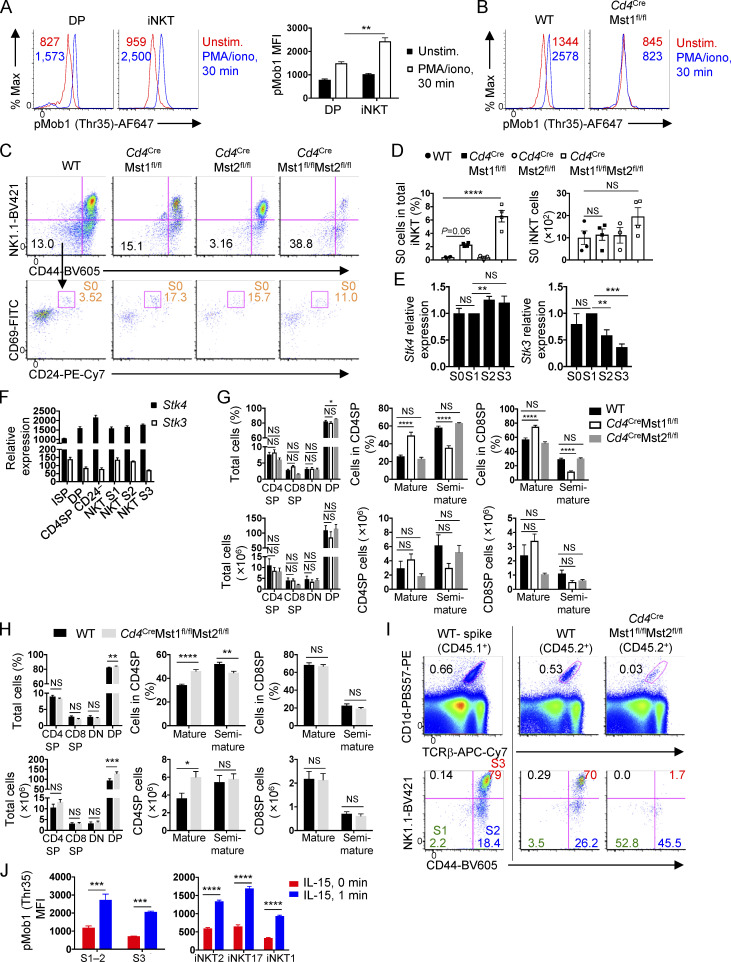
To address the role of Hippo signaling in iNKT cell development, we first examined Hippo signaling activity in thymic DP cells and iNKT cells by assessing the phosphorylation of Mob1, a well-defined downstream Mst target (Praskova et al., 2008). Phosphorylation of Mob1 (Thr35) was increased in iNKT cells compared with DP cells (Fig. S1 A), and was lost in the absence of Mst1 (Fig. S1 B), suggesting that Hippo signaling may be involved in iNKT cell development. To systematically dissect the role of Hippo signaling, we deleted multiple molecules in this pathway. First, we crossed mice with floxed alleles for Mst1 (encoded by Stk4) and Mst2 (encoded by Stk3) with Cd4Cre mice to generate mice with deletion of Mst1 and/or Mst2 at the DP stage of thymocyte development when iNKT cells arise (denoted Cd4CreMst1fl/fl, Cd4CreMst2fl/fl, or Cd4CreMst1fl/flMst2fl/fl). In the absence of Mst1, there was a significant loss in the frequency and number of iNKT cells in the thymus, spleen, and liver compared with WT mice (Fig. 1, A and B). In the thymus, Mst1-deficient mice showed no change in the number of early progenitor S0 iNKT cells (TCRβintCD1d-PBS57tet+CD69+CD24+CD44–NK1.1–; Fig. S1, C and D). However, analysis of the subsequent stages showed that Mst1 deficiency altered iNKT cell developmental stages, with reduced frequency of S3 (TCRβintCD1d-PBS57tet+CD24–CD44+NK1.1+) cells and increased frequency of S2 (TCRβintCD1d-PBS57tet+CD24–CD44+NK1.1–) cells, but no change in S1 (TCRβintCD1d-PBS57tet+CD24–CD44–NK1.1–) cells (Fig. 1, C and D). Further, total number analysis revealed that the loss of iNKT cells in Mst1-deficient mice was specific for S3 cells (Fig. 1 D). Together, these data uncover a role for Mst1 in iNKT cell development.
Figure S1.
Requirement of Mst1 and Mst2 in iNKT cell and conventional T cell development. (A) Analysis of pMob1 (Thr35) with or without PMA/ionomycin stimulation for 30 min in WT DP thymocytes and total iNKT cells (n = 2 mice/group, with two technical replicates). (B) Analysis of pMob1 (Thr35) with or without PMA/ionomycin stimulation for 30 min in WT and Cd4CreMst1fl/fl S3 iNKT cells. (C and D) Flow cytometry analysis (C) and percentage and number (D) of S0 iNKT cells in the thymus of WT, Cd4CreMst1fl/fl, Cd4CreMst2fl/fl, and Cd4CreMst1fl/flMst2fl/fl mice. S0 iNKT cells were defined as TCRβintCD1d-PBS57tet+CD44–NK1.1–CD24+CD69+. S0 iNKT cell percentage was analyzed from CD8-depleted thymocytes, while total number was analyzed from CD1d-PBS57tet-enriched thymocytes (n = 3 or 4 mice/group). (E) Real-time PCR analysis of Stk4 (Mst1) and Stk3 (Mst2) relative expression in thymic S0, S1, S2, and S3 WT iNKT cells (n = 6 or 7 samples/group). (F) Stk4 and Stk3 expression in thymocyte T cell subsets curated from the Immunological Genome Project (GSE15907) microarray public dataset. (G) Flow cytometry analysis of CD4SP (CD4+CD8–TCRβ+), CD8SP (CD4–CD8+TCRβ+), DN (CD4–CD8–), and DP (CD4+CD8+) cells (left); mature (CD62LhiCD69loTCRβ+) and semi-mature (CD62LloCD69hiTCRβ+) CD4SP (middle) and CD8SP cells (right) in the thymus from WT, Cd4CreMst1fl/fl, Cd4CreMst2fl/fl mice (n = 6–8 mice/group). (H) Flow cytometry analysis of CD4SP (CD4+CD8–TCRβ+), CD8SP (CD4–CD8+TCRβ+), DN (CD4–CD8–), and DP (CD4+CD8+) cells (left); mature (CD62LhiCD69loTCRβ+) and semi-mature (CD62LloCD69hiTCRβ+) CD4SP (middle) and CD8SP cells (right) in the thymus from WT and Cd4CreMst1fl/flMst2fl/fl mice (n = 5–8 mice/group). (I) Flow cytometry analysis of thymic iNKT cells and developmental stages (S1, S2, and S3) from mixed bone marrow chimera mice. (J) Flow cytometry analysis of IL-15 induction of pMob1 (Thr35) in thymic iNKT cell developmental stages (S1–2 and S3; left) and effector subsets (iNKT2, iNKT17, and iNKT1; right; n = 3–5 mice/group). Numbers indicate MFI (A and B) or percentage of cells in gates or quadrants (C and I). Data are mean ± SEM. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. One-way ANOVA with Tukey’s test (A, D, and E) or two-way ANOVA with Bonferroni’s test (G, H, and J). Data are representative of two (A, B, I, and J) or three (C), or combined from two (H) or three (D, E, and G) independent experiments.
Figure 1.
Mst1 and Mst2 promote iNKT cell development and generation of S3 mature cells, with Mst1 playing a dominant role. (A–D) WT, Cd4CreMst1fl/fl, Cd4CreMst2fl/fl, and Cd4CreMst1fl/flMst2fl/fl mice were analyzed. (A) Flow cytometry analysis of iNKT cells from the thymus, spleen, and liver. iNKT cells from the thymus and spleen are gated from 7-AAD–B220– cells, and iNKT cells from the liver are gated from 7-AAD– cells. (B) Percentage (as gated in A) and total number of iNKT cells in the thymus, spleen, and liver (n = 6–13 mice/group). (C) Flow cytometry analysis of iNKT cell developmental stages from the thymus, gated from 7-AAD–B220–TCRβintCD1d-PBS57+ cells. S1 = CD24–CD44–NK1.1–, S2 = CD24–CD44+NK1.1–, S3 = CD24–CD44+NK1.1+. (D) Percentages (as gated in C) and numbers of iNKT cells at S1, S2, and S3 in the thymus (n = 6–10 mice/group). Numbers indicate percentage of cells in gates or quadrants (A and C). Data in plots indicate mean ± SEM (B and D). *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. One-way ANOVA with Tukey’s test (B and D). Data are combined from seven (A–D) independent experiments.
Unlike Mst1, genetic deletion of Mst2 alone did not significantly reduce the frequency or number of total iNKT cells in the thymus and liver (Fig. 1, A and B), although there was a modestly decreased number of iNKT cells in the spleen (Fig. 1 B). Also, Mst2-deficient mice showed normal frequencies and numbers of iNKT cell developmental stages in the thymus (Fig. 1, C and D; and Fig. S1, C and D). Consistent with these observations, Mst2 expression was reduced at late-stage iNKT cells (Fig. S1, E and F) and was expressed at much lower levels compared with Mst1 (Fig. S1 F). However, in the absence of Mst1, Mst2 expression was modestly increased in S2 and S3 iNKT cells (data not shown), suggesting that Mst2 may promote iNKT cell development when Mst1 is limiting. Consistent with this hypothesis, deletion of both Mst1 and Mst2 resulted in a trend toward further reduced frequency and number of iNKT cells in the thymus, spleen, and liver compared with Mst1-deficient mice (Fig. 1, A and B). Moreover, Mst1/2-deficient mice had elevated frequencies of S0–S2 iNKT cells but reduced S3 iNKT cells compared with Mst1-deficient mice, indicative of a more severe defect in maturation (Fig. 1, C and D; and Fig. S1, C and D ). These data indicate a dominant role for Mst1 in iNKT cell developmental maturation, and also partial genetic redundancy between Mst1 and Mst2.
In contrast to defective iNKT cell development, analysis of conventional T cells in the thymi of WT, Mst1-deficient, Mst2-deficient, and Mst1/2-deficient mice revealed no major reductions in the frequencies and numbers of CD4 single positive (SP), CD8SP, DN, and DP cells (Fig. S1, G and H). There was an increase in the frequency but not number of mature (CD62LhiCD69lo) CD4SP and CD8SP cells (and a corresponding reduction of semi-mature cells) in Mst1-deficient mice, which did not occur in Mst2-deficient mice (Fig. S1 G). The combined absence of both Mst1 and Mst2 promoted the accumulation of mature CD4SP but not CD8SP T cells in the thymus (Fig. S1 H). These data suggest that the expression of both Mst1 and Mst2 may promote the egress of mature T cells from the thymus, which is in line with the findings using related genetic models (Dong et al., 2009; Mou et al., 2012).
Unique and cell-intrinsic role of Mst1 signaling in iNKT cell development
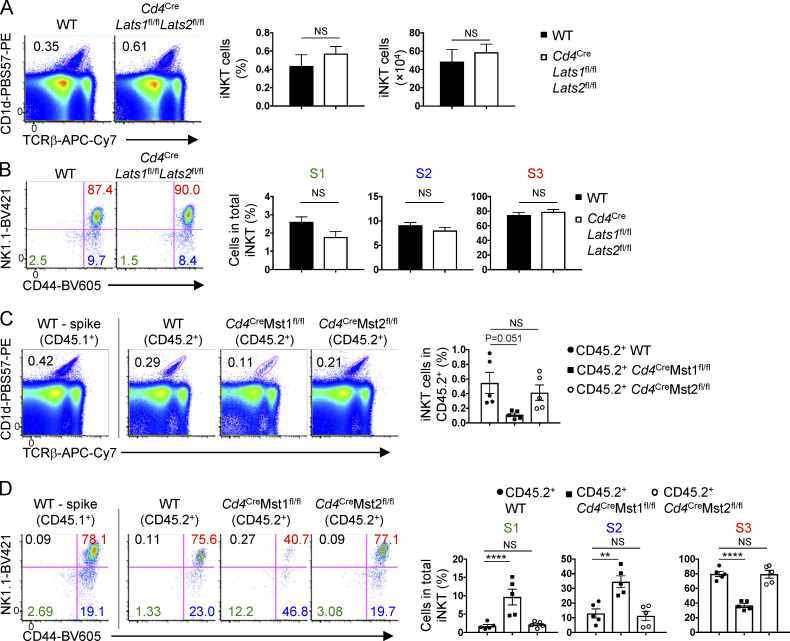
In the canonical Hippo signaling pathway, Lats1 and Lats2 are the downstream mediators of Mst signaling and negatively regulate the nuclear cofactors Yap (encoded by Yap1) and Taz (Wwtr1; Yu et al., 2015). To determine if the canonical Hippo pathway is required for iNKT cell development, we crossed Cd4Cre mice with Lats1fl/fl and Lats2fl/fl mice (denoted Cd4CreLats1fl/flLats2fl/fl) to generate mice with T cells deficient for Lats1/2 signaling. Lats1/2 deficiency did not affect the frequencies or numbers of total iNKT cells and developmental stages (S1–S3; Fig. 2, A and B). Similarly, Yap/Taz-deficient mice had largely normal iNKT cell development and maturation (data not shown). Therefore, iNKT cell development does not require Lats1/2 or Yap/Taz, suggesting that Mst1/2 regulate iNKT cell development through a noncanonical Hippo signaling pathway.
Figure 2.
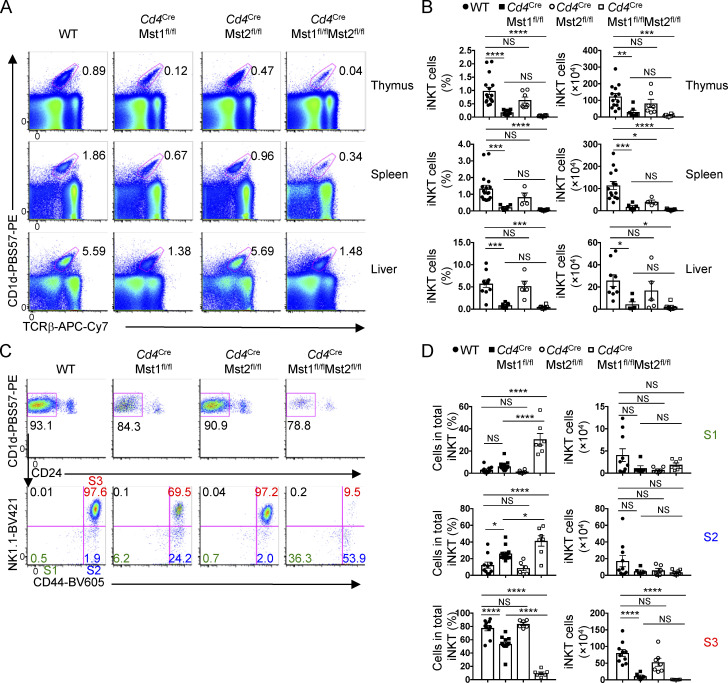
Unique and cell-intrinsic role of Mst1 in iNKT cell development. (A) Flow cytometry analysis (left), percentage (middle), and number (right) of iNKT cells from the thymus of WT and Cd4CreLats1fl/flLats2fl/fl mice (n = 3 or 4 mice/group). (B) Flow cytometry analysis (left) and percentages (right) of S1, S2, and S3 iNKT cells in the thymus of indicated mice (n = 3 or 4 mice/group). (C) Flow cytometry analysis (left) and percentage (right) of total iNKT cells from WT, Cd4CreMst1fl/fl, and Cd4CreMst2fl/fl mixed bone marrow chimera mice (n = 5 mice/group). (D) Flow cytometry analysis (left) and percentages (right) of S1, S2, and S3 iNKT cells in the thymus of indicated mice (n = 5 mice/group). Numbers indicate percentage of cells in gates or quadrants. Data in plots indicate mean ± SEM (A–D). **, P < 0.01, ****, P < 0.0001. Two-tailed unpaired Student’s t test (A and B) or one-way ANOVA with Tukey’s test (C and D). Data are combined from two (A and B) or three (C and D) independent experiments.
To determine if Mst-dependent regulation of iNKT cell development is cell-intrinsic, we generated mixed bone marrow chimera mice, in which Rag1−/− mice were reconstituted with CD45.1+ WT (“spike”) bone marrow cells mixed at a 1:1 ratio with bone marrow cells from CD45.2+ WT, Mst1-deficient, Mst2-deficient, or Mst1/2-deficient mice. Because WT DP cells are present, the bone marrow chimera system also allows us to rule out any potential defects in antigen presentation by Mst1/2-deficient DP cells that could account for the alterations in iNKT cell development. Mixed bone marrow chimera mice revealed a reduction in total iNKT cells and altered frequencies of iNKT cell developmental stages (i.e., increased S1 and S2 and decreased S3 cells) in the absence of Mst1 (Fig. 2, C and D) or both Mst1 and Mst2 (Fig. S1 I), but not in the absence of Mst2 alone (Fig. 2, C and D). Altogether, these data reveal Mst signaling as a unique and cell-intrinsic regulator for iNKT cell development and maturation.
Mst1 is activated by IL-15 and mediates IL-15–mediated iNKT cell survival
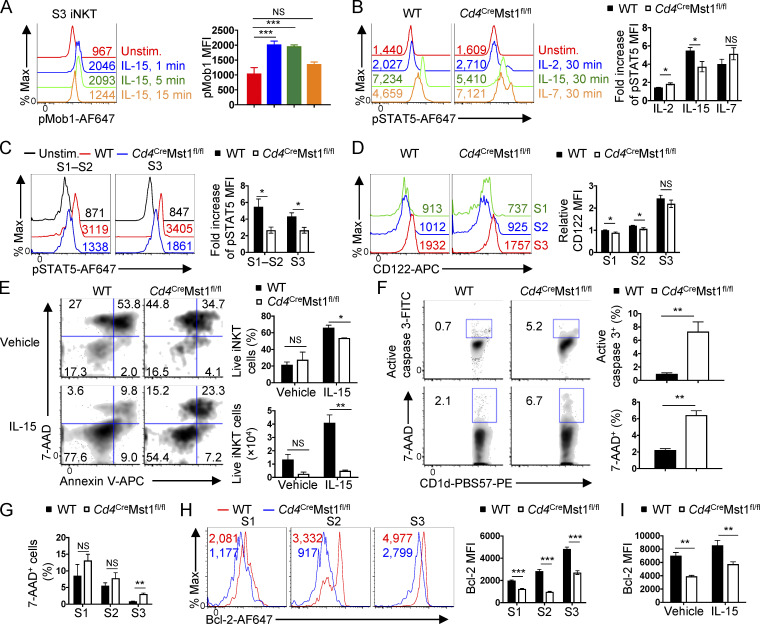
The common gamma chain (γc) cytokines IL-2, IL-7, and IL-15 are critical regulators of T cell homeostasis. While iNKT cell stages and effector subsets have differential requirements for cytokine-mediated development and survival, S3 iNKT cells are predominantly dependent on IL-15 (Crosby and Kronenberg, 2018; Matsuda et al., 2002; Wang and Hogquist, 2018). To determine whether γc-cytokine signaling serves as an upstream signal for Mst1 in iNKT cells, we assessed the induction of Mob1 phosphorylation by IL-15 and found that IL-15 transiently up-regulated pMob1 (Thr35; Fig. 3 A), which occurred in each iNKT cell developmental stage and effector subset examined (Fig. S1 J). Further, STAT5 phosphorylation in response to IL-15 but not IL-2 or IL-7 was reduced in Mst1-deficient iNKT cells, indicating that Mst1 amplifies IL-15–STAT5 signaling (Fig. 3 B). Importantly, the regulation of IL-15 signaling was not secondary to altered iNKT cell subsets caused by Mst1 deficiency, as impaired IL-15–induced STAT5 phosphorylation was observed in both S1–S2 and S3 Mst1-deficient iNKT cells (Fig. 3 C). We also found a modest reduction of CD122 (IL-2/IL-15Rβ) expression on S1 and S2 but not S3 cells lacking Mst1 (Fig. 3 D). Together, these data indicate that Mst1 is activated by IL-15 and contributes to IL-15–STAT5 signaling in iNKT cells.
Figure 3.
Mst1 promotes IL-15–mediated iNKT cell survival. (A) Flow cytometry analysis (left) and mean fluorescence intensity (MFI, right) of pMob1 (Thr35) induced by IL-15 (10 ng/ml) in WT thymic S3 iNKT cells (n = 2 mice/group with two technical replicates) at the indicated time points. Max, maximum; unstim, unstimulated. (B) Flow cytometry analysis (left) and MFI fold increase (right) of pSTAT5 induced by IL-2, IL-15, or IL-7 (10 ng/ml) in total thymic iNKT cells from WT and Cd4CreMst1fl/fl mice (n = 3 mice/group). Fold change is relative to unstimulated WT controls. (C) Flow cytometry analysis (left) and MFI fold increase (right) of pSTAT5 induced by IL-15 (10 ng/ml, 30 min stimulation) in thymic S1–S2 (combined) and S3 iNKT cells from WT and Cd4CreMst1fl/fl mice (n = 2 or 3 mice/group with two technical replicates). (D) Flow cytometry analysis (left) and relative MFI (right) of CD122 expression on thymic S1, S2, and S3 iNKT cells (n = 4 or 5 mice/group). Data are relative to the average MFI for WT S1 cells. (E) Flow cytometry analysis (left), percentages (right, upper), and numbers (right, lower) of iNKT cell survival, assessed by 7-AAD and annexin V staining after in vitro culture with vehicle or IL-15 (20 ng/ml) for 60–72 h (n = 3–5 mice/group). (F) Flow cytometry analysis (left) and percentages (right) of active caspase 3 and 7-AAD staining in total thymic iNKT cells (n = 3–5 mice/group). (G) Quantification of 7-AAD+ thymic S1, S2, and S3 iNKT cells (n = 3 mice/group). (H) Flow cytometry analysis (left) and MFI (right) of Bcl-2 expression in thymic S1, S2, and S3 iNKT cells (n = 3 mice/group). (I) Bcl-2 expression in total thymic iNKT cells after overnight culture with vehicle or IL-15 (10 ng/ml; n = 3 mice/group). Numbers indicate percentage of cells in gates or quadrants (E and F), or MFI (A–D and H). Data in plots indicate mean ± SEM (A–I). *, P < 0.05, **, P < 0.01, ***, P < 0.001. Two-tailed unpaired Student’s t test (A–I). Data are representative of or combined from two (A–G and I) or three (H) independent experiments.
As IL-15 plays a crucial role in iNKT cell homeostasis (Crosby and Kronenberg, 2018; Matsuda et al., 2002; Wang and Hogquist, 2018), we next explored the effect of Mst1 deficiency on iNKT cell survival. Compared with WT iNKT cells, Mst1-deficient cells had reduced survival when cultured with IL-15 in vitro (Fig. 3 E). Moreover, the frequency of Mst1-deficient iNKT cells stained with active caspase 3 or 7-AAD ex vivo was considerably increased compared with that of WT counterparts (Fig. 3 F), with S3 iNKT cells showing the most pronounced elevation of 7-AAD staining (Fig. 3 G). Together, these data reveal a role for Mst1 in promoting IL-15–mediated iNKT cell survival.
Survival of T cells is mediated by the cell-intrinsic apoptotic pathway and the balance of anti- and pro-apoptotic Bcl-2 family members. The anti-apoptotic protein Bcl-2 promotes T cell survival by combating Bim-mediated cell apoptosis, and expression of Bcl-2 can be regulated by γc cytokine signaling (Surh and Sprent, 2008). Mst1-deficient iNKT cells had reduced expression of Bcl-2 in S1–S3 cells (Fig. 3 H), and this defect persisted after culture with IL-15 (Fig. 3 I). Thus, associated with Mst1-dependent iNKT cell survival, Mst1 has an important role in promoting Bcl-2 expression.
Dynamic gene expression during iNKT cell maturation and key role of Mst1 in establishing quiescence
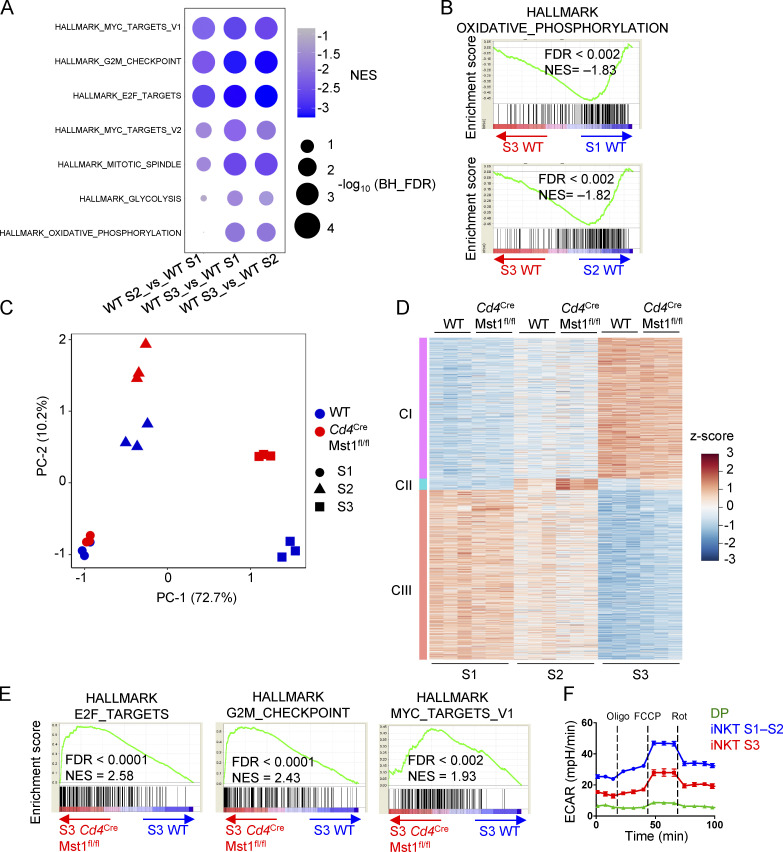
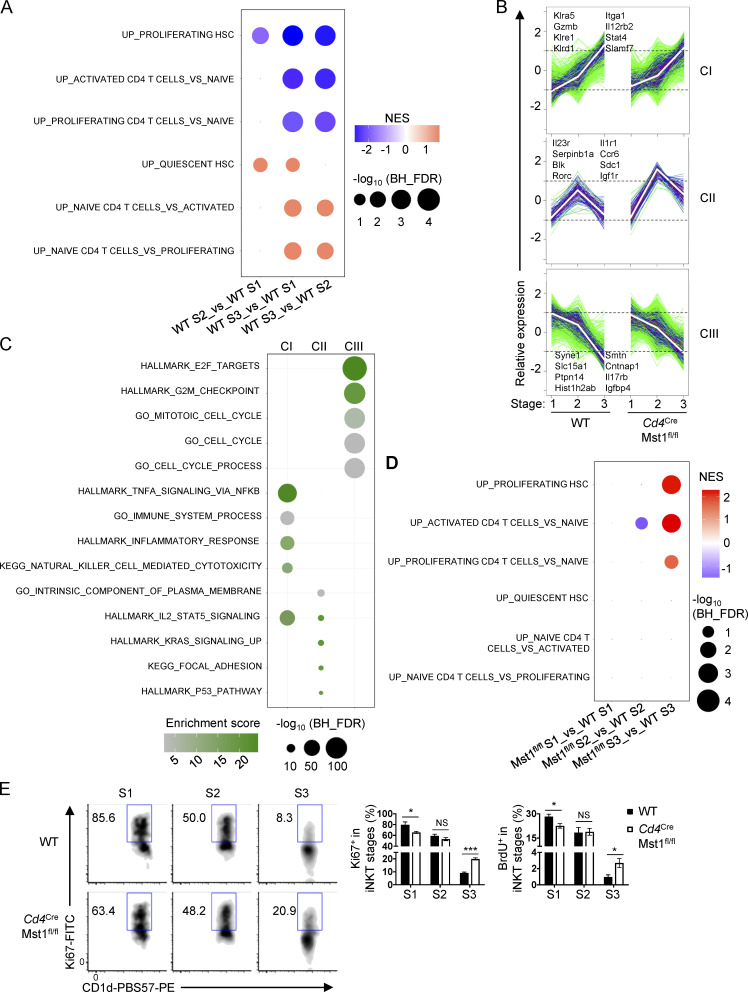
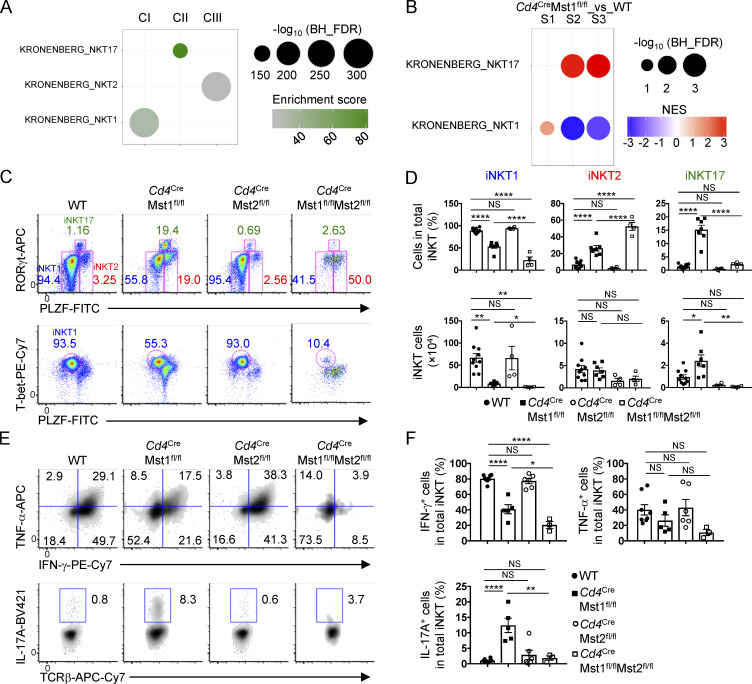
These observations prompted us to explore the transcriptional programs during iNKT cell development. To investigate the physiological regulation and molecular pathways involved in this process, we first analyzed the gene expression profiles of iNKT cells at S1, S2, and S3 from WT mice, and performed gene set enrichment analysis (GSEA) using the HALLMARK database. We found that the top negatively enriched pathways in S3 cells compared with S1 and S2 cells were related to cell cycle (e.g., Myc targets, G2M checkpoint, and E2F targets) and metabolic programs (e.g., glycolysis and oxidative phosphorylation [OXPHOS]; Fig. S2, A and B), indicating that transition into S3 cells is marked by down-regulation of proliferation and metabolic programs. Because such processes are reminiscent of cells becoming more quiescent, we next examined if quiescence programs were altered during iNKT cell development. To this end, we curated three gene sets that were up-regulated in quiescent hematopoietic stem cells (HSC) and naive CD4+ T cells and three gene sets that were up-regulated in activated and proliferating HSC and CD4+ T cells (Fig. 4 A; Elo et al., 2010; Venezia et al., 2004). Activated and proliferating CD4+ T cells are defined by the stimulation time points 24 h and 72 h, respectively, with the former time point indicating cells that have exited quiescence but are not yet under active proliferation (Chapman and Chi, 2018). GSEA using these six gene sets revealed that, relative to WT S1 and S2 iNKT cells, S3 cells were positively enriched for the naive/quiescence transcriptional programs (Fig. 4 A, lower three rows), and negatively enriched for the activation/proliferation transcriptional programs (Fig. 4 A, upper three rows), indicating that iNKT cells acquire quiescence signatures when they become S3 cells.
Figure S2.
WGCNA identifies developmentally regulated and Mst1-dependent gene expression clusters. (A) GSEA using the public HALLMARK gene sets in WT S1, WT S2, and WT S3 iNKT cells. The top most enriched pathways (upper five rows) and top enriched metabolic pathways (lower two rows) are presented. (B) GSEA enrichment plots of the HALLMARK OXPHOS gene set from comparison in A. (C) Principal component analysis of genes differentially expressed (with >0.5 log2 fold change) between WT S1, WT S2, WT S3, Cd4CreMst1fl/fl S1, Cd4CreMst1fl/fl S2, and Cd4CreMst1fl/fl S3 iNKT cells. (D) WGCNA clustering of 3,230 genes differentially expressed (with >1.5 fold change and <0.05 FDR). Heatmap of cluster 1 (CI), 2 (CII), and 3 (CIII) DE genes. (E) GSEA enrichment plots of three HALLMARK gene sets in Cd4CreMst1fl/fl S3 iNKT cells compared with WT S3 iNKT cells. (F) ECAR analysis after the addition of oligomycin (Oligo), FCCP, and rotenone (Rot) in thymic WT DP, S1–S2 iNKT (combined), and S3 iNKT cells. NES, normalized enrichment score. Data are representative of two (F) independent experiments.
Figure 4.
Mst1 enforces iNKT cellular quiescence during terminal maturation. (A) GSEA using gene sets up-regulated in proliferating and activated HSC and CD4+ T cells (upper three rows) or up-regulated in quiescent HSC and naive CD4+ T cells (lower three rows) for pairwise comparisons of WT S1, S2, and S3 thymic iNKT cells. (B) WGCNA clustering of 3,230 genes differentially expressed among the developmental stages (S1, S2, and S3) in WT and Mst1-deficient iNKT cells (with FDR <0.05 and fold-change >1.5). (C) Functional enrichment analysis of WGCNA clusters using Fisher’s exact test with FDR <0.05. Analysis was done using the public HALLMARK, GO, and KEGG databases. Plot shows the top five enriched pathways for clusters II and III (CII and CIII), and the top four enriched pathways with the addition of the KEGG NK cell–mediated cytotoxicity pathway for cluster I (CI). (D) GSEA using the gene sets described in A for pairwise comparisons of WT and Mst1-deficient S1, S2, and S3 iNKT cells. (E) Flow cytometry analysis of Ki67 expression in S1, S2, and S3 iNKT cells (left), and percentages of Ki67+ (middle) or BrdU+ (right) cells in S1, S2, and S3 iNKT cells in the thymus of WT and Cd4CreMst1fl/fl mice (n = 4 or 5 mice/group). NES, normalized enrichment score. Numbers indicate percentage of cells in gates (E). Data in plots indicate mean ± SEM (E). *, P < 0.05, ***, P < 0.001. Two-tailed unpaired Student’s t test (E). Data are combined from two (E, BrdU) or three (E, Ki67) independent experiments.
The effect of Mst1 deficiency on iNKT cell maturation presents a unique tool to perturb the transcriptional regulation of iNKT cell development. Thus, we compared the gene expression profiles of WT and Mst1-deficient iNKT cells in developmental stages S1, S2, and S3. Principal component analysis showed that the variance in gene expression was low between WT and Mst1-deficient iNKT cells at S1, but Mst1 deficiency accounted for increased variance at S2, and to an even greater extent at S3 (Fig. S2 C), suggesting temporally regulated and context-dependent roles of Mst1. To further examine the gene expression network during iNKT cell development, we first identified genes that were significantly changed between any stage or genotype using two-way ANOVA, and then defined 3,230 genes that were differentially expressed (DE; fold-change >1.5, false discovery rate [FDR] <0.05) in at least one of the pairwise comparisons between WT and Mst1-deficient cells at the same stage. Next, we performed weighted gene correlation network analysis (WGCNA) to identify gene clusters of highly correlated genes from the 3,230 DE genes (Tan et al., 2017). WGCNA identified three gene clusters: I, II, and III, which accounted for 1,411, 114, and 1,705 of the total DE genes, respectively (Fig. 4 B and Fig. S2 D). Cluster I genes were progressively increased in expression during iNKT cell maturation (Fig. 4 B and Fig. S2 D). Cluster II genes were up-regulated at S2 and then down-regulated at S3, and there was clear increased expression of these genes in Mst1-deficient S2 and S3 iNKT cells (Fig. 4 B and Fig. S2 D). Finally, cluster III genes were progressively decreased during iNKT cell maturation, and expression of these genes was increased in Mst1-deficient S3 compared with WT S3 iNKT cells (Fig. 4 B and Fig. S2 D). Thus, these data reveal the dynamic regulation of gene expression programs during iNKT cell maturation, with Mst1 controlling gene expression primarily at the late stages of iNKT cell development.
To identify the functional pathways associated with each WGCNA cluster, we performed functional enrichment analysis using the HALLMARK, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO) databases. The top enriched pathways for cluster I genes were related to natural killer (NK) cell function (Fig. 4 C), consistent with previous studies showing that iNKT cells progressively up-regulate expression of NK-related genes during development into S3 (Cohen et al., 2013). Cluster II genes were most enriched for pathways related to adhesion and cytokine signaling, while the highest enriched pathways for cluster III genes were associated with cell cycle regulation (Fig. 4 C). The increased expression of cluster III genes in Mst1-deficient S3 iNKT cells (Fig. 4 B and Fig. S2 D) suggests that Mst1 may be a critical regulator of the cell cycle–related transcriptional programs at the late developmental stage.
We therefore examined the role of Mst1 in modulating the proliferation and quiescence programs during iNKT cell development. Using the curated quiescence/naive and activation/proliferation gene sets, we found that Mst1-deficient S3 iNKT cells were positively enriched for activation/proliferation transcriptional programs (Fig. 4 D, top three rows) compared with WT S3 iNKT cells, and this alteration was not observed in S1 or S2 cells (Fig. 4 D). These data raised the possibility that Mst1-deficient iNKT cells are less quiescent at S3. Consistent with these data, GSEA showed that Mst1-deficient S3 iNKT cells up-regulated transcriptional programs related to cell cycle (e.g., E2F targets and G2M checkpoint), as well as Myc signaling, a central regulator of cell cycle (Chi, 2012; Dose et al., 2009; Wang et al., 2011; Fig. S2 E). Indeed, analysis of Ki67 expression and BrdU incorporation in iNKT cells at each developmental stage revealed that Mst1-deficient S3 iNKT cells had increased frequencies of Ki67+ and BrdU+ cells compared with WT S3 iNKT cells (Fig. 4 E). Together, these data demonstrate that Mst1 enforces the suppression of cell cycle programs associated with the establishment of cellular quiescence during iNKT cell maturation into S3 cells.
Mst1 regulates metabolic and mitochondrial homeostasis during iNKT cell development
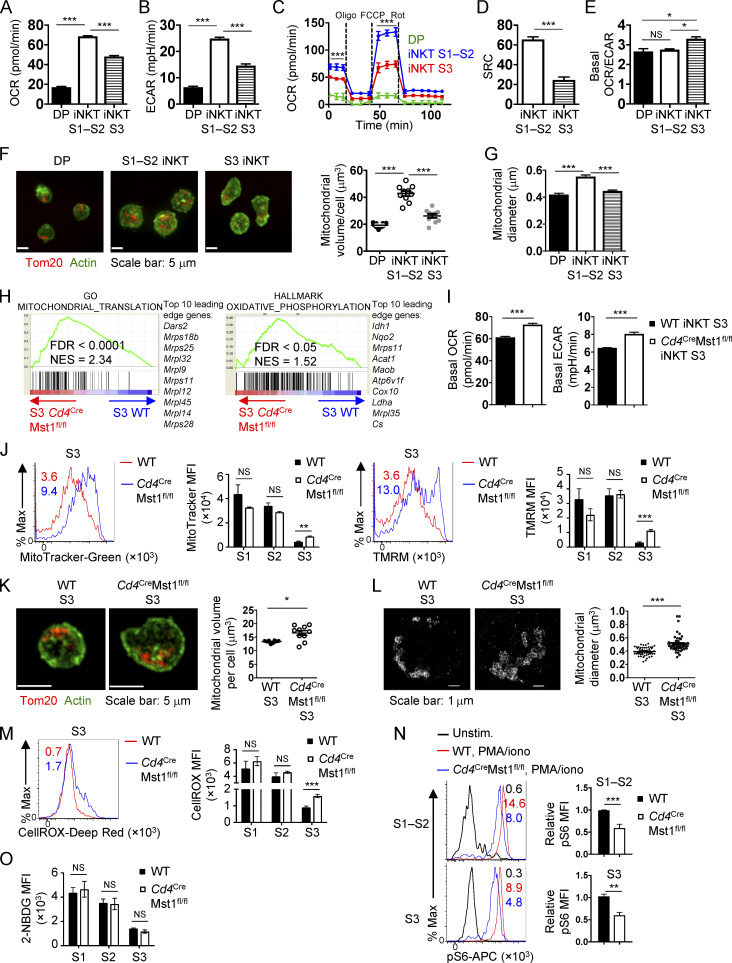
We next determined the mechanisms underpinning cellular quiescence during iNKT cell development. Of note, GSEA of the WT S1, S2, and S3 iNKT cells indicated that transcriptional programs associated with metabolism (glycolysis and OXPHOS) were dynamically regulated during iNKT cell development (Fig. S2, A and B). We therefore measured the oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) in WT DP cells, and immature (S1–S2) and mature (S3) iNKT cells. Interestingly, thymocytes transitioning from the DP stage into immature iNKT cells up-regulated OXPHOS and glycolytic activity, as indicated by elevated OCR and ECAR, respectively (Fig. 5, A–C; and Fig. S2 F). Also, compared with S1–S2 iNKT cells, S3 iNKT cells had reduced OCR, ECAR, and spare respiratory capacity (SRC), which is a measure of the mitochondrial capacity to generate energy under stressed conditions (Fig. 5, A–D; and Fig. S2 F ). The OCR/ECAR ratio was increased in S3 iNKT cells (Fig. 5 E), indicating that iNKT cells acquire metabolic features similar to quiescent naive and memory T cells (i.e., OXPHOS activity is increased compared with glycolysis; Geltink et al., 2018) at the terminal developmental stage. We also imaged and quantified the mitochondrial volume and diameter in DP, S1–S2 iNKT, and S3 iNKT cells. Up-regulation of OCR and ECAR in S1–S2 iNKT cells compared with DP cells (Fig. 5, A and B) was accompanied by increased mitochondrial volume and diameter (Fig. 5, F and G). Furthermore, S3 iNKT cells showed reduced mitochondrial volume and diameter relative to S1–S2 iNKT cells (Fig. 5, F and G), which correlated with a reduction in OCR, ECAR, and SRC (Fig. 5, A–D). Collectively, these data show that metabolic activity and mitochondrial content are dynamically regulated during iNKT cell development.
Figure 5.
Dynamic regulation of iNKT cell metabolism during development and the Mst1 dependence. (A and B) Basal OCR (A) and ECAR (B) in WT thymic CD4+CD8+ (DP), S1–S2 iNKT (combined), and S3 iNKT cells. (C) OCR in WT DP, S1–S2 iNKT, and S3 iNKT cells after the addition of oligomycin (Oligo), FCCP, and rotenone (Rot). (D) SRC (= maximum OCR – baseline OCR) in WT S1–S2 iNKT and S3 iNKT cells. (E) Basal OCR/ECAR ratio in WT DP, S1–S2 iNKT, and S3 iNKT cells. (F) Confocal microscopy analysis of Tom20 (red, staining mitochondria) and actin (green) in WT DP, S1–S2 iNKT, and S3 iNKT cells (left). Scale bar, 5 µm. Right, quantification of mitochondrial volume per cell (μm3; n = 10 cells/group). (G) Quantification of mitochondrial diameter (μm) using STORM imaging of WT DP, S1–S2 iNKT, and S3 iNKT cells. (H) Enriched pathways identified by GSEA from GO and HALLMARK gene sets in Mst1-deficient S3 iNKT cells compared with WT S3 iNKT cells. (I) Basal OCR and ECAR in thymic WT and Cd4CreMst1fl/fl S3 iNKT cells (n = 3 wells/group combined from 15 mice/group). (J) MFI of MitoTracker (left) and TMRM (right) in thymic S1, S2, and S3 iNKT cells from WT and Cd4CreMst1fl/fl mice (n = 3 or 4 mice/group). Histogram plots represent flow cytometry analysis of MitoTracker and TMRM in thymic S3 iNKT cells. (K) Confocal microscopy analysis (left) of Tom20 (red, staining mitochondria) and actin (green) in thymic WT and Cd4CreMst1fl/fl S3 iNKT cells and quantification (right) of mitochondrial volume per cell (μm3; n = 2 mice/group; n = 269–461 total cells from 10 fields). Scale bar, 5 µm. (L) STORM imaging (left) and quantification (right) of mitochondrial diameter (μm) in thymic WT and Cd4CreMst1fl/fl S3 iNKT cells (n = 2 mice/group; n = 35–37 cells imaged/group). Scale bar, 1 µm. (M) Flow cytometry analysis (left) and MFI (right) of CellROX (cellular ROS) in thymic S1, S2, and S3 iNKT cells from WT and Cd4CreMst1fl/fl mice (n = 4 or 5 mice/group). (N) Flow cytometry analysis of phosphorylated S6 (pS6) in thymic S1–S2 and S3 iNKT cells from WT and Cd4CreMst1fl/fl mice stimulated with or without PMA/ionomycin (PMA/iono) for 30 min (n = 4–6 mice/group). Data are relative to the average pS6 MFI for WT cells. (O) MFI of ex vivo 2-NBDG uptake in S1, S2, and S3 iNKT cells from WT and Cd4CreMst1fl/fl mice (n = 5 mice/group). Numbers in the plots indicate MFI (J, M, and N). Data in plots indicate mean ± SEM (A–G and I–O). *, P < 0.05, **, P < 0.01, ***, P < 0.001. One-way ANOVA with Tukey’s test (A, B, and E–G), two-way ANOVA with Bonferroni’s test (C), or two-tailed unpaired Student’s t test (D, I, and J–O). Data are representative of two independent experiments (A–E, I, K, and L), or combined from two (F, G, and L) or three (J and M–O) independent experiments.
Given that Mst1-deficient iNKT cells had reduced features of quiescence (Fig. 4, D and E), we next explored if these defects were also associated with changes in cellular metabolism. GSEA showed that transcriptional programs for mitochondrial translation and OXPHOS were enriched in Mst1-deficient S3 iNKT cells (Fig. 5 H). We therefore measured OCR and ECAR and found that these parameters were elevated in Mst1-deficient S3 iNKT cells compared with WT cells (Fig. 5 I). Next, we characterized the dynamics of mitochondrial features during iNKT cell maturation and the dependence upon Mst1. Consistent with the imaging data of mitochondria (Fig. 5 F), flow-cytometric analyses by MitoTracker and TMRM staining revealed that WT S3 iNKT cells down-regulated mitochondrial mass and membrane potential, respectively, compared with WT S1 and S2 iNKT cells (Fig. 5 J), as previously reported (Salio et al., 2014). However, compared with WT S3 iNKT cells, Mst1-deficient S3 iNKT cells had elevated MitoTracker and tetramethylrhodamine, methyl ester (TMRM) staining (Fig. 5 J), as well as increased mitochondrial volume and diameter as measured by high-resolution imaging (Fig. 5, K and L), in line with elevated OCR (Fig. 5 I). We also found that intracellular ROS levels (as indicated by CellROX staining) were down-regulated during development in WT iNKT cells, whereas Mst1-deficient S3 iNKT cells had higher levels of ROS (Fig. 5 M). Although mTORC1 promotes mitochondrial metabolism in conventional T cells (Tan et al., 2017), mTORC1 signaling was modestly reduced in Mst1-deficient S1–S2 and S3 iNKT cells, as determined by the phosphorylation of S6 (Fig. 5 N), suggesting that the increased mitochondrial parameters were unlikely due to increased mTORC1 activity. Altogether, these data reveal Mst1 as a crucial regulator of mitochondrial homeostasis during late-stage iNKT cell development.
Aside from mitochondrial features, glycolysis was also dynamically regulated during iNKT cell development (Fig. 5 B and Fig. S2 F; Salio et al., 2014). We therefore used flow cytometry to analyze the uptake of 2-NBDG, a glucose analogue (Zeng et al., 2016), to examine if glucose utilization is altered in Mst1-deficient iNKT cells. We found that WT iNKT cells down-regulated the uptake of 2-NBDG at S3 (Fig. 5 O). However, in contrast to the altered mitochondrial profiles and ECAR (Fig. 5 I), the uptake of 2-NBDG was similar between WT and Mst1-deficient iNKT cells at all stages examined (Fig. 5 O). Thus, Mst1 is not required to regulate glucose uptake during iNKT cell development, although it does restrain ECAR in S3 iNKT cells. Altogether, these data indicate that Mst1 predominately regulates mitochondrial homeostasis that is associated with the establishment of quiescence programs during late-stage iNKT cell development.
Mst signaling regulates reciprocal iNKT1 and iNKT17 cell development
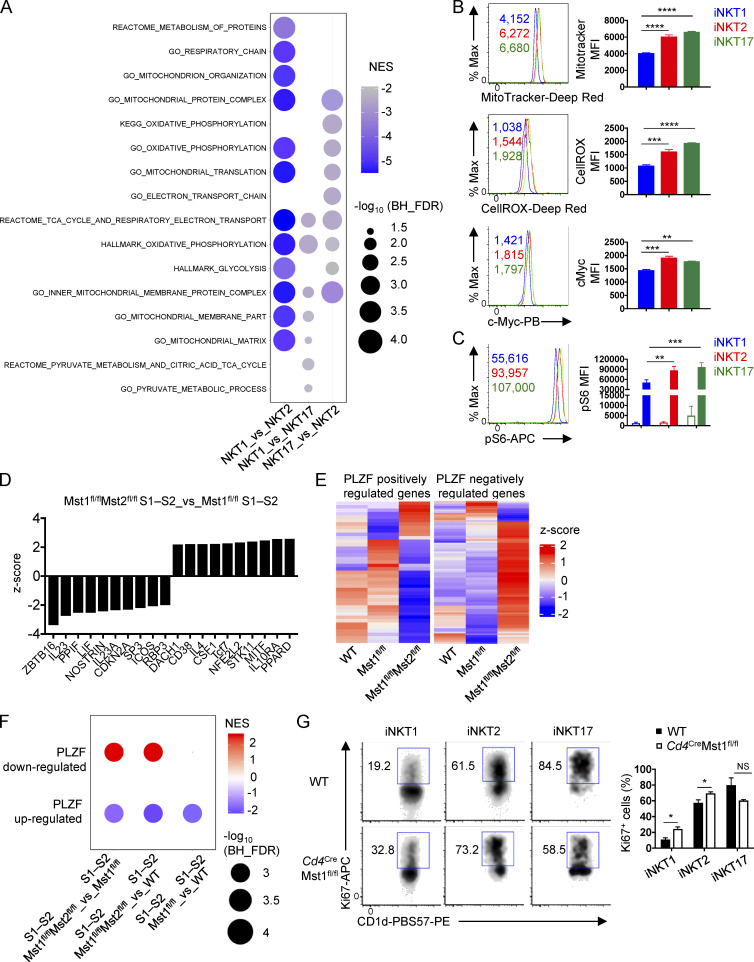
The iNKT developmental stages are comprised of three effector subsets (iNKT1, iNKT2, and iNKT17), with iNKT2 cells enriched in S1 and S2, iNKT17 cells in S2, and iNKT1 in S3 (Lee et al., 2013). Therefore, we hypothesized that iNKT effector cell differentiation is linked to specific metabolic programs. First, we examined metabolism-associated transcriptional programs in iNKT1, iNKT2, and iNKT17 cells using a published RNA sequencing (RNA-seq) dataset (Engel et al., 2016). GSEA using the HALLMARK, GO, and canonical datasets revealed that, relative to iNKT2 and iNKT17 cells, iNKT1 cells showed negative enrichments for transcriptional programs related to mitochondrial metabolism (e.g., TCA cycle and OXPHOS) and glycolysis (Fig. S3 A). Second, we examined metabolic parameters by flow cytometry and found that iNKT1 cells had decreased mitochondrial quantity, cellular ROS, and c-Myc expression compared with iNKT2 and iNKT17 cells (Fig. S3 B). Third, S6 phosphorylation, indicative of mTORC1 signaling that correlates with anabolic metabolism, was lower in iNKT1 cells compared with iNKT2 and iNKT17 cells following PMA and ionomycin stimulation (Fig. S3 C). Together, these data indicate that iNKT1 cells are more metabolically quiescent compared with iNKT2 and iNKT17 cells, suggesting dynamic regulation of cellular metabolism during iNKT effector cell differentiation.
Figure S3.
Transcriptional programs and flow cytometry analysis of metabolic processes, mitochondrial activity, and PLZF signaling in iNKT cell effector subsets and developmental stages. (A) GSEA using the public HALLMARK, GO, and canonical gene sets for iNKT effector cells (iNKT1, iNKT2, and iNKT17) from RNA-seq datasets previously published (Engel et al., 2016). Top enriched pathways related to mitochondrial metabolism or glycolysis are represented for each pairwise comparison. None of these metabolic pathways were positively enriched. (B) Flow cytometry analysis (left) and MFI (right) of MitoTracker, CellRox, and c-Myc in iNKT effector cells from the thymus of WT mice (n = 3 mice/group). (C) pS6 analysis in iNKT effector subsets unstimulated (open bar) or stimulated (closed bar) with PMA/ionomycin for 30 min (n = 3 mice/group). (D) IPA of the transcriptomes of thymic S1–S2 iNKT cells from Cd4CreMst1fl/flMst2fl/fl and Cd4CreMst1fl/fl mice. Data show the top 10 up-regulated and down-regulated pathways related to complexes, transcriptional regulators, enzymes, transporters, receptors, and cytokines. (E) Heatmap showing PLZF target genes differentially expressed between Cd4CreMst1fl/fl S1–S2 iNKT cells and Cd4CreMst1fl/flMst2fl/fl S1–S2 iNKT cells (with nominal P < 0.05) in thymic S1–S2 iNKT cells from WT, Cd4CreMst1fl/fl, and Cd4CreMst1fl/flMst2fl/fl mice. (F) GSEA analysis using the curated PLZF-regulated gene sets (with log2 fold change >0.5 and FDR <0.05; Mao et al., 2016) in WT S1–S2, Cd4CreMst1fl/fl S1–S2, and Cd4CreMst1fl/flMst2fl/fl S1–S2 iNKT cells. NES, normalized enrichment score. Differentially expressed transcripts were identified using two-way ANOVA followed by multiple testing correction using the Benjamini–Hochberg method to estimate FDR. (G) Flow cytometry analysis (left) and percentages (right) of Ki67+ iNKT effector cells from the thymus of WT and Cd4CreMst1fl/fl mice (n = 3 mice/group). Numbers indicate MFI or percentage of cells in gates. Data are mean ± SEM. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. One-way ANOVA with Tukey’s test (B), two-way ANOVA with Tukey’s test (C), two-tailed unpaired Student’s t test (G). Data are representative of two (B and C) or three (G) independent experiments.
To examine the role of Mst1 in iNKT effector fate decisions, we first performed functional enrichment analysis of the WGCNA clusters using the curated gene signatures for the iNKT1, iNKT2, and iNKT17 effector cell subsets (Engel et al., 2016). Cluster I, II, and III genes were enriched for iNKT1, iNKT17, and iNKT2 effector cell gene signatures, respectively (Fig. 6 A). Moreover, GSEA of the iNKT effector cell gene signatures revealed up-regulation of the iNKT17 gene signature in Mst1-deficient S2 and S3 iNKT cells, while the iNKT1 gene signature was down-regulated (Fig. 6 B), suggesting a possible role for Mst1 in shaping transcriptional programing related to iNKT effector cell lineages.
Figure 6.
Mst1 reciprocally regulates iNKT1 and iNKT17 effector cell subsets. (A) Functional enrichment analysis of WGCNA clusters using Fisher’s exact test with FDR <0.05. Analysis was done using the published gene sets for iNKT17, iNKT2, and iNKT1 cells (Engel et al., 2016). (B) GSEA using the iNKT1 and iNKT17 gene sets described in A for pairwise comparison of Mst1-deficient and WT S1, S2, and S3 iNKT cells. Data are representative of the enrichment in Mst1-deficient S1, S2, and S3 iNKT cells. (C and D) Flow cytometry analysis (C), percentages (D, upper), and numbers (D, lower) of iNKT effector cell subsets in the thymus of WT, Cd4CreMst1fl/fl, Cd4CreMst2fl/fl, and Cd4CreMst1fl/flMst2fl/fl mice (n = 4–10 mice/group). iNKT1: T-bethiPLZFlo, iNKT2: RORγtloPLZFhi, and iNKT17: RORγthiPLZFint. (E and F) Flow cytometry analysis (E) and percentages (F) of intracellular cytokine expression in thymic iNKT cells from indicated mice (n = 3–8 mice/group). Numbers indicate percentage of cells in gates or quadrants (C and E). Data in plots indicate mean ± SEM (D and F). *, P < 0.05, **, P < 0.01, ****, P < 0.0001. One-way ANOVA with Tukey’s test (D and F). Data are combined from four (E and F) or five (C and D) independent experiments.
To directly test the functional importance of Mst1 in iNKT effector cell development, we analyzed iNKT cell subsets in the thymus of WT or Mst1-deficient mice based on transcription factor and cytokine profiles: iNKT1 (T-bethiPLZFlo or RORγtloPLZFlo; IFN-γ–producing), iNKT2 (RORγtloPLZFhi; IL-4–producing), and iNKT17 (RORγthiPLZFint; IL-17A–producing; Crosby and Kronenberg, 2018; Wang and Hogquist, 2018). Transcription factor analysis revealed that compared with WT controls, Mst1-deficient mice had reduced frequency and number of iNKT1 cells but markedly increased frequency and number of iNKT17 cells (Fig. 6, C and D). Mst1-deficient mice also showed a modestly increased frequency but not number of iNKT2 cells (Fig. 6, C and D), suggesting that Mst1 may not be an important regulator of iNKT2 cells. The altered frequencies and numbers of iNKT1 and iNKT17 cell subsets in Mst1-deficient mice correlated with changes in cytokine production, with decreased IFN-γ– and increased IL-17A–producing iNKT cells upon PMA/ionomycin stimulation (Fig. 6, E and F). Therefore, Mst1 is a critical positive regulator of iNKT1 cell development and reciprocally regulates iNKT1 and iNKT17 cell differentiation.
To understand the individual and combined roles of Mst1 and Mst2 in iNKT effector cell development, we analyzed the frequencies and numbers of iNKT1, iNKT2, and iNKT17 cells in Mst2- and Mst1/2-deficient mice. Mst2-deficient mice showed no differences in iNKT effector cell subsets or cytokine production (Fig. 6, C–F). However, Mst1/2-deficient mice had lower frequency and number of iNKT1 cells compared with WT or Mst1-deficient iNKT cells based on transcription factor analysis (Fig. 6, C and D). Consistent with these observations, there was a more pronounced reduction of IFN-γ expression in Mst1/2-deficient iNKT cells than Mst1-deficient cells (Fig. 6, E and F), indicating that both Mst1 and Mst2 contribute to IFN-γ production by iNKT1 cells. Surprisingly, the increased frequency of iNKT17 cells and increased IL-17A production in Mst1-deficient mice were largely blocked with the additional deletion of Mst2 (Fig. 6, C–F), suggesting that Mst2 promotes iNKT17 cell development in the absence of Mst1. Altogether, these data suggest partially redundant functions for Mst1 and Mst2 in iNKT1 cell differentiation but opposing roles in iNKT17 cell development.
To identify putative mechanisms underlying altered effector subset development in Mst1- and Mst1/2-deficient mice, we compared the gene expression profiles of WT, Mst1-deficient, and Mst1/2-deficient iNKT cells in developmental stages S1–S2, collectively comprised of iNKT2 and iNKT17 cells, and S3, predominately comprised of iNKT1 cells (Lee et al., 2013). Ingenuity pathway analysis (IPA) showed that the activity of PLZF (encoded by Zbtb16), a master transcription factor critical for iNKT cell expansion and effector lineage differentiation during thymic development (Kovalovsky et al., 2008; Savage et al., 2008), was reduced in Mst1/2-deficient S1–S2 iNKT cells compared with Mst1-deficient S1–S2 iNKT cells (Fig. S3 D), but not in S3 cells (data not shown). Importantly, compared with iNKT1 cells, iNKT17 cells are more dependent on increased PLZF protein expression and activity for development (Park et al., 2019). Consistent with the IPA, the expression of many positively and negatively regulated PLZF target genes (Mao et al., 2016) was decreased or increased, respectively, in Mst1/2-deficient S1–S2 iNKT cells (Fig. S3 E), even though PLZF protein expression itself was not altered (data not shown). Therefore, we performed GSEA and observed that Mst1/2-deficient S1–S2 iNKT cells were enriched for the PLZF down-regulated target genes, and negatively enriched for PLZF up-regulated target genes (Fig. S3 F). Together, these data show that PLZF activity is impaired in Mst1/2-deficient S1–S2 iNKT cells, which may contribute to reduced iNKT17 cell development in Mst1/2-deficient mice compared with Mst1-deficient mice.
Mitochondrial homeostasis controls iNKT1 cell differentiation and quiescence
S3 iNKT cells and iNKT1 cells are transcriptionally and metabolically quiescent (Fig. 4 A, Fig. 5, A–E; Fig. S2, A and B; and Fig. S3, A–C) and can establish long-lived thymic residency (Berzins et al., 2006). To determine whether impaired quiescence in Mst1-deficient S3 cells was associated with a particular effector subset, we examined the cell cycle state in iNKT1, iNKT2, and iNKT17 cells. Importantly, Ki67 expression was up-regulated in Mst1-deficient iNKT1 cells and to a lesser extent iNKT2 cells (Fig. S3 G), while iNKT17 cell proliferation was not significantly altered (Fig. S3 G). Thus, Mst1 enforces quiescence in iNKT1 cells.
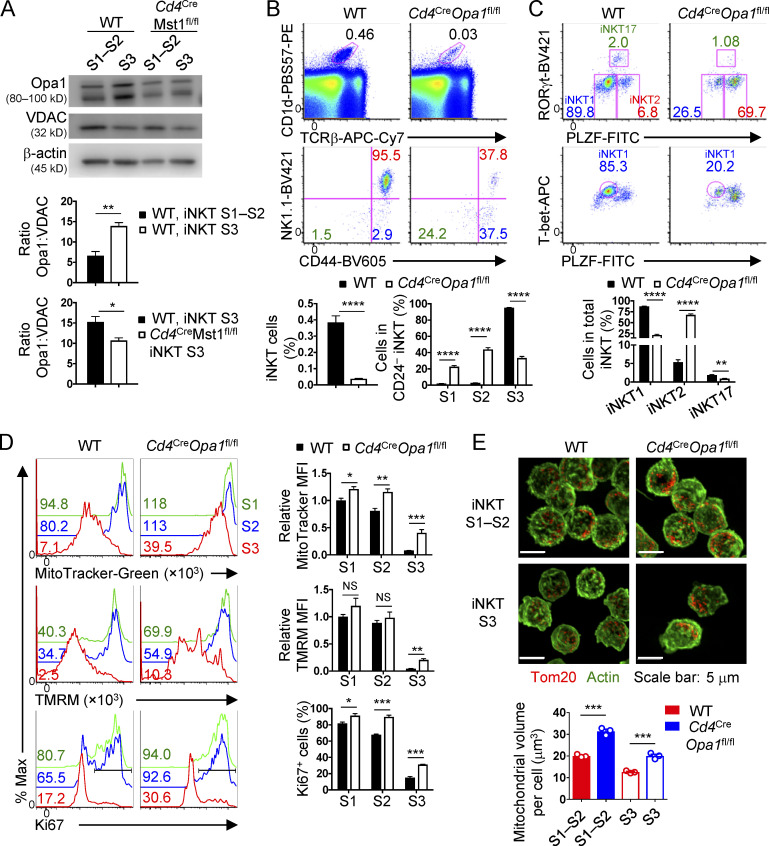
Mitochondrial homeostasis is critical for metabolic reprogramming and cell fate decisions (Geltink et al., 2018), but how this process is regulated in iNKT cell development is poorly understood. Opa1, a protein that promotes fusion of the inner mitochondrial membrane, is necessary for the generation of metabolically quiescent memory CD8+ T cells (Buck et al., 2016). Therefore, we examined the regulation of Opa1 protein expression by Mst1 during iNKT cell development. Relative to the mitochondrial loading control voltage-dependent anion channel (VDAC, which exhibits stable protein expression when Opa1 protein is reduced; Kushnareva et al., 2016), Opa1 expression was increased in WT S3 iNKT cells (which are more quiescent) compared with WT S1–S2 iNKT cells (Fig. 7 A). Further, compared with WT cells, Mst1-deficient S3 iNKT cells (which are less quiescent) had reduced Opa1 expression (Fig. 7 A). These observations prompted us to examine the role of Opa1 in iNKT effector subset development and regulation of cellular quiescence. We crossed Cd4Cre mice with Opa1fl/fl mice (denoted Cd4CreOpa1fl/fl) to generate mice with T cell–specific deletion of Opa1. Opa1-deficient iNKT cells had reduced S3 cells and elevated frequencies of S1 and S2 cells (Fig. 7 B). Further, development of iNKT1 cells was impaired, while the frequencies of iNKT2 and iNKT17 cells were significantly elevated or slightly reduced, respectively (Fig. 7 C). Opa1-deficient iNKT cells had elevated mitochondrial mass, mitochondrial membrane potential, Ki67+ proliferative cells, and mitochondrial volume by imaging, with the majority of these parameters showing a more pronounced change in S3 iNKT cells (Fig. 7, D and E), suggesting that Opa1 enforces iNKT cellular quiescence during development. Altogether, these data indicate that the regulation of mitochondrial homeostasis is critical for the development of S3 iNKT cells and iNKT1 cells and establishment of cellular quiescence.
Figure 7.
Opa1 enforces S3 iNKT and iNKT1 cell development and quiescence-associated features. (A) Immunoblot analysis of Opa1, VDAC (mitochondrial loading control), and β-actin (total protein loading control) in thymic S1–S2 and S3 iNKT cells from WT and Cd4CreMst1fl/fl mice. Opa1 expression was quantified relatively to VDAC expression (n = 3–6 samples/group). (B and C) Flow cytometry analysis (upper) and percentages (lower) of total thymic iNKT cells and developmental stages (S1, S2, and S3; B) and effector cell subsets (C) in the thymus of WT and Cd4CreOpa1fl/fl mice. (D) Flow cytometry analysis (left) and quantification of relative MFI (right) of MitoTracker (upper) or TMRM (middle) and percentages of Ki67+ cells (lower) in thymic S1, S2, and S3 iNKT cells from WT and Cd4CreOpa1fl/fl mice (n = 5 mice/group). MitoTracker and TMRM expression is relative to average MFI of WT S1 iNKT cells. (E) Confocal microscopy analysis (upper) of Tom20 (red, staining mitochondria) and actin (green) in S1–S2 and S3 iNKT cells from the thymus of WT and Cd4CreOpa1fl/fl mice and quantification (lower) of mitochondrial volume per cell (μm3; n = 2 mice/group; n = 20–91 total cells from three fields). Numbers indicate MFI or percentage of cells in gates or quadrants (B–D). Data in plots indicate mean ± SEM (A–E). *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. Two-tailed unpaired Student’s t test (A–E). Data are combined from three (A) or four (B–D) or representative of two (E) independent experiments.
iNKT17 cell differentiation in the absence of Mst1 requires ICOS–mTORC2 signaling
Recent evidence has identified altered TCR signal strength as a regulator of iNKT cell effector subset development, with decreased TCR signal strength leading to a predominance of iNKT1 cells, while strong TCR signal strength enhances iNKT2 and iNKT17 cell development (Tuttle et al., 2018; Zhao et al., 2018). The induction of Mob1 (Thr35) phosphorylation downstream of TCR/CD28 signaling occurred in an Mst1-dependent manner (Fig. S4, A and B), suggesting that TCR can activate Mst1 signaling without altering Mst1 expression (Fig. S4 C). To determine whether Mst1 signaling is linked to the regulation of TCR signal strength, we examined the expression of signature molecules correlated directly with TCR signal strength (TCRβ, CD5, CD6, and Egr2) or indirectly with TCR signal strength and avidity for self-antigen (Ly6C; Martin et al., 2013; Tuttle et al., 2018) in iNKT effector subsets from WT and Mst1-deficient mice. Mst1-deficient iNKT17 cells showed largely unaltered expression of these molecules, except for a slight reduction of Egr2 (Fig. S4 D). In addition, Mst1-deficient iNKT1 cells had a modest increase of Egr2 and Ly6C and normal expression of other markers (Fig. S4 D). As iNKT17 and iNKT1 cell development is respectively associated with strong and weak TCR signal strength (Tuttle et al., 2018; Zhao et al., 2018), these changes are unlikely to account for the increased iNKT17 and reduced iNKT1 cell development in the absence of Mst1. Therefore, Mst1 is likely not a major regulator of TCR signal strength in iNKT cells, which is in agreement with a previous report that Mst1 deficiency does not alter the activation of effector molecules (e.g., CD3ζ, ZAP70, and Lck) downstream of TCR in conventional T cells (Zhou et al., 2008).
Figure S4.
ICOS–mTORC2 signaling in iNKT cell development. (A) Flow cytometry analysis of pMob1 (Thr35) MFI in thymic S1–S2 and S3 iNKT cells from WT mice (n = 4 samples/group). (B) Flow cytometry analysis of pMob1 (Thr35) MFI in thymic WT and Cd4CreMst1fl/fl iNKT cells. (C) Immunoblot analysis of total Mst1 and β-actin protein in thymic S1–S2 and S3 iNKT cells from WT mice after stimulation by cross-linked CD3 and CD28. (D) Flow cytometry analysis of relative MFI of indicated molecule expression on thymic iNKT cell effector subsets from WT and Cd4CreMst1fl/fl mice (n = 3 or 4 mice/group). Data are relative to the average MFI of each WT iNKT effector subset. (E) Flow cytometry analysis (upper) and statistics of MFI (lower) of ICOS expression on total iNKT cells in the thymus of WT and Cd4CreMst1fl/fl mice (n = 4 mice/group). (F) Flow cytometry analysis (upper) and relative MFI (lower) of 4-1BB, OX40 and GITR expression on total iNKT cells in the thymus of WT and Cd4CreMst1fl/fl mice (n = 3–6 mice/group). Data are relative to the average MFI of WT iNKT cells. (G) Flow cytometry analysis (left) and percentages (right) of iNKT developmental stages (S1, S2, and S3) from the thymus of WT, ICOS−/− (Cd4CreMst1fl/+ or +/+), Cd4CreMst1fl/fl, and Cd4CreMst1fl/flICOS−/− mice (n = 4 or 5 mice/group). (H) Induction of Akt phosphorylation (Ser473) in WT S1–S2 and S3 iNKT cells after cross-linking ICOS, CD3, or ICOS and CD3. (I) Flow cytometry analysis of ICOS expression on WT S1, S2, and S3 iNKT cells. (J) Flow cytometry analysis (left) and percentages (right) of iNKT developmental stages from the thymus of WT, Cd4CreRictorfl/fl (Mst1fl/+), Cd4CreMst1fl/fl (Rictorfl/+ or +/+), and Cd4CreMst1fl/flRictorfl/fl mice (n = 4 or 5 mice/group). Numbers indicate percentage of cells in gates or quadrants or MFI. Data are mean ± SEM. *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. Two-way ANOVA with Bonferroni’s test (A), two-tailed unpaired Student’s t test (D and E), one-way ANOVA with Tukey’s test (F, G, and J). Data are representative of two (A), three (B), or four (H and I) or combined from two (E and F) or three (D, G, and J) independent experiments.
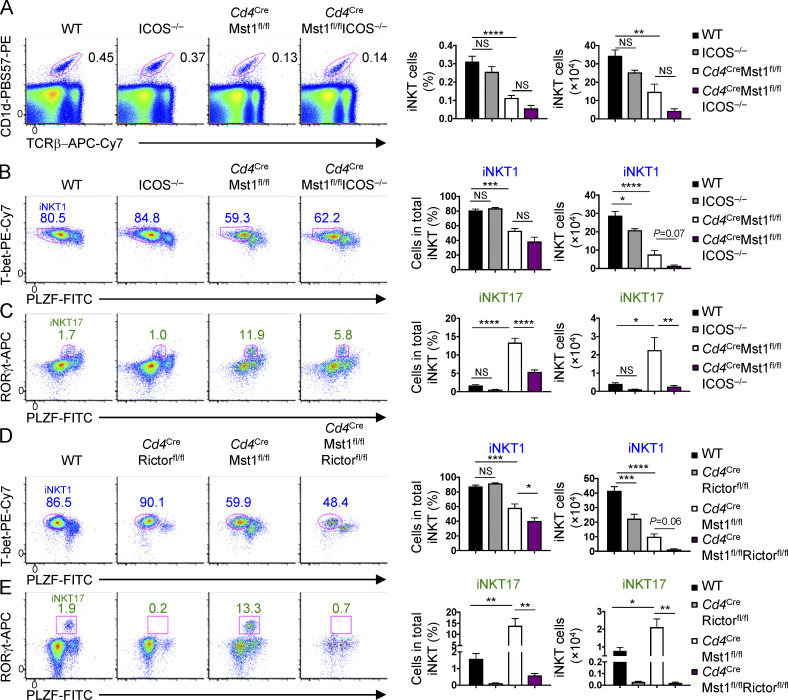
In addition to TCR, costimulatory molecules provide critical secondary signals to promote the development of iNKT cells, as well as iNKT effector cell differentiation (Constantinides and Bendelac, 2013). Among costimulatory molecules expressed by iNKT cells, we found an increase in ICOS expression but not 4-1BB, OX40, or glucocorticoid-induced tumor necrosis factor receptor (GITR; which had a modest decrease of expression) on Mst1-deficient iNKT cells (Fig. S4, E and F). To test whether the elevated ICOS expression is functionally relevant, we crossed mice with germline deletion of Icos (denoted ICOS−/−) with Cd4CreMst1fl/fl mice (Cd4CreMst1fl/flICOS−/− mice) and assessed iNKT cell development and effector lineage differentiation. The frequencies and numbers of total iNKT cells (Fig. 8 A) and developmental stages (S1–S3; Fig. S4 G) were similar between WT and ICOS-deficient mice. While Cd4CreMst1fl/flICOS−/− mice showed no major alterations in the frequency of total iNKT cells compared with Mst1-deficient mice (Fig. 8 A) or other costimulatory molecules (Fig. S4 F), deletion of ICOS had modest effects on iNKT cell maturation, as Cd4CreMst1fl/flICOS−/− mice had increased S1 iNKT cells and decreased S2 iNKT cells compared with Mst1-deficient mice (Fig. S4 G). In terms of iNKT effector cell subsets, the deletion of ICOS alone modestly reduced iNKT1 cell number and had no significant effect on iNKT17 cells (Fig. 8, B and C). However, in the absence of Mst1, ICOS deletion largely blocked the excessive iNKT17 cell development (Fig. 8 C). Together, these data show that abberant iNKT17 cell differentiation in the absence of Mst1 requires ICOS signaling.
Figure 8.
iNKT17 cell development in Mst1-deficient mice depends upon ICOS–mTORC2 signaling. (A) Flow cytometry analysis (left), percentage (middle), and number (right) of iNKT cells in the thymus of WT, ICOS−/− (Cd4CreMst1fl/+ or +/+), Cd4CreMst1fl/fl, and Cd4CreMst1fl/flICOS−/− mice (n = 5–7 mice/group). (B) Flow cytometry analysis (left), percentage (middle), and number (right) of the iNKT1 (T-bethiPLZFlo) effector cell subset in the thymus of WT, ICOS−/− (Cd4CreMst1fl/+ or +/+), Cd4CreMst1fl/fl, and Cd4CreMst1fl/flICOS−/− mice (n = 5–7 mice/group). (C) Flow cytometry analysis (left), percentage (middle), and number (right) of the iNKT17 (RORγthiPLZFint) effector cell subset in the thymus of WT, ICOS−/−, Cd4CreMst1fl/fl, and Cd4CreMst1fl/flICOS−/− mice (n = 5–7 mice/group). (D) Flow cytometry analysis (left), percentage (middle), and number (right) of the iNKT1 (T-bethiPLZFlo) effector cell subset in the thymus of WT, Cd4CreRictorfl/fl (Mst1fl/+), Cd4CreMst1fl/fl (Rictorfl/+ or +/+), and Cd4CreMst1fl/flRictorfl/fl mice (n = 4 or 5 mice/group). (E) Flow cytometry analysis (left), percentage (middle), and number (right) of the iNKT17 (RORγthiPLZFint) effector cell subset in the thymus of WT, Cd4CreRictorfl/fl (Mst1fl/+), Cd4CreMst1fl/fl (Rictorfl/+ or +/+), and Cd4CreMst1fl/flRictorfl/fl mice (n = 4 or 5 mice/group). Numbers indicate percentage of cells in gates (A–E). Data in plots indicate mean ± SEM (A–E). *, P < 0.05, **, P < 0.01, ***, P < 0.001, ****, P < 0.0001. One-way ANOVA with Tukey’s test (A–E). Data are combined from three (D and E) or four (A–C) independent experiments.
ICOS engagement drives PI3K–Akt signaling in activated conventional T cells (Zeng et al., 2016). Indeed, ICOS-mediated stimulation of S1–S2 or S3 iNKT cells promoted Akt phosphorylation at Ser473 (pAkt Ser473), an mTORC2-dependent phosphorylation site (Fig. S4 H). The induction of pAkt Ser473 by ICOS was more pronounced in S1–S2 iNKT cells compared with S3 cells (Fig. S4 H), in line with the increased ICOS expression on S1 and S2 iNKT cells (Fig. S4 I). Deletion of Rictor, the obligate adaptor of mTORC2 (Lee et al., 2010; Wei et al., 2014) failed to rescue iNKT1 effector cell differentiation and further impaired S3 iNKT cell development in Mst1-deficient mice (Fig. 8 D and Fig. S4 J). However, similar to the deficiency of ICOS, deletion of Rictor in Mst1-deficient mice blocked excessive iNKT17 cell development (Fig. 8 E). Together, these data support that the ICOS–mTORC2 signaling axis selectively contributes to the altered iNKT17 cell development in Mst1-deficient mice.
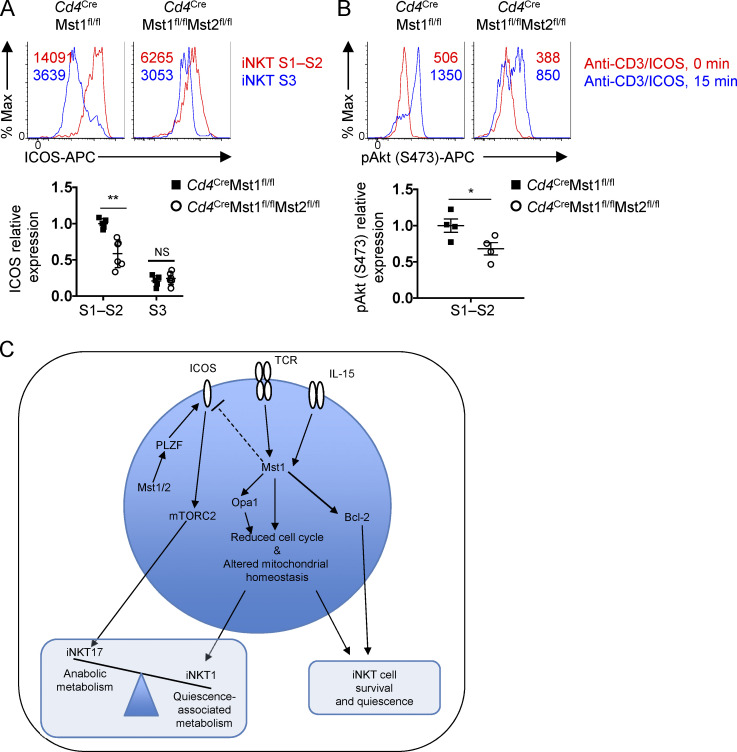
We observed from the IPA of Mst1/2-deficient relative to Mst1-deficient S1–S2 iNKT cells that aside from PLZF (Fig. S3, D–F), signaling by ICOS, a positively regulated gene target for PLZF (Mao et al., 2016), was among those significantly reduced in Mst1/2-deficient cells (Fig. S3 D). Also, ICOS expression was reduced on Mst1/2-deficient S1–S2 iNKT cells compared with Mst1-deficient S1–S2 iNKT cells (Fig. S5 A), which was associated with impaired TCR/ICOS-induced mTORC2 signaling (Fig. S5 B). These data suggest that, in the absence of Mst1, Mst2 likely aids iNKT17 cell development by promoting ICOS–mTORC2 signaling.
Figure S5.
Regulation of ICOS–mTORC2 signaling in iNKT cells by Mst1 and Mst2. (A) Flow cytometry analysis (upper) and relative MFI (lower) of ICOS on thymic iNKT developmental stages S1–S2 and S3 (n = 5 or 6 mice/group). Data are relative to the average MFI of ICOS on Cd4CreMst1fl/fl S1–S2 iNKT cells. (B) Flow cytometry analysis (upper) and relative MFI (lower) of pAkt (Ser473) in thymic S1–S2 iNKT cells (n = 4 mice/group). Data are relative to the average MFI of pAkt (Ser473) in Cd4CreMst1fl/fl S1–S2 iNKT cells after anti-CD3/ICOS cross-linking. (C) Model depicting the role of Mst1 and Mst2 in iNKT cell development in the thymus. Numbers in plots indicate MFI. *, P < 0.05, **, P < 0.01. Two-tailed unpaired Student’s t test (A and B). Data are combined from two (A and B) independent experiments.
Discussion
Despite the emerging interest of immunometabolism as a regulatory node for T cell responses, how signaling and metabolic pathways integrate to control iNKT cell developmental maturation remains largely unclear. In this study, we showed that Mst signaling acts independently of the canonical Hippo signaling pathway and orchestrates transcriptional programs and mitochondrial homeostasis to coordinate cellular quiescence with the development of S3 iNKT cells. Mst1 deficiency alone or combined with Mst2 results in significantly impaired iNKT cell survival and S3 iNKT cell development. These alterations are associated with defective IL-15–STAT5 signaling. We also demonstrate that Mst1 is required for the establishment of metabolic quiescence during the transition into terminally mature S3 iNKT and iNKT1 effector cells, which are characterized by reduced anabolic metabolism and metabolic gene signatures. Further, we define Mst1 as a reciprocal regulator of the iNKT1 and iNKT17 effector cell subsets, which are dependent on Opa1-mediated mitochondrial homeostasis and ICOS–mTORC2 signaling, respectively. These findings establish previously unknown roles of Mst1 as a key molecular determinant of iNKT cell development, by linking metabolic quiescence to mitochondrial homeostasis and immune signaling (Fig. S5 C).
Dynamic regulation of entry and exit from cellular quiescence influences T cell fate. Exit from cellular quiescence is characterized by unique features, including the transition from G0 into G2-S-M phases of cell cycle, increased nutrient uptake, activation of anabolic metabolism, and reprogramming of mitochondrial metabolism (Chapman and Chi, 2018), with the supposition that cells must down-regulate these features to revert back to a quiescent state. Several studies have revealed certain features of cellular and metabolic quiescence in S3 iNKT cells (Benlagha et al., 2005; Engel et al., 2016; Finlay et al., 2010; Salio et al., 2014; Thapa et al., 2017b), which also exhibit long-lived residency within the thymus (Berzins et al., 2006). However, the molecular drivers for cell reentry into a quiescent state during iNKT cell development have remained poorly understood. Here, we identified Mst1 as a positive regulator of quiescence establishment in terminally mature S3 iNKT cells through the suppression of proliferation and mitochondrial programs. As Lats1/2 are dispensable for iNKT cell development, our data suggest that Mst1 regulates iNKT cellular quiescence independent of the canonical Hippo/Mst signaling pathway that can regulate cell proliferation by Lats1/2–YAP/TAZ (Yu et al., 2015). Instead, we provide evidence that this noncanoncial Mst signaling pathway partly involves regulation of mitochondrial homeostasis by Opa1, consistent with the roles of metabolites, such as ATP, and mitochondrial activity in the regulation of cell cycle machinery and progression (Mandal et al., 2005). Beyond Opa1, how Mst1 modulates mitochondrial homeostasis in iNKT cells requires additional investigation, but may be attributed to altered mitochondrial turnover (Pei et al., 2015; Salio et al., 2014; Zhu et al., 2018) or control of mitochondrial dynamics as recently reported in dendritic cells (Du et al., 2018).
Unlike conventional αβ T cells, innate-like lymphocytes, such as mucosal-associated invariant T cells and iNKT cells, differentiate into effector cells during development in the thymus. This unique feature provides early functional heterogeneity and tissue distribution to promote antitumor and pathogen immunity (Crosby and Kronenberg, 2018; Lee et al., 2015), but could have deleterious effects in certain diseases such as allergy (Crosby and Kronenberg, 2018). Here, we revealed Mst1 as a critical reciprocal regulator of iNKT1 and iNKT17 cell development, and also identified Mst2 as a positive regulator for iNKT17 cell development when Mst1 is absent, establishing for the first time, to our knowledge, opposing roles for Mst1 and Mst2. The differential regulation of iNKT cell development by Mst1 and Mst2 when one or the other is absent could be associated with the differential kinase activities between Mst kinase heterodimers and homodimers (Rawat et al., 2016). Reduced iNKT1 cell development in Mst1-deficient mice may be due to impaired IL-15 signaling, as IL-15 promotes functional maturation of iNKT1 cells at S3 and induction of T-bet gene expression (Gordy et al., 2011; Matsuda et al., 2002; Townsend et al., 2004). T-bet can additionally repress Runx1, a coactivator of the RORγt gene that promotes iNKT17 cell development (Lazarevic et al., 2011; Thapa et al., 2017a). Further, our data show that the accumulation of iNKT17 thymocytes is dependent upon the ICOS–mTORC2 signaling axis. While ICOS–mTORC2 signaling can suppress Foxo1 (Zeng et al., 2016), a key transcription factor implicated in conventional T cell quiescence, metabolic homeostasis, and T helper 17 (Th17) cell differentiation (Hamilton and Jameson, 2012; Hedrick et al., 2012; Luo and Li, 2018; Newton et al., 2018; Niu et al., 2018; Wu et al., 2013), Mst1-deficient iNKT cells have no clear alteration in Foxo1 protein expression or activity (data not shown). Thus, the link between failure to enter quiescence and increased iNKT17 cell development in the absence of Mst1 remains to be further explored.
Aside from immunological signals, how metabolic programs govern conventional T cell fate decisions remains an outstanding question. Our observations that the maturation of S3 iNKT cells is linked with the reentry into a metabolically quiescent state and is associated with mitochondrial fusion are reminiscent of the differentiation of effector into memory CD8+ T cells. Specifically, both iNKT and CD8+ T cells transition from proliferative expansion, which is associated with aerobic glycolysis and mitochondrial remodeling, into quiescent cells with poised effector functions (Buck et al., 2016; Geltink et al., 2018). This transition into a quiescent state correlates with reduced mTORC1 signaling, which favors elevated mitochondrial mass and the generation of quiescent memory CD8+ T cells (Araki et al., 2009; Buck et al., 2016; Pollizzi et al., 2015; van der Windt et al., 2012). These features are in contrast to TCR-induced quiescence exit that requires mTORC1 signaling for mitochondrial biogenesis and oxidative function (Tan et al., 2017; Yang et al., 2013), suggesting context-dependent effects orchestrated by mTORC1 downstream of cytokines such as IL-15 or TCR. We also show that iNKT effector subsets have distinct metabolic transcriptional profiles and differential requirements for Opa1-driven mitochondrial fusion, indicating that metabolic pathways may direct iNKT cell fate. Indeed, emerging evidence reveals that specific metabolites and metabolic enzymes can modulate immune cell fate decisions. For instance, glutaminase-dependent metabolism reciprocally regulates Th1 and Th17 cell differentiation (Johnson et al., 2018). Our study suggests that development of iNKT1, but not iNKT17 cells, is dependent upon mitochondrial fusion, which may be further linked to the roles of intracellular ROS in iNKT cell subset differentiation (Pyaram et al., 2019). Thus, the reciprocal regulation of iNKT1 and iNKT17 cell development is likely a complex interplay of signaling and metabolic pathways, with biochemical details requiring further investigation. Understanding how specific metabolites and metabolic programs control iNKT cell subset development may allow selective modulation of iNKT cell immunity to infections and cancer.
In summary, our data establish Mst signaling as a molecular rheostat to tune immunological signals and cellular and metabolic programs, which coordinately regulate the differentiation and effector programming of iNKT cells. Given that terminally mature S3 iNKT cells, which can establish residency within the thymus (Berzins et al., 2006), and memory CD8+ T cells both appear to favor OXPHOS over glycolysis, these findings highlight a conserved metabolic program that allows T cells to maintain their survival and long-term persistence, but also allows for rapid functional remodeling upon reactivation (Kumar et al., 2019). Further, the impaired ability of Mst1-deficient iNKT thymocytes to enter into a quiescent state is consistent with the accumulating evidence for dysregulated Hippo pathways with human cancers (Moroishi et al., 2015; Yu et al., 2015), which may be attributed to defective quiescence programs. The identification of Mst signaling as a molecular rheostat to enforce cellular quiescence opens possible new avenues to target this signaling pathway or the quiescence programs for therapeutic intervention of immune-mediated disorders.
Materials and methods
Mice
C57BL/6, CD45.1+, Lats1fl, Lats2fl, Mst1fl (Stk4fl), Mst2fl (Stk3fl), ICOS−/−, and Rag1−/− mice were purchased from the Jackson Laboratory. Rictorfl mice were previously described (Yang et al., 2011). Opa1fl mice were previously described (Zhang et al., 2011). All conditional floxed mice were crossed with Cd4Cre mice. Age- and gender-matched WT controls were littermates that were Cre– mice that contained floxed alleles, or Cd4Cre nonfloxed, and both genders were used. Mice were backcrossed to the C57BL/6 background and used at 6 to 12 wk of age. For mixed bone marrow chimera generation, T cell–depleted bone marrow cells from WT, Cd4creMst1fl/fl, Cd4CreMst2fl/fl, or Cd4CreMst1fl/flMst2fl/fl mice (all CD45.2+) and CD45.1+ WT spike controls were mixed at a 1:1 ratio and transferred into sub-lethally irradiated (5 Gy) Rag1−/− mice, followed by reconstitution for 6–8 wk. All mice were kept in specific pathogen–free conditions in the Animal Resource Center at St. Jude Children’s Research Hospital. All animal protocols were approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
Flow cytometry
Lymphocytes were isolated from thymus, spleen, or liver tissues. For analysis of surface markers, cells were stained in PBS containing 2% FBS with appropriate surface antibodies: mCD1d-PBS57 tetramer (provided by the National Institutes of Health Tetramer Facility), anti-TCRβ (Tonbo Biosciences, H57-597), anti-CD24 (M1/69), anti-NK1.1 (PK136), anti-CD44 (1M7), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD69 (H1.2F3), anti-CD122 (TM-b1), anti-ICOS (C398.4A), anti-B220 (RA3-6B2), anti-CD5 (53–7.3), anti-CD6 (OX-129), anti-Egr2 (erongr2), anti-Ly6C (HK1.4), anti-4-1BB (17B5), anti-GITR (DTA-1), anti-OX40 (OX-86), and 7-AAD (Sigma-Aldrich, A9400). For analysis of intracellular molecules, unless otherwise indicated, the Foxp3 fixation/permeabilization buffers were used per the manufacturer’s instructions (Thermo Fisher Scientific, 00–5523-00) with the following antibodies: Bcl-2 (BCL/10C4), Ki67 (SolA15), RORγt (B2D), T-bet (4B10), PLZF (Mags.21F7), TNF-α (MP6-XT22), IL-17A (TC11-18H10.1), IFN-γ (XMG1.2), and c-Myc (D84C12). BrdU, active caspase 3, and annexin V staining was performed per the manufacturer’s instructions (BD Biosciences, 552598, 550480, and 550474). For detection of phosphorylated proteins, cells were stimulated and immediately fixed with Phosflow Lyse/Fix buffer (BD Biosciences, 558049), followed by permeabilization with Phosflow Perm buffer III (BD Biosciences, 558050), and staining with antibodies to phospho-Akt (Ser473; BD Biosciences, M89-61), phospho-S6 (Ser235/236; Cell Signaling Technology, D57.2.2E), phospho-Mob1 (Thr35; Cell Signaling Technology, D2F10), or phospho-STAT5 (Tyr694; BD Biosciences, 612599). For staining mitochondria, intracellular ROS, and 2-NBDG uptake, unfixed lymphocytes were incubated for 30 min at 37°C with 10 nM MitoTracker Green or MitoTracker Deep Red (Life Technologies, M7514 and MM22426), 10 nM TMRM (ImmunoChemistry Technologies, 9105), or 2.5 µM CellROX Deep Red (Life Technologies, C10422), or for 1 h at 37°C with 30 µM 2-NBDG (Thermo Fisher Scientific, N13195) before staining surface markers. For assessment of mitochondria and cellular ROS in fixed cells, MitoTracker Deep Red and CellROX Deep Red were used per the manufacturer’s instructions (according to Life Technologies, these dyes can be used on fixed and permeabilized cells, albeit with reduced sensitivity). Flow cytometry data were acquired on LSRII or LSR Fortessa instruments (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Enrichment of iNKT cells and thymic developmental stage analysis
For analysis of S0 iNKT cells, mCD1d-PBS57+ thymocytes were enriched using anti-PE microbeads (Miltenyi), as previously described (Benlagha et al., 2005). Alternatively, to avoid TCR cross-linking by tetramer staining, total thymocytes were enriched for iNKT cells by depleting CD8+ cells using the CD8+ microbeads (Miltenyi), as previously described (Benlagha et al., 2005).
iNKT stages were defined as follows: S0 = TCRβintCD1d-PBS57tet+CD69+CD24+CD44–NK1.1–; S1 = TCRβintCD1d-PBS57tet+CD24–CD44–NK1.1–; S2 = TCRβintCD1d-PBS57tet+CD24–CD44+NK1.1–; and S3 = TCRβintCD1d-PBS57tet+CD24–CD44+NK1.1+.
Conventional T cell developmental stages were defined as follows: DN = CD4–CD8–; DP = CD4+CD8+; CD4SP = CD4+CD8–TCRβ+; CD8SP = CD4–CD8+TCRβ+; mature CD4SP or CD8SP = CD62LhiCD69loCD4/8+/−TCRβ+; and semi-mature CD4SP or CD8SP = CD62LloCD69hiCD4/8+/−TCRβ+.
iNKT cell in vitro cultures
For in vitro cultures to assess cell survival, thymocytes depleted of CD8+ cells were cultured in complete Click’s medium for 1–3 d at a concentration of 2 × 106 cells/well in a 48-well plate, with or without the addition of rmIL-15 (20 ng/ml; Peprotech, 210–15).
In vitro stimulation for cytokine and phospho-flow
For short-term stimulation of iNKT cells to assess cytokine production, thymocytes depleted of CD8+ cells were stimulated with PMA (1 µM; Sigma-Aldrich, P8139) and ionomycin (1 µM; Sigma-Aldrich, 10634) along with monensin (BD Biosciences, BDB554724) for 4–5 h at 37°C. For assessment of phosphorylation of S6 (Ser235/236) and Mob1 (Thr35) by PMA and ionomycin stimulation, phosphorylation of Mob1 (Thr35) by rmIL-15 (10 ng/ml), and phosphorylation of STAT5 (Tyr694) by cytokine stimulation (rmIL-2 [Peprotech, 212–12; 10 ng/ml], rmIL-15 [10 ng/ml], or rmIL-7 [Peprotech, 217–17; 10 ng/ml]), thymocyte subsets were sorted, rested in complete medium for 1–2 h at 37°C, and then stimulated at 37°C for indicated time points. For anti-ICOS and anti-CD3 stimulation to assess pAkt (Ser473), or anti-CD28 and anti-CD3 stimulation to assess pMob1 (Thr35) or Mst1, sorted thymic iNKT cells were incubated with anti-ICOS antibody (5 µg/ml; Biolegend, C398.4A) or biotinylation anti-CD28 (5 µg/ml; BD Biosciences, 37.51) alone or in combination with biotinylated anti-CD3 antibody for 20 min on ice (5 µg/ml; BD Biosciences, 500A2) followed by cross-linking with goat anti-hamster antibody (10 µg/ml; Jackson ImmunoResearch, 127–005-160) and/or streptavidin (10 µg/ml; Invitrogen, 434301) at 37°C for indicated time points.
Cell purification
From the thymus, DP (CD4+CD8+), CD4SP (CD4+CD8–), total iNKT (TCRβintCD1d-PBS57-tet+) or iNKT cell developmental subsets (S1, S2, or S3) were sorted using the Sony SY3200 sorter (Sony). Purity for all cells used was >95%.
Fluorescence microscopy
To analyze mitochondria, sorted DP cells and iNKT cell subsets were fixed in PBS containing 2% paraformaldehyde for 10 min. Cells were permeabilized with 0.1% Triton X-100 in PBS for 3 min before incubation with blocking buffer comprised of PBS containing 2% bovine serum albumin and 5% donkey serum. Cells were stained overnight in blocking buffer containing rabbit anti-Tom20 (1 µg/ml; Abcam, 186735). Cells were subsequently stained with AF647-conjugated donkey anti-rabbit (1 µg/ml; Jackson Immunoresearch, 711–605-152) and AF488-conjugated phalloidin (1 U/ml; Thermo Fisher Scientific, A12379). High-resolution images were acquired using a Marianis spinning disk confocal microscope (Intelligent Imaging Innovations) equipped with a 100× 1.4 numerical aperture objective, 488 nm and 647 nm laser lines, and Delta Evolve EMCCD camera (Photometrics), and analyzed using Slidebook software (Intelligent Imaging Innovations). To analyze mitochondrial diameter, stochastic optical reconstruction microscopy (STORM) was performed as previously described (Du et al., 2018). Briefly, sorted DP and iNKT cell subsets were seeded onto poly-L-lysine–coated chamber coverslips (Ibidi USA) and allowed to settle for 30 min before fixation with 2% paraformaldehyde followed by treatment with sodium borohydride to quench free reactive groups. Cells were permeabilized and stained with rabbit anti-Tom20 as outlined above. Super resolution microscopy was performed using an N-STORM system (Nikon Instruments) comprised of a 100× 1.45 numerical aperture objective, DU897 EMCCD camera (Andor), high-powered 647 nm laser launch (Agilent), and NIS Elements STORM software.
Seahorse analysis of glycolytic and mitochondrial oxidative activities
FACS-sorted iNKT cells at various development stages from the thymi of 15–20 C57BL/6 (Fig. 5, A–E) or Cd4CreMst1fl/fl mice and WT littermate controls (Fig. 5 I) were combined, rested for 1 h, and then plated in a Seahorse 96-well plate with 2.5 × 105 cells/well in 3 or 4 replicate wells. OCR and ECAR were measured in DMEM Seahorse XF media supplemented with 10 mM D-glucose under basal conditions and in response to 1 µM oligomycin, 1.5 µM fluoro-carbonyl cyanide phenylhydrazone (FCCP), and 500 nM rotenone using an XF96 Extracellular Flux Analyzer (Seahorse Bioscience).
Immunoblot analysis
Immunoblots were performed as described (Shi et al., 2019), using the following antibodies: Opa1 (Novus, NB110), VDAC (CST, D73D12), Mst1 (CST, 3682), and β-actin (CST, 8H10D10). Relative Opa1 expression was calculated by the ratio between Opa1 versus VDAC intensity in the immunoblot. Immunoblots were imaged by LI-COR Odyssey FC, and band densities were quantified by Image Studio Software (LI-COR).
RNA purification and real-time PCR
RNA was isolated using the RNeasy Micro Kit according to the manufacturer’s instructions (Qiagen). cDNA was generated using the High Capacity cDNA Reverse Transcription Kit according to the manufacturer’s instructions (Applied Biosystems, 4368814). For real-time PCR analysis, TaqMan primers and probes (Applied Biosystems) were used with the Fast Start Universal Probe Master (Rox; Sigma-Aldrich, 4913949001) per the manufacturer’s instructions. The following primers/probes from Applied Biosystems were used: Stk4 (Mm01251755_m1), Stk3 (Mm00490478_m1), and Actb (Mm00607939_s1).
Gene expression profiling and bioinformatic analysis
iNKT cell developmental stages were sorted from the thymus of WT (n = 3) and Cd4CreMst1fl/fl (n = 3) mice or WT (n = 6 or 7), Cd4CreMst1fl/fl (n = 4 or 5), and Cd4CreMst1fl/flMst1fl/fl (n = 1 or 2 samples; cells combined from six mice/sample) mice, as described above. RNA samples were analyzed using the Mouse Gene 2.0 ST Signals array or Clariom S Mouse array. Differentially expressed transcripts were identified using two-way ANOVA followed by multiple testing correction using the Benjamini–Hochberg method to estimate FDR. Pairwise fold-change was calculated using R package limma as previously described (Zeng et al., 2013). GSEA was performed using public gene sets (HALLMARK, KEGG, and GO) and curated gene sets from publications as previously described (Subramanian et al., 2005; Zeng et al., 2013). We used the iNKT effector subset gene sets (KRONENBERG_NKT1, KRONENBERG_NKT2, KRONENBERG_NKT17) that were previously defined by the RNA-seq dataset (GSE74597; Engel et al., 2016). We used the Immunological Genome Project public dataset to analyze relative expression of Stk4 and Stk3 among thymocyte subsets by RMA normalization (GSE15907). We used public gene sets curated by the Broad Institute (C7, GSE17974), which were generated by comparing gene expression among naive, activated, and proliferating human CD4+ T cells (Elo et al., 2010). “UP_Activated CD4 T cells_vs_naive” and “UP_Naive CD4 T cells_vs_activated” gene sets contain the genes that were up-regulated and down-regulated in CD4+ T cells at 24 h after anti-CD3/CD28 activation compared with naive CD4+ T cells, respectively; “UP_Proliferating CD4 T cells_vs_naive” and “UP_Naive CD4 T cells_vs_proliferating” gene sets contain the genes that were up-regulated and down-regulated in CD4+ T cells at 72 h post-CD3/CD28 activation compared with naive CD4+ T cells, respectively. For the proliferating and quiescent HSC signatures, we used gene sets that were previously defined (Venezia et al., 2004). Specifically, “UP_Proliferating HSC” contains the up-regulated genes that were shared among proliferating HSC and fetal-liver HSC; “UP_Quiescent HSC” contains the shared genes that were down-regulated in proliferating HSC and up-regulated in quiescent HSC. For the PLZF gene signatures, we used the previously published microarray dataset (GSE81772; Mao et al., 2016), and defined the differentially regulated genes in Luxoid mutant iNKT cells compared with WT iNKT cells with FDR <0.05 and log2 fold-change >0.5. IPA (Qiagen) was used to analyze activity of upstream regulators of gene expression. For WGCNA, we first identified genes that were significantly changed between any stage or genotype using two-way ANOVA, of which 3,230 genes were DE in at least one pairwise comparison out of the three (Cd4CreMst1fl/fl S1 vs. WT S1, Cd4CreMst1fl/fl S2 vs. WT S2, Cd4CreMst1fl/fl S3 vs. WT S3), with FDR <0.05 and fold-change >1.5, as previously described (Tan et al., 2017). The WGCNA R package (Langfelder et al., 2008) was then used to identify coexpression clusters from the 3,230 DE genes, as previously described (Tan et al., 2017). Functional enrichment of each coexpression cluster was determined using right-tailed Fisher’s exact test and the Benjamini–Hochberg method (FDR <0.05).
Statistical analysis for biological experiments
P values were calculated with two-tailed unpaired Student’s t test, one-way ANOVA, and two-way ANOVA with post-test analysis as indicated in the figure legends (GraphPad Prism). Statistical significance was as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. All data are displayed as mean ± SEM.
Data availability
The microarray data that support the findings of this study have been deposited in GEO under accession no. GSE130043.
Online supplemental material
Fig. S1 provides characterization of pMob1 (downstream target of Mst) signaling in thymocytes, and the contribution of Mst1 and Mst2 on S0 iNKT cell and conventional T cell development in the thymus. Fig. S2 provides additional bioinformatic analysis examining the expression of gene programs associated with cellular and metabolic quiescence in iNKT cells. Fig. S3 characterizes by gene expression and flow cytometry the metabolic programs, mitochondria, and cell cycle of iNKT effector subsets, and the contribution of Mst1 and Mst2 in PLZF signaling in iNKT cell developmental stages. Fig. S4 shows TCR induction of pMob in iNKT cell developmental stages, the regulation of TCR signal strength and costimulatory molecule expression by Mst1, and the impact of ICOS or Rictor deficiency on iNKT cell development stages. Fig. S5 shows the regulation of ICOS–mTORC2 signaling by Mst1 and Mst2 and provides a model depicting the role of Mst1 and Mst2 in iNKT cell development.
Acknowledgments
The authors acknowledge X. Du for critical discussion of the data, Y. Wang for critical reading and editing of the manuscript, M. Hendren, A. KC, and S. Rankin for animal colony management, the St. Jude Immunology FACS core facility for cell sorting, and the Tetramer Facility of the National Institutes of Health for mCD1d-PBS57.
This work was supported by National Institutes of Health grants AI105887, AI131703, AI140761, AI150241, AI150514, CA221290, and CA250533 (to H. Chi).
Author contributions: J.L. Raynor designed, performed, and analyzed experiments and wrote the manuscript; C. Liu designed, performed, and analyzed experiments; C. Guy performed imaging assays; N.M. Chapman provided insights for bioinformatic analysis and co-wrote the manuscript; H. Shi contributed to biochemical experiments; Y. Dhungana and G. Neale performed bioinformatic analyses; H. Sesaki provided critical reagents and insight; and H. Chi designed experiments, co-wrote the manuscript, and provided overall direction.
References
- Araki, K., Turner A.P., Shaffer V.O., Gangappa S., Keller S.A., Bachmann M.F., Larsen C.P., and Ahmed R.. 2009. mTOR regulates memory CD8 T-cell differentiation. Nature. 460:108–112. 10.1038/nature08155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardestani, A., Lupse B., and Maedler K.. 2018. Hippo Signaling: Key Emerging Pathway in Cellular and Whole-Body Metabolism. Trends Endocrinol. Metab. 29:492–509. 10.1016/j.tem.2018.04.006 [DOI] [PubMed] [Google Scholar]
- Benlagha, K., Kyin T., Beavis A., Teyton L., and Bendelac A.. 2002. A thymic precursor to the NK T cell lineage. Science. 296:553–555. 10.1126/science.1069017 [DOI] [PubMed] [Google Scholar]
- Benlagha, K., Wei D.G., Veiga J., Teyton L., and Bendelac A.. 2005. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 202:485–492. 10.1084/jem.20050456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzins, S.P., McNab F.W., Jones C.M., Smyth M.J., and Godfrey D.I.. 2006. Long-term retention of mature NK1.1+ NKT cells in the thymus. J. Immunol. 176:4059–4065. 10.4049/jimmunol.176.7.4059 [DOI] [PubMed] [Google Scholar]
- Buck, M.D., O’Sullivan D., Klein Geltink R.I., Curtis J.D., Chang C.H., Sanin D.E., Qiu J., Kretz O., Braas D., van der Windt G.J., et al. 2016. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell. 166:63–76. 10.1016/j.cell.2016.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, N.M., and Chi H.. 2018. Hallmarks of T-cell Exit from Quiescence. Cancer Immunol. Res. 6:502–508. 10.1158/2326-6066.CIR-17-0605 [DOI] [PubMed] [Google Scholar]
- Chi, H. 2012. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 12:325–338. 10.1038/nri3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, N.R., Brennan P.J., Shay T., Watts G.F., Brigl M., Kang J., and Brenner M.B.. ImmGen Project Consortium . 2013. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat. Immunol. 14:90–99. 10.1038/ni.2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides, M.G., and Bendelac A.. 2013. Transcriptional regulation of the NKT cell lineage. Curr. Opin. Immunol. 25:161–167. 10.1016/j.coi.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby, C.M., and Kronenberg M.. 2018. Tissue-specific functions of invariant natural killer T cells. Nat. Rev. Immunol. 18:559–574. 10.1038/s41577-018-0034-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashtsoodol, N., Shigeura T., Aihara M., Ozawa R., Kojo S., Harada M., Endo T.A., Watanabe T., Ohara O., and Taniguchi M.. 2017. Alternative pathway for the development of Vα14+ NKT cells directly from CD4-CD8- thymocytes that bypasses the CD4+CD8+ stage. Nat. Immunol. 18:274–282. 10.1038/ni.3668 [DOI] [PubMed] [Google Scholar]
- Dong, Y., Du X., Ye J., Han M., Xu T., Zhuang Y., and Tao W.. 2009. A cell-intrinsic role for Mst1 in regulating thymocyte egress. J. Immunol. 183:3865–3872. 10.4049/jimmunol.0900678 [DOI] [PubMed] [Google Scholar]
- Dose, M., Sleckman B.P., Han J., Bredemeyer A.L., Bendelac A., and Gounari F.. 2009. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. USA. 106:8641–8646. 10.1073/pnas.0812255106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, X., Wen J., Wang Y., Karmaus P.W.F., Khatamian A., Tan H., Li Y., Guy C., Nguyen T.M., Dhungana Y., et al. 2018. Hippo/Mst signalling couples metabolic state and immune function of CD8α+ dendritic cells. Nature. 558:141–145. 10.1038/s41586-018-0177-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elo, L.L., Järvenpää H., Tuomela S., Raghav S., Ahlfors H., Laurila K., Gupta B., Lund R.J., Tahvanainen J., Hawkins R.D., et al. 2010. Genome-wide profiling of interleukin-4 and STAT6 transcription factor regulation of human Th2 cell programming. Immunity. 32:852–862. 10.1016/j.immuni.2010.06.011 [DOI] [PubMed] [Google Scholar]
- Engel, I., Seumois G., Chavez L., Samaniego-Castruita D., White B., Chawla A., Mock D., Vijayanand P., and Kronenberg M.. 2016. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol. 17:728–739. 10.1038/ni.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay, D.K., Kelly A.P., Clarke R., Sinclair L.V., Deak M., Alessi D.R., and Cantrell D.A.. 2010. Temporal differences in the dependency on phosphoinositide-dependent kinase 1 distinguish the development of invariant Valpha14 NKT cells and conventional T cells. J. Immunol. 185:5973–5982. 10.4049/jimmunol.1000827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geltink, R.I.K., Kyle R.L., and Pearce E.L.. 2018. Unraveling the Complex Interplay Between T Cell Metabolism and Function. Annu. Rev. Immunol. 36:461–488. 10.1146/annurev-immunol-042617-053019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordy, L.E., Bezbradica J.S., Flyak A.I., Spencer C.T., Dunkle A., Sun J., Stanic A.K., Boothby M.R., He Y.W., Zhao Z., et al. 2011. IL-15 regulates homeostasis and terminal maturation of NKT cells. J. Immunol. 187:6335–6345. 10.4049/jimmunol.1003965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, S.E., and Jameson S.C.. 2012. CD8 T cell quiescence revisited. Trends Immunol. 33:224–230. 10.1016/j.it.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, S.M., Hess Michelini R., Doedens A.L., Goldrath A.W., and Stone E.L.. 2012. FOXO transcription factors throughout T cell biology. Nat. Rev. Immunol. 12:649–661. 10.1038/nri3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia, J., Williams A., Goff L.A., Staron M., Licona-Limón P., Kaech S.M., Nakayama M., Rinn J.L., and Flavell R.A.. 2013. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity. 38:984–997. 10.1016/j.immuni.2013.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L., Li X., Zhou D., Geng J., and Chen L.. 2018. Role of Hippo signaling in regulating immunity. Cell. Mol. Immunol. 15:1003–1009. 10.1038/s41423-018-0007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M.O., Wolf M.M., Madden M.Z., Andrejeva G., Sugiura A., Contreras D.C., Maseda D., Liberti M.V., Paz K., Kishton R.J., et al. 2018. Distinct Regulation of Th17 and Th1 Cell Differentiation by Glutaminase-Dependent Metabolism. Cell. 175:1780–1795.e19. 10.1016/j.cell.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri, K., Katakai T., Ebisuno Y., Ueda Y., Okada T., and Kinashi T.. 2009. Mst1 controls lymphocyte trafficking and interstitial motility within lymph nodes. EMBO J. 28:1319–1331. 10.1038/emboj.2009.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, J.H., and Guan K.L.. 2018. Interplay between YAP/TAZ and Metabolism. Cell Metab. 28:196–206. 10.1016/j.cmet.2018.07.010 [DOI] [PubMed] [Google Scholar]
- Kovalovsky, D., Uche O.U., Eladad S., Hobbs R.M., Yi W., Alonzo E., Chua K., Eidson M., Kim H.J., Im J.S., et al. 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9:1055–1064. 10.1038/ni.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., Pyaram K., Yarosz E.L., Hong H., Lyssiotis C.A., Giri S., and Chang C.H.. 2019. Enhanced oxidative phosphorylation in NKT cells is essential for their survival and function. Proc. Natl. Acad. Sci. USA. 116:7439–7448. 10.1073/pnas.1901376116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnareva, Y., Seong Y., Andreyev A.Y., Kuwana T., Kiosses W.B., Votruba M., and Newmeyer D.D.. 2016. Mitochondrial dysfunction in an Opa1(Q285STOP) mouse model of dominant optic atrophy results from Opa1 haploinsufficiency. Cell Death Dis. 7:e2309. 10.1038/cddis.2016.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P., Zhang B., and Horvath S.. 2008. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 24:719–720. 10.1093/bioinformatics/btm563 [DOI] [PubMed] [Google Scholar]
- Lazarevic, V., Chen X., Shim J.H., Hwang E.S., Jang E., Bolm A.N., Oukka M., Kuchroo V.K., and Glimcher L.H.. 2011. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol. 12:96–104. 10.1038/ni.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Gudapati P., Dragovic S., Spencer C., Joyce S., Killeen N., Magnuson M.A., and Boothby M.. 2010. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 32:743–753. 10.1016/j.immuni.2010.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.J., Holzapfel K.L., Zhu J., Jameson S.C., and Hogquist K.A.. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14:1146–1154. 10.1038/ni.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.J., Wang H., Starrett G.J., Phuong V., Jameson S.C., and Hogquist K.A.. 2015. Tissue-Specific Distribution of iNKT Cells Impacts Their Cytokine Response. Immunity. 43:566–578. 10.1016/j.immuni.2015.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, C.T., and Li M.O.. 2018. Foxo transcription factors in T cell biology and tumor immunity. Semin. Cancer Biol. 50:13–20. 10.1016/j.semcancer.2018.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, S., Guptan P., Owusu-Ansah E., and Banerjee U.. 2005. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell. 9:843–854. 10.1016/j.devcel.2005.11.006 [DOI] [PubMed] [Google Scholar]
- Mao, A.P., Constantinides M.G., Mathew R., Zuo Z., Chen X., Weirauch M.T., and Bendelac A.. 2016. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc. Natl. Acad. Sci. USA. 113:7602–7607. 10.1073/pnas.1601504113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, B., Auffray C., Delpoux A., Pommier A., Durand A., Charvet C., Yakonowsky P., de Boysson H., Bonilla N., Audemard A., et al. 2013. Highly self-reactive naive CD4 T cells are prone to differentiate into regulatory T cells. Nat. Commun. 4:2209. 10.1038/ncomms3209 [DOI] [PubMed] [Google Scholar]
- Matsuda, J.L., Gapin L., Sidobre S., Kieper W.C., Tan J.T., Ceredig R., Surh C.D., and Kronenberg M.. 2002. Homeostasis of V alpha 14i NKT cells. Nat. Immunol. 3:966–974. 10.1038/ni837 [DOI] [PubMed] [Google Scholar]
- Moroishi, T., Hansen C.G., and Guan K.L.. 2015. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer. 15:73–79. 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, F., Praskova M., Xia F., Van Buren D., Hock H., Avruch J., and Zhou D.. 2012. The Mst1 and Mst2 kinases control activation of rho family GTPases and thymic egress of mature thymocytes. J. Exp. Med. 209:741–759. 10.1084/jem.20111692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, R.H., Shrestha S., Sullivan J.M., Yates K.B., Compeer E.B., Ron-Harel N., Blazar B.R., Bensinger S.J., Haining W.N., Dustin M.L., et al. 2018. Maintenance of CD4 T cell fitness through regulation of Foxo1. Nat. Immunol. 19:838–848. 10.1038/s41590-018-0157-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, L., Xuan X., Wang J., Li L., Yang D., Jing Y., Westerberg L.S., and Liu C.. 2018. Akt2 Regulates the Differentiation and Function of NKT17 Cells via FoxO-1-ICOS Axis. Front. Immunol. 9:1940. 10.3389/fimmu.2018.01940 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Park, H., Tsang M., Iritani B.M., and Bevan M.J.. 2014. Metabolic regulator Fnip1 is crucial for iNKT lymphocyte development. Proc. Natl. Acad. Sci. USA. 111:7066–7071. 10.1073/pnas.1406473111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.Y., DiPalma D.T., Kwon J., Fink J., and Park J.H.. 2019. Quantitative Difference in PLZF Protein Expression Determines iNKT Lineage Fate and Controls Innate CD8 T Cell Generation. Cell Rep. 27:2548–2557.e4. 10.1016/j.celrep.2019.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, B., Zhao M., Miller B.C., Véla J.L., Bruinsma M.W., Virgin H.W., and Kronenberg M.. 2015. Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation. J. Immunol. 194:5872–5884. 10.4049/jimmunol.1402154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi, K.N., Patel C.H., Sun I.H., Oh M.H., Waickman A.T., Wen J., Delgoffe G.M., and Powell J.D.. 2015. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. J. Clin. Invest. 125:2090–2108. 10.1172/JCI77746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova, M., Xia F., and Avruch J.. 2008. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18:311–321. 10.1016/j.cub.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyaram, K., Kumar A., Kim Y.H., Noel S., Reddy S.P., Rabb H., and Chang C.H.. 2019. Keap1-Nrf2 System Plays an Important Role in Invariant Natural Killer T Cell Development and Homeostasis. Cell Rep. 27:699–707.e4. 10.1016/j.celrep.2019.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat, S.J., Araiza-Olivera D., Arias-Romero L.E., Villamar-Cruz O., Prudnikova T.Y., Roder H., and Chernoff J.. 2016. H-ras Inhibits the Hippo Pathway by Promoting Mst1/Mst2 Heterodimerization. Curr. Biol. 26:1556–1563. 10.1016/j.cub.2016.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salio, M., Puleston D.J., Mathan T.S., Shepherd D., Stranks A.J., Adamopoulou E., Veerapen N., Besra G.S., Hollander G.A., Simon A.K., and Cerundolo V.. 2014. Essential role for autophagy during invariant NKT cell development. Proc. Natl. Acad. Sci. USA. 111:E5678–E5687. 10.1073/pnas.1413935112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403. 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Liu C., Tan H., Li Y., Nguyen T.M., Dhungana Y., Guy C., Vogel P., Neale G., Rankin S., et al. 2018. Hippo Kinases Mst1 and Mst2 Sense and Amplify IL-2R-STAT5 Signaling in Regulatory T Cells to Establish Stable Regulatory Activity. Immunity. 49:899–914.e6. 10.1016/j.immuni.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Chapman N.M., Wen J., Guy C., Long L., Dhungana Y., Rankin S., Pelletier S., Vogel P., Wang H., et al. 2019. Amino Acids License Kinase mTORC1 Activity and Treg Cell Function via Small G Proteins Rag and Rheb. Immunity. 51:1012–1027.e7. 10.1016/j.immuni.2019.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J., Wang S., Deng W., Wu J., Gao J., and Zhong X.P.. 2014. Mechanistic target of rapamycin complex 1 is critical for invariant natural killer T-cell development and effector function. Proc. Natl. Acad. Sci. USA. 111:E776–E783. 10.1073/pnas.1315435111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surh, C.D., and Sprent J.. 2008. Homeostasis of naive and memory T cells. Immunity. 29:848–862. 10.1016/j.immuni.2008.11.002 [DOI] [PubMed] [Google Scholar]
- Tan, H., Yang K., Li Y., Shaw T.I., Wang Y., Blanco D.B., Wang X., Cho J.H., Wang H., Rankin S., et al. 2017. Integrative Proteomics and Phosphoproteomics Profiling Reveals Dynamic Signaling Networks and Bioenergetics Pathways Underlying T Cell Activation. Immunity. 46:488–503. 10.1016/j.immuni.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa, P., Manso B., Chung J.Y., Romera Arocha S., Xue H.H., Angelo D.B.S., and Shapiro V.S.. 2017a. The differentiation of ROR-γt expressing iNKT17 cells is orchestrated by Runx1. Sci. Rep. 7:7018. 10.1038/s41598-017-07365-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa, P., Romero Arocha S., Chung J.Y., Sant’Angelo D.B., and Shapiro V.S.. 2017b. Histone deacetylase 3 is required for iNKT cell development. Sci. Rep. 7:5784. 10.1038/s41598-017-06102-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, M.J., Weinmann A.S., Matsuda J.L., Salomon R., Farnham P.J., Biron C.A., Gapin L., and Glimcher L.H.. 2004. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 20:477–494. 10.1016/S1074-7613(04)00076-7 [DOI] [PubMed] [Google Scholar]
- Tuttle, K.D., Krovi S.H., Zhang J., Bedel R., Harmacek L., Peterson L.K., Dragone L.L., Lefferts A., Halluszczak C., Riemondy K., et al. 2018. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat. Commun. 9:2650. 10.1038/s41467-018-05026-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt, G.J., Everts B., Chang C.H., Curtis J.D., Freitas T.C., Amiel E., Pearce E.J., and Pearce E.L.. 2012. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 36:68–78. 10.1016/j.immuni.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venezia, T.A., Merchant A.A., Ramos C.A., Whitehouse N.L., Young A.S., Shaw C.A., and Goodell M.A.. 2004. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2:e301. 10.1371/journal.pbio.0020301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., and Hogquist K.A.. 2018. How Lipid-Specific T Cells Become Effectors: The Differentiation of iNKT Subsets. Front. Immunol. 9:1450. 10.3389/fimmu.2018.01450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., McCormick L.L., Fitzgerald P., Chi H., Munger J., and Green D.R.. 2011. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 35:871–882. 10.1016/j.immuni.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J., Yang K., and Chi H.. 2014. Cutting edge: Discrete functions of mTOR signaling in invariant NKT cell development and NKT17 fate decision. J. Immunol. 193:4297–4301. 10.4049/jimmunol.1402042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., Yosef N., Thalhamer T., Zhu C., Xiao S., Kishi Y., Regev A., and Kuchroo V.K.. 2013. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 496:513–517. 10.1038/nature11984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Yang J., Yang K., Wang H., Gorentla B., Shin J., Qiu Y., Que L.G., Foster W.M., Xia Z., et al. 2014. iNKT cells require TSC1 for terminal maturation and effector lineage fate decisions. J. Clin. Invest. 124:1685–1698. 10.1172/JCI69780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K., Neale G., Green D.R., He W., and Chi H.. 2011. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat. Immunol. 12:888–897. 10.1038/ni.2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K., Shrestha S., Zeng H., Karmaus P.W., Neale G., Vogel P., Guertin D.A., Lamb R.F., and Chi H.. 2013. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity. 39:1043–1056. 10.1016/j.immuni.2013.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F.X., Zhao B., and Guan K.L.. 2015. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 163:811–828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, H., Yang K., Cloer C., Neale G., Vogel P., and Chi H.. 2013. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 499:485–490. 10.1038/nature12297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, H., Cohen S., Guy C., Shrestha S., Neale G., Brown S.A., Cloer C., Kishton R.J., Gao X., Youngblood B., et al. 2016. mTORC1 and mTORC2 Kinase Signaling and Glucose Metabolism Drive Follicular Helper T Cell Differentiation. Immunity. 45:540–554. 10.1016/j.immuni.2016.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Wakabayashi N., Wakabayashi J., Tamura Y., Song W.J., Sereda S., Clerc P., Polster B.M., Aja S.M., Pletnikov M.V., et al. 2011. The dynamin-related GTPase Opa1 is required for glucose-stimulated ATP production in pancreatic beta cells. Mol. Biol. Cell. 22:2235–2245. 10.1091/mbc.e10-12-0933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M., Svensson M.N.D., Venken K., Chawla A., Liang S., Engel I., Mydel P., Day J., Elewaut D., Bottini N., and Kronenberg M.. 2018. Altered thymic differentiation and modulation of arthritis by invariant NKT cells expressing mutant ZAP70. Nat. Commun. 9:2627. 10.1038/s41467-018-05095-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D., Medoff B.D., Chen L., Li L., Zhang X.F., Praskova M., Liu M., Landry A., Blumberg R.S., Boussiotis V.A., et al. 2008. The Nore1B/Mst1 complex restrains antigen receptor-induced proliferation of naïve T cells. Proc. Natl. Acad. Sci. USA. 105:20321–20326. 10.1073/pnas.0810773105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L., Xie X., Zhang L., Wang H., Jie Z., Zhou X., Shi J., Zhao S., Zhang B., Cheng X., and Sun S.C.. 2018. TBK-binding protein 1 regulates IL-15-induced autophagy and NKT cell survival. Nat. Commun. 9:2812. 10.1038/s41467-018-05097-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The microarray data that support the findings of this study have been deposited in GEO under accession no. GSE130043.