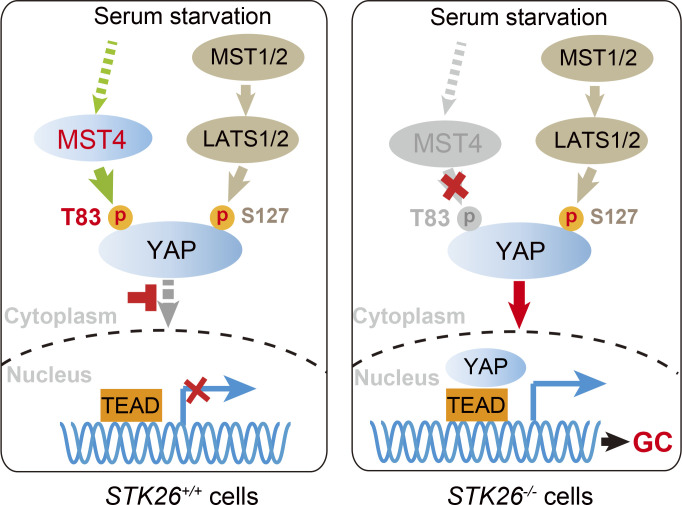
An et al. identify a novel MST4-mediated YAP inactivation signaling pathway and demonstrate that the MST4–YAP axis is required to suppress gastric tumorigenesis in vivo.
Abstract
Hyperactivation of YAP has been commonly associated with tumorigenesis, and emerging evidence hints at multilayered Hippo-independent regulations of YAP. In this study, we identified a new MST4–YAP axis, which acts as a noncanonical Hippo signaling pathway that limits stress-induced YAP activation. MST4 kinase directly phosphorylated YAP at Thr83 to block its binding with importin α, therefore leading to YAP cytoplasmic retention and inactivation. Due to a consequential interplay between MST4-mediated YAP phospho-Thr83 signaling and the classical YAP phospho-Ser127 signaling, the phosphorylation level of YAP at Thr83 was correlated to that at Ser127. Mutation of T83E mimicking MST4-mediated alternative signaling restrained the activity of both wild-type YAP and its S127A mutant mimicking loss of classical Hippo signal. Depletion of MST4 in mice promoted gastric tumorigenesis with diminished Thr83 phosphorylation and hyperactivation of YAP. Moreover, loss of MST4–YAP signaling was associated with poor prognosis of human gastric cancer. Collectively, our study uncovered a noncanonical MST4–YAP signaling axis essential for suppressing gastric tumorigenesis.
Graphical Abstract
Introduction
The Hippo signaling pathway has been extensively studied for its essential roles in organ size control, tissue homeostasis, and tumorigenesis (Lin et al., 2018; Pan, 2010; Yu et al., 2015). In the well-characterized Hippo kinase cascade, MST1/2 kinases phosphorylate LATS1/2 kinases, which in turn phosphorylate the downstream transcriptional coactivator Yes-associated protein (YAP) or transcriptional coactivator with PDZ-binding motif (TAZ; Lin et al., 2018; Moya and Halder, 2019; Pan, 2010; Yu et al., 2015). Phosphorylation of YAP/TAZ, thus causing its cytoplasmic retention and subsequent degradation, is required to avoid otherwise aberrant cell proliferation (Misra and Irvine, 2018; Zhang et al., 2008, 2015; Zhao et al., 2007). In the absence of Hippo signal, YAP/TAZ undergo dephosphorylation and enter the nucleus, where they bind to the TEA domain family of transcription factors (TEAD1–4) to regulate target gene expression, leading to increased cell proliferation and decreased apoptosis (Totaro et al., 2018).
Dysregulation of Hippo pathway contributes to tumorigenesis and has been closely associated with a wide range of human cancers including liver, lung, breast, and gastric cancers (GCs; Chen et al., 2012; Cottini et al., 2014; Harvey et al., 2013; Jiao et al., 2014). Hyperactivation of YAP is frequently observed in most types of cancers. However, it is rare or less common to observe mutation or abnormal expression of the upstream tumor suppressor kinases (Zheng and Pan, 2019). For example, we and others have shown that YAP is hyperactivated in GC (Jiao et al., 2014; Kang et al., 2011; Zhang et al., 2020) and that constitutive activation of YAP drives GC development (Choi et al., 2018; Huang et al., 2020; Jiao et al., 2018); yet MST1/2 and LATS2 kinases seem to be normally expressed without any mutation in GC. These phenomena hint at the possibility of additional or alternative regulation of YAP activity beyond the canonical Hippo kinase cascade.
Despite various types of posttranslational modifications (PTMs) including dephosphorylation (Huang et al., 2013; Liu et al., 2013; Wang et al., 2012; Wilson et al., 2014), ubiquitination (Cho et al., 2020; Sun et al., 2019; Yao et al., 2018), methylation (Fang et al., 2018; Oudhoff et al., 2013), and O-GlcNAcylation (Peng et al., 2017; Zhang et al., 2017) that have been implicated in fine-tuning YAP activity, phosphorylation is regarded as a dominant manner of YAP regulation. The classic Hippo-YAP signaling features sequential phosphorylation of YAP at Ser127 (Zhang et al., 2008, 2015; Zhao et al., 2007) and Ser381/Ser384 (Zhao et al., 2010). Currently, how phosphorylation and thus activation of YAP respond to distinct stimuli is undergoing intensive investigation, especially in various tissue-specific contexts. For instance, cellular energy stress stimulates YAP phosphorylation at S94 by AMP-activated protein kinase, thereby inhibiting YAP activities (Mo et al., 2015; Wang et al., 2015), whereas osmotic stress-induced phosphorylation by Nemo-like kinase at Ser128 activates YAP (Hong et al., 2017; Moon et al., 2017).
The mammalian sterile20-like serine/threonine kinase (STK) family consists of MST1, 2, 3, 4, and STK25, among which MST1/2 kinases represent the canonical Hippo. For the rest members of this family, MST4 is relatively well studied, and is involved in cell proliferation, polarity, and cancer metastasis (Madsen et al., 2015; ten Klooster et al., 2009; Thompson and Sahai, 2015). With its expression dynamically responding to bacterial infection, MST4 kinase can directly phosphorylate TRAF6 to confine macrophage-mediated inflammation, therefore avoiding tissue damage caused by immune overaction (Jiao et al., 2015). Recently, MST4 has been found to orchestrate gastric acid secretion by phosphorylating ACAP4 and Ezrin (Jiang et al., 2015; Yuan et al., 2017). In addition, MST4 stimulates autophagy via phosphorylation-dependent activation of ATG4B in glioblastoma (Huang et al., 2017). Thus, MST4 seems to regulate diverse biological processes via phosphorylating different substrates. Despite comprehensive profiling of features for MST4 substrate targeting and signaling outputs (Miller et al., 2019), the potential function of MST4 in the Hippo pathway remains elusive.
Previously, we and others have discovered VGLL4 as a natural antagonist of YAP (Koontz et al., 2013; Zhang et al., 2014), and developed a peptide inhibitor mimicking VGLL4 to treat GC (Jiao et al., 2014). Recently, we further identified IRF3 as an agonist of YAP in the context of viral infection, and showed that pharmacological inhibition of IRF3 suppressed YAP-driven gastric tumorigenesis (Jiao et al., 2018). In this current study, we characterized MST4 as a direct bona fide kinase of YAP, which phosphorylates YAP at Thr83 upon serum starvation, thereby causing impaired nuclear importing of YAP. We further demonstrated this MST4–YAP axis functions in parallel with the canonical Hippo kinase cascade. Moreover, depletion of MST4 exacerbated GC growth by promoting YAP activity. Consistently, both MST4 expression and YAP Thr83 phosphorylation were found to be markedly decreased in GC patients and strongly associated with poor prognosis. Overall, this work uncovered a noncanonical MST4–YAP signaling axis and demonstrated its pathological function in gastric tumorigenesis.
Results
Identification of MST4 as an inhibitor of stress-induced YAP activation
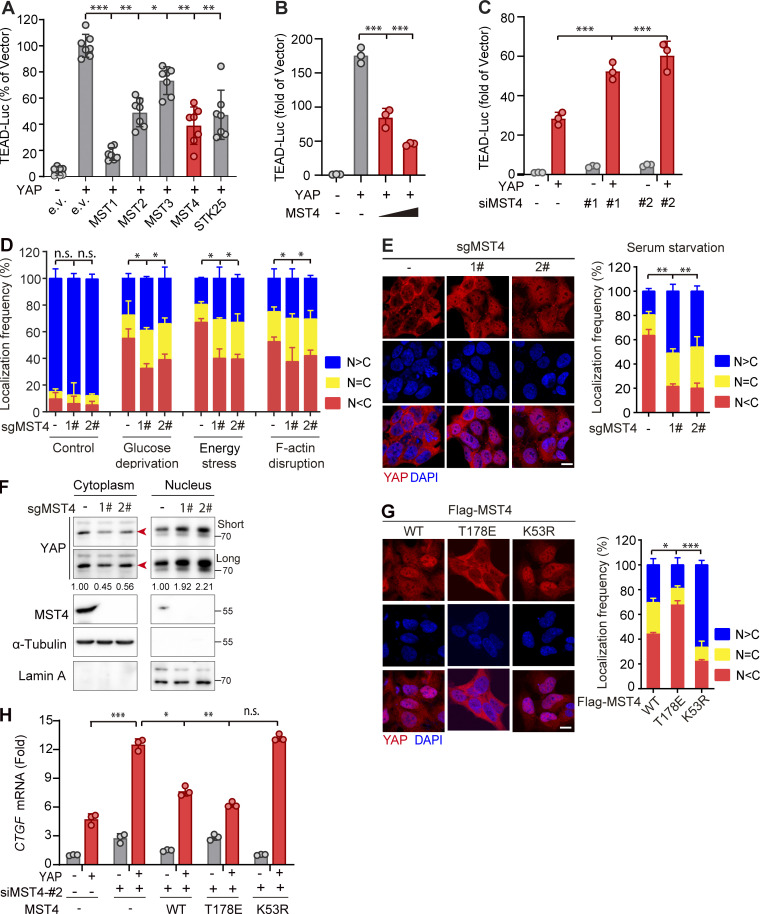
Given that the MST family of kinases share relatively conserved three-dimensional structures but only MST1/2 kinases are regarded as mammalian Hippo homologues, we hypothesized that members other than MST1/2 might also relate to the Hippo pathway such as the activation of YAP. To test this possibility, we used a luciferase-based reporter assay in HEK293FT cells to monitor YAP-induced transactivation of TEAD4 in response to overexpression of various MST family members. Confirming the suitability of our screen, ectopic expression of YAP induced robust activation of TEAD4 promoter–mediated luciferase activity (Fig. 1 A). Consistent with the known role of Hippo kinases in suppressing YAP activation, overexpression of MST1 or MST2 kinase dramatically decreased the reporter activity (Fig. 1 A and Fig. S1 A). Intriguingly, we found all other MST members including MST3, MST4, and STK25 significantly also attenuated YAP-stimulated reporter activities, among which MST4 showed the strongest inhibitory effects and was thus chosen for further characterization (Fig. 1 A and Fig. S1 A).
Figure 1.
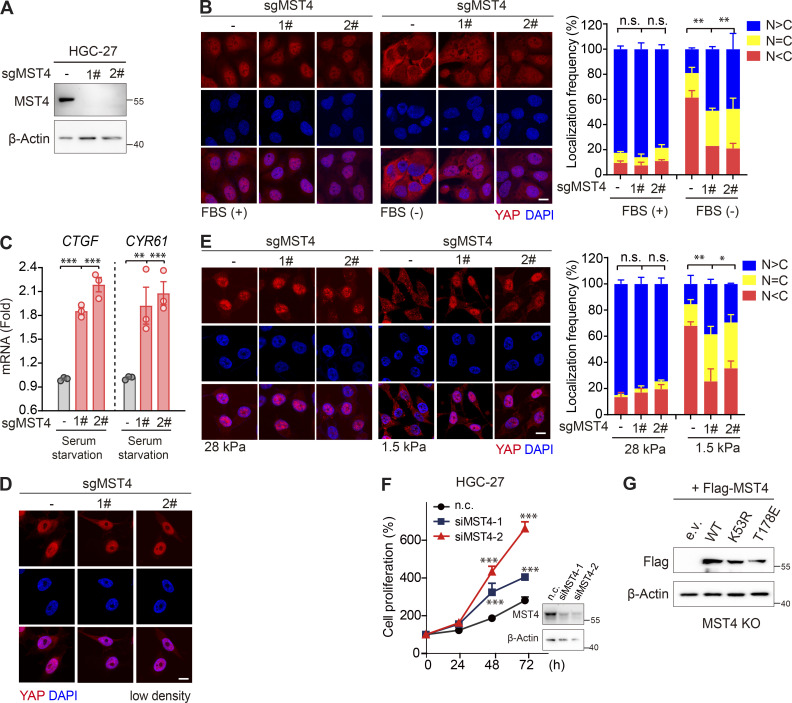
Identification of MST4 as a negative regulator of YAP activation. (A) Mini-screen identifies MST3, MST4, and STK25 as negative regulators of YAP activation. HEK293FT cells were transiently transfected with either empty or Flag-tagged YAP and indicated constructs, together with TEAD-luciferase reporters. Luciferase assay was performed 48 h after transfection (n = 2 within a total of 7 replicates, *, P < 0.05; **, P < 0.01; ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with column 2). (B) MST4 dose-dependently inhibits YAP transcriptional activity. HEK293FT cells were cotransfected with YAP and an increasing amount of MST4 expression construct (0, 1, and 3 µg) for 48 h before performing a luciferase assay (n = 3, ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with column 2). (C) MST4 depletion activates YAP-mediated transcription. HEK293FT cells were transfected with indicated siRNAs for 24 h. Cells were further transfected with either empty or YAP construct for another 24 h before harvest for luciferase assay (n = 3, *, P < 0.05; **, P < 0.01, one-way ANOVA with Dunnett’s post hoc analysis, compared with column 2). (D and E) MST4 depletion increases YAP nuclear localization in response to stimuli. 293A cells (WT and KO) were untreated or treated with glucose-free medium (4 h), 2-DG (50 mM, 2 h), dihydrocytochalasin B (5 µM, 1 h), and serum-free medium (12 h) before processing for IFA assay using anti-YAP antibodies. N > C, YAP is enriched in nucleus; N = C, YAP is evenly distributed in cytoplasm and nucleus; N < C, YAP is enriched in cytoplasm (scale bar, 5 µm; n = 3, *, P < 0.05; **, P < 0.01, one-way ANOVA with Dunnett’s post hoc analysis, compared with control gRNA). (F) 293A cells (WT and KO) were treated with serum-free medium for 12 h. Fractionation assay was performed to examine YAP protein distribution via Western blotting analysis using indicated antibodies. Short, short exposure; Long, long exposure (n = 3, quantification values represent the mean from three repeats). (G and H) MST4-mediated YAP inactivation requires its kinase activity. (G) 293A cells stably expressing Flag-tagged MST4WT, MST4T178E, and MST4K53R were processed for IFA assay to examine YAP subcellular localization (scale bar, 5 µm; n = 3, *, P < 0.05; ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with MST4WT). (H) 293A cells pretreated with MST4 siRNA#2 were transfected with indicated MST4 constructs, together with or without YAP plasmid. qPCR analysis of CTGF mRNA expression were performed in each treatment (n = 3, *, P < 0.05; **, P < 0.01; ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with column 4). All data are presented as mean ± SEM. e.v., empty vector; n.s., not significant. See also Fig. S1.
Figure S1.
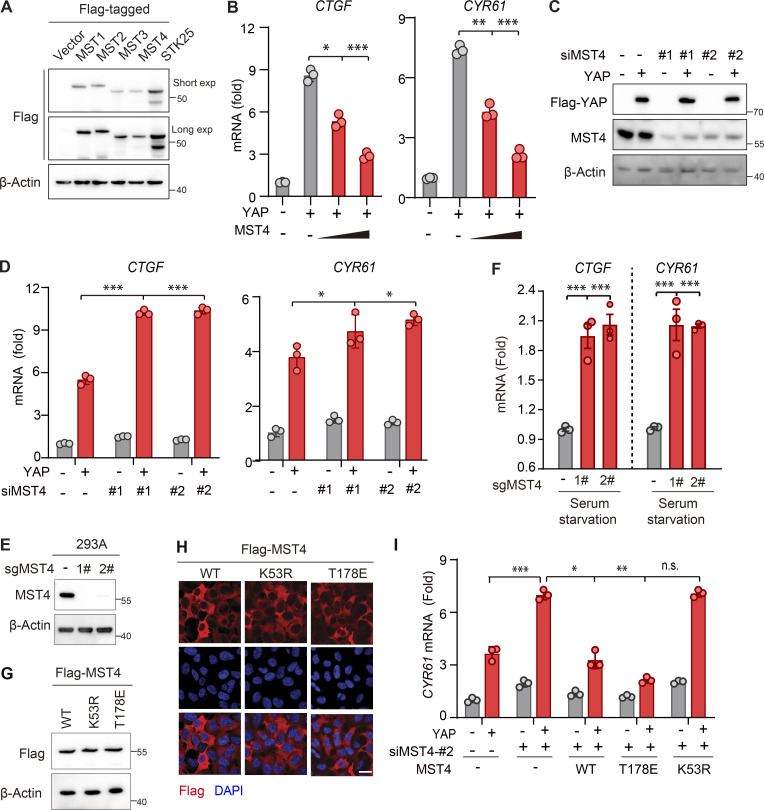
MST4 negatively regulates YAP activation. (A) Western blotting analysis of exogenously expressed indicated constructs using anti-Flag antibodies (n = 2). (B) MST4 dose-dependently inhibits YAP target gene expression. HEK293FT cells were treated as described in Fig. 1 B followed by qPCR analysis of CTGF and CYR61 mRNA expression (n = 3, *, P < 0.05; **, P < 0.01; ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with column 2). (C) Validation of the siRNA-mediated MST4 knockdown efficiency by Western blotting using indicated antibodies (n = 2 biological repeats). (D) MST4 depletion activates YAP target gene expression. HEK293FT cells were treated as described in Fig. 1 C followed by qPCR analysis of CTGF and CYR61 mRNA expression (n = 3, *, P < 0.05; ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with column 2). (E) Generation and validation of the 293A MST4 KO cells using CRISPR-Cas9 technology. (F) MST4 depletion promotes YAP target gene expression. 293A cells (WT and KO) were treated with serum-free medium for 12 h. mRNA was extracted, and transcriptional levels of CTGF and CYR61 were examined by real-time qPCR (n = 3, ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with control gRNA). (G and H) Generation and validation of the 293A KO cells with reconstitution with indicated constructs as used in Fig. 1 G by either IFA (G; n = 1 biological repeat) or Western blotting (H; n = 1 biological repeat). (I) 293A cells pretreated with MST4 siRNA#2 were transfected with indicated MST4 constructs, together with or without YAP plasmid. qPCR analysis of CYR61 mRNA expression were performed in each treatment (n = 3, n.s., not significant, *, P < 0.05; **, P < 0.01; ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with column 4). Data are presented as the mean ± SEM. Related to Fig. 1.
Confirming MST4’s negative regulation of YAP activation, overexpression of MST4 substantially inhibited YAP transactivation, as well as the expression of downstream target genes CTGF and CYR61, in a dose-dependent manner (Fig. 1 B and Fig. S1 B). Conversely, siRNA-mediated MST4 depletion greatly stimulated YAP-induced TEAD4-luciferase activity and target genes' transcription (Fig. 1 C and Fig. S1, C and D). To further explore MST4’s regulation of YAP activity, we generated MST4 KO cells using CRISPR-Cas9 methodology in 293A cells (Fig. S1 E), and examined subcellular localization of YAP under various conditions, including serum starvation, glucose deprivation, energy stress, and F-actin disruption (Fig. 1, D and E). As expected, these stimuli led to decreased localization of YAP in the nucleus (Fig. 1, D and E). To our surprise, depletion of MST4 resulted in a robust increase of YAP nuclear localization; we observed this under all tested conditions, but note that the most efficient nuclear translocation occurred under serum starvation (Fig. 1, D and E).
We then went on to investigate MST4 regulation of YAP in a context of using serum starvation as a stress signal. To further verify the above observations of MST4 depletion-induced nuclear localization of YAP, we separated and analyzed by Western blotting nuclear and cytoplasmic YAP under the stress of serum starvation. Consistently, more YAP was present in the nuclear fractions of MST4-deficient cells compared with control cells (Fig. 1 F). Meanwhile, we found that transcriptions of YAP target genes CTGF and CYR61 were significantly up-regulated in MST4 KO cells in response to serum starvation (Fig. S1 F). These lines of evidence together strongly suggest that MST4 limits YAP activity in response to various Hippo-triggering stress signals.
MST4 limits YAP activation depending on its kinase activity
To evaluate whether the observed MST4-mediated YAP inactivation requires its kinase activity, we stably reconstituted MST4 KO cells (gRNA2) with MST4 alleles encoding WT MST4 (hereafter as MST4WT), its kinase inactive K53R mutant (MST4K53R), or its phosphomimic T178E mutant (MST4T178E; Fig. S1, G and H). In the cells expressing MST4WT, YAP was distributed throughout the cell but more sequestered in the cytoplasm (Fig. 1 G). Moreover, the cytoplasmic localization of YAP was further enhanced in the cells expressing the constitutively activated MST4T178E mutant, which showed <20% frequency for nuclear localization of YAP (Fig. 1 G). In contrast, YAP was mostly localized in the nucleus (∼70% frequency) in the cells expressing the kinase-dead MST4K53R mutant (Fig. 1 G). These observations clearly indicate that MST4 inhibits nuclear translocation of YAP in a manner dependent on its kinase activity.
We also measured the mRNA levels of the YAP target genes including CTGF and CYR61 in MST4-depleted cells reconstituted with MST4WT, MST4T178E, or MST4K53R. Consistently, we found that overexpression of YAP led to increased transcription of CTGF and CYR61, and observed that this increase was further stimulated in MST4-depleted cells (Fig. 1 H and Fig. S1 I). Moreover, ectopic expression of MST4WT and MST4T178E, but not MST4K53R, markedly reversed this increase in YAP-induced transcription of the target genes (Fig. 1 H and Fig. S1 I). Collectively, these results demonstrate that MST4, via its kinase activity, limits stress-related YAP activation, thereby defining a previously unknown MST4-regulated branch of the YAP signaling pathway.
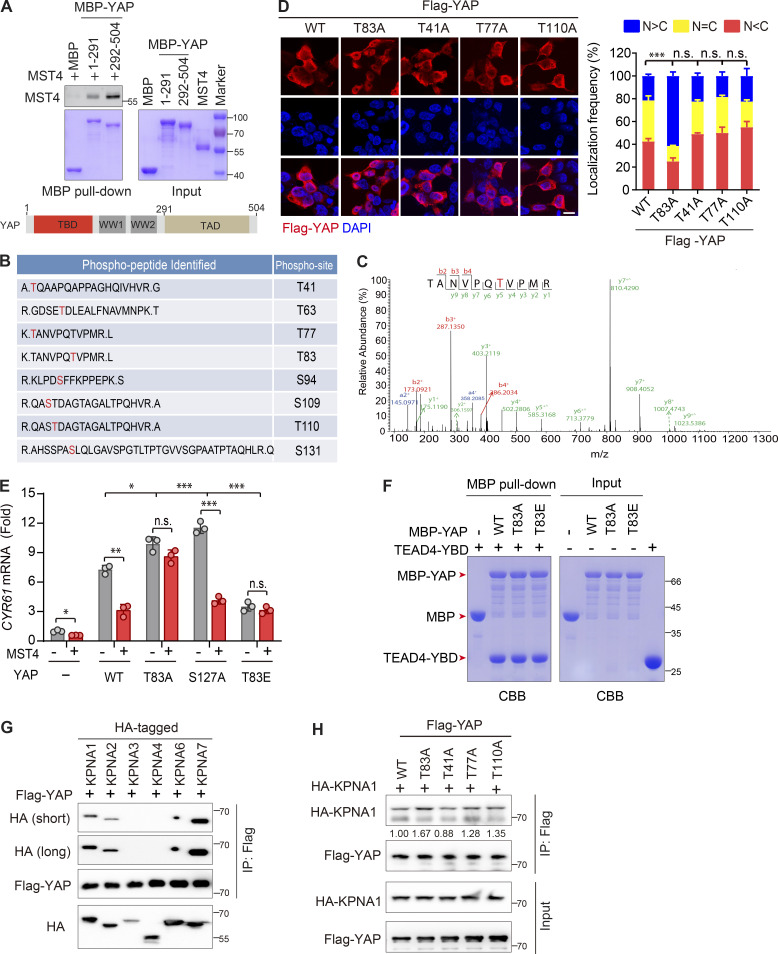
MST4 directly interacts with and phosphorylates YAP at Thr83
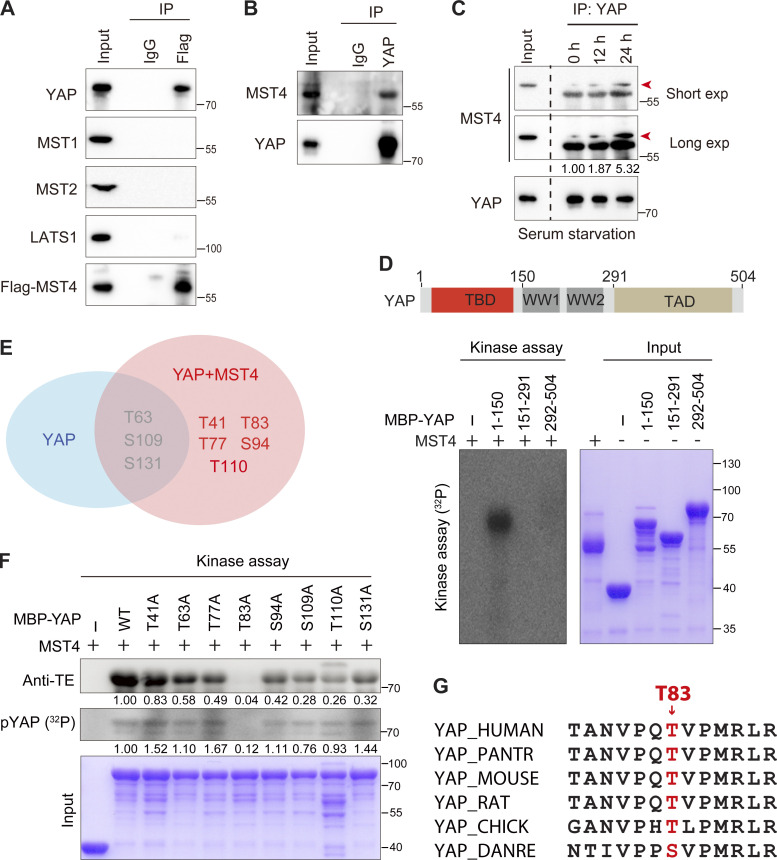
Seeking potential mechanism(s) through which MST4 limits YAP activation, we examined whether MST4 may interact with core Hippo signaling components. To this end, Flag-tagged MST4 was overexpressed, and coimmunoprecipitation (Co-IP) assays were performed in HEK293FT cells (Fig. 2 A). We failed to detect any interaction between MST4 and upstream kinases including MST1, MST2, and LATS1; however, this analysis revealed an interaction of MST4 with YAP (Fig. 2 A). We then confirmed that endogenous YAP also interacts with endogenous MST4 in separate immunoprecipitation experiments (Fig. 2 B). Further, and consistent with our aforementioned finding that MST4 inactivates YAP in response to serum starvation, this stress condition enhanced the YAP–MST4 interaction (Fig. 2 C). To further dissect the MST4–YAP interaction, we purified proteins of MST4 and maltose-binding protein (MBP)–tagged YAP fragments and analyzed their interaction using a pull-down approach, and our results indicated that YAP directly interacts with MST4 (Fig. S2 A).
Figure 2.
MST4 interacts with and phosphorylates YAP at Thr83. (A) Exogenously expressed MST4 interacts with YAP. HEK293FT cells were transiently transfected with plasmid encoding Flag-MST4 for 48 h. Cells lysates were incubated with either control IgG or Flag antibodies together with protein A beads. Immunoprecipitates were analyzed by Western blotting using indicated antibodies (n = 3). (B) Endogenous MST4 interacts with YAP. Cell lysates derived from HEK293FT cells were incubated with protein A agarose beads conjugated with YAP antibody or IgG. Immunoprecipitates were detected using indicated antibodies (n = 2). (C) The MST4–YAP interaction is enhanced upon serum starvation. HEK293FT cells were treated with serum-free medium for the indicated time before processing for immunoprecipitation assay (n = 2, quantification values represent the mean from two repeats). (D) MST4 phosphorylates YAP N terminus. Purified MBP-tagged YAP fragmental fusion proteins were incubated with purified MST4 protein in the kinase assay buffer supplemented with 0.1 mM [γ32P]-ATP for 30 min. Samples were subjected to autoradiography analysis or Coomassie Brilliant Blue staining. MBP was used as a negative control. Schematic illustration of the domain structures of YAP is shown (n = 3). (E) MS analysis of YAP phosphorylation sites mediated by MST4. Purified YAP N-terminal proteins (1–150 aa) were coincubated with or without MST4 protein in the kinase reaction buffer in vitro. Samples were subsequently processed for MS analysis (n = 1). (F) MST4 mainly phosphorylates YAP at Thr83. Purified YAP N-terminal proteins (1–291 aa) harboring the indicated point mutation were coincubated with MST4 protein in vitro and were subjected to either radioactive kinase assay or ATPγS-based kinase assay. Samples were separated by SDS-PAGE and analyzed by autoradiography or Western blotting using anti–thiophosphate ester (anti-TE) antibody or Coomassie Brilliant Blue staining (n = 3, quantification values represent the mean from three repeats). (G) Sequence alignment of the putative YAP phosphorylated site Thr83 among indicated species. IP, immunoprecipitation; exp, exposure. See also Fig. S2.
Figure S2.
MST4-mediated YAP Thr83 phosphorylation impairs its nuclear import. (A) MST4 directly interacts with YAP. Purified MST4 protein and MBP-tagged YAP fragmental fusion proteins were subjected to MBP–pull down assay. The immunoprecipitates were analyzed by Western blotting using anti-MST4 antibody or Coomassie Blue staining. MBP tag was used as negative control (n = 3). (B) List of YAP phospho-peptides identified by MS (n = 1). (C) The pink map of phosphorylation of YAP Thr83 identified by MS. (D) Identification of the key MS-identified phosphorylation sites required for MST4-mediated YAP inactivation. 293A cells were transfected with indicated YAP constructs for 24 h before processing to IFA assay using Flag antibody (scale bar, 5 µm, n = 3, n.s., not significant, ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with YAPWT). (E) YAP Thr83 phosphorylation is required for MST4-mediated YAP inactivation. HEK293FT cells were transfected with indicated YAP constructs together with or without MST4 for 48 h. Cells were processed for real-time qPCR analysis of CYR61 transcriptional levels (n = 3, n.s, not significant; *, P< 0.05; **, P< 0.01; ***, P< 0.001, unpaired t test and one-way ANOVA with Dunnett’s post hoc analysis as compared with column 3). (F) YAP Thr83 phosphorylation does not affect its TEAD4-binding affinity. Purified TEAD4-YBD protein and indicated MBP-tagged YAP proteins (1–150 aa) were subjected to MBP–pull down assay. The immunoprecipitates were analyzed by Coomassie Brilliant Blue staining. MBP tag was used as negative control (n = 3). (G) YAP interacts with KPNA1, KPNA2, and KPNA7. HEK293FT cells were cotransfected with constructs encoding Flag-YAP and HA-tagged KPNA(s) for 48 h. Cell lysates were incubated with Flag beads for 4 h and analyzed by Western blotting using indicated antibodies (n = 3). (H) YAP Thr83 mutation but not the other MS-identified sites increases YAP-KPNA1 association. HEK293FT cells were cotransfected with constructs encoding HA-KPNA1 and Flag-tagged YAP mutants for 48 h. Cell lysates were incubated with Flag beads for 4 h and analyzed by Western blotting using indicated antibodies (n = 3, quantification values represent the mean from three repeats). Data are presented as the mean ± SEM. Related to Fig. 2 and Fig. 3.
Our findings that MST4 interacts with and limits YAP activation in a kinase activity-dependent manner prompted us to speculate that YAP may be a substrate of MST4. Indeed, an in vitro kinase assay using purified MST4 and MBP-tagged YAP protein fragments revealed that the N-terminal domain of YAP (1–150 aa) was robustly phosphorylated when coincubated with MST4 (Fig. 2 D), thereby defining YAP as a novel MST4 phosphorylation target. To identify the specific residue(s) of YAP phosphorylated by MST4, purified MBP-tagged YAP N-terminal proteins were coincubated with or without MST4 in vitro and were subsequently subjected to mass spectrometry (MS)–based phosphoproteomics analysis. Results indicated that YAP was phosphorylated at several residues including Thr63, Ser109, and Ser131 without adding MST4 in the system; however, coincubation with MST4 further phosphorylated YAP at Thr41, Thr77, Thr83, Ser94, and Thr110 (Fig. 2 E and Fig. S2, B and C).
Next, we adopted a point mutation strategy to identify which residue of YAP is the key phosphorylation site mediated by MST4. To this end, we mutated each phosphorylation site to alanine (A), purified the corresponding MBP-tagged YAP N-terminal proteins, and performed in vitro kinase assays. Strikingly, mutation of Thr83, but not the other putative residues, completely blocked YAP phosphorylation by MST4 as assessed via both anti-thiophosphate ester specific antibody and radio-labeled 32P analyses (Fig. 2 F). Sequence alignment of YAP Thr83 revealed this threonine residue is conserved from chicken to human (Fig. 2 G). Taken together, these results establish that YAP is a bona fide substrate of MST4 kinase and indicate that Thr83 of YAP is the major site targeted for phosphorylation by MST4.
Phosphorylation at Thr83 is required for MST4-mediated YAP inactivation
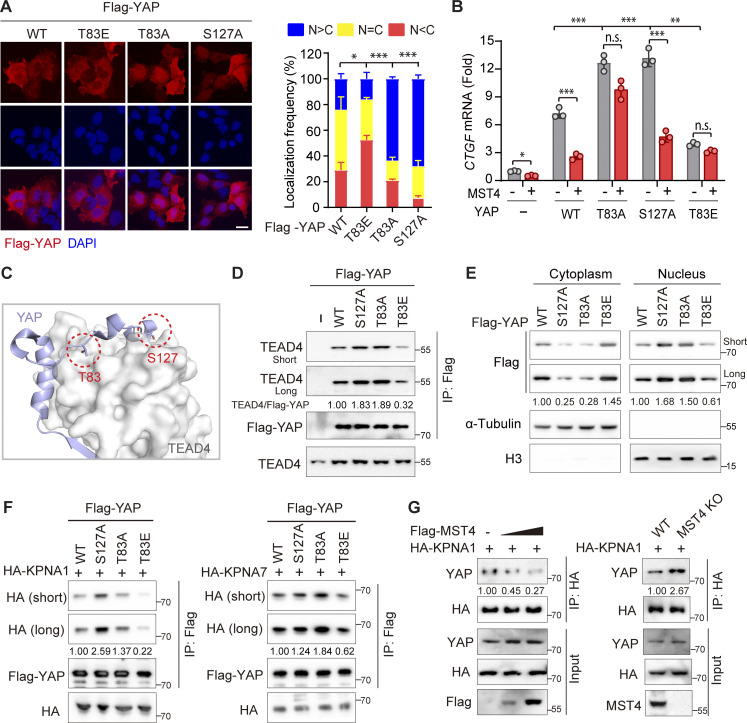
It is known that phosphorylation of YAP at Ser127 by the classic Hippo kinase cascade is essential for its cytoplasmic localization and inactivation. Since MST4 can attenuate YAP activation and phosphorylate YAP at Thr83, we hypothesized that phosphorylation of YAP at Thr83 (hereafter referred as pYAPT83) is responsible for the MST4-mediated inhibitory effect. To test this possibility, we replaced Thr83 with a glutamic acid (T83E, hereafter referred as YAPT83E) to mimic MST4-mediated Thr83 phosphorylation, and examined the subcellular localization of the YAPT83E mutant. Consistent with our finding for the phosphomimetic MST4T178E mutant variant (Fig. 1 G), we found whereas WT YAP tended to be evenly distributed in the cells, the YAPT83E mutant was mainly localized in the cytoplasm (Fig. 3 A). The YAPT83A mutant that is resistant to MST4-mediated phosphorylation, on the contrary, showed a clear nuclear localization pattern, comparable to the pattern for the classic constitutively activated mutant variant YAPS127A (Fig. 3 A). Meanwhile, mutation of other candidate sites including Thr41, Thr77, and Thr110 did not significantly alter the subcellular distribution patterns of YAP (Fig. S2 D).
Figure 3.
YAP Thr83 phosphorylation impairs its nuclear import and activation. (A) YAPT83A mutant mainly localizes into nucleus. 293A cells transfected with indicated YAP constructs were subjected to IFA assay using Flag antibody (scale bar, 5 µm; n = 3, *, P < 0.05; ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with YAPWT). (B) YAP Thr83 phosphorylation is required for MST4-mediated YAP inactivation. HEK293FT cells were transfected with indicated YAP constructs together with or without MST4 for 48 h. Cells were processed for real-time qPCR analysis of CTGF transcriptional levels (n = 3, n.s., not significant, *, P < 0.05; **, P< 0.01; ***, P < 0.001, unpaired t test and one-way ANOVA with Dunnett’s post hoc analysis as compared with column 4). (C) Cartoon views of the YAP Thr83 position at the YAP-TEAD4 interface (PDB ID: 3KYS). (D) YAPT83E mutant impairs its association with TEAD4. Cell lysates derived from HEK293FT cells transfected with indicated YAP constructs were subjected to Flag immunoprecipitation. Immunoprecipitates were detected using anti-Flag or anti-TEAD4 antibodies (n = 3, quantification values represent the mean from three repeats). (E) YAPT83E was mainly enriched in the cytoplasm. HEK293FT cells transfected with indicated YAP constructs were subjected to fractionation assay, and YAP cytoplasmic/nucleus distributions were examined via Western blotting using indicated antibodies (n = 3, quantification values represent the mean from three repeats). (F) Phosphorylation of YAP at Thr83 impairs YAP–KPNA(s) interactions. HEK293FT cells were cotransfected constructs encoding Flag-YAP (WT and mutants) and HA-KPNA1 (left panel) or HA-KPNA7 (right panel) for 48 h. Cell lysates were incubated with Flag beads for 4 h and analyzed by Western blotting using indicated antibodies (n = 3, quantification values represent the mean from three repeats). (G) MST4 negatively regulates YAP-KPNA1 binding. 293A cells were cotransfected with HA-KPNA1 and increased amount of Flag-MST4 plasmid (0, 1, and 3 µg; left panel). Or 293A cells (WT and MST4 KO) were transfected with HA-KPNA1 (right panel). Cell lysates were subjected to HA immunoprecipitation and analyzed using indicated antibodies (n = 3, quantification values represent the mean from three repeats). Data are presented as the mean ± SEM. See also Fig. S2.
We next measured the abilities of WT and various YAP mutants to stimulate CTGF and CYR61 mRNA transcription and found that overexpression of YAPWT, as well as YAPT83A and YAPS127A, all significantly increased target gene transcription (Fig. 3 B and Fig. S2 E). Notably, the YAPS127A and YAPT83A mutants showed more robust induction compared with YAPWT, findings consistent with our observation of their increased nuclear localization (Fig. 3 A). In contrast, the cells expressing the phosphomimetic YAPT83E mutant had lower CTGF and CYR61 expression than cells expressing YAPWT as well as cells expressing YAPT83A or YAPS127A (Fig. 3 B and Fig. S2 E). Furthermore, overexpression of MST4 efficiently suppressed YAPWT-induced gene expression but did not affect the expression-inducing activities of YAPT83A or YAPT83E (Fig. 3 B and Fig. S2 E), thereby demonstrating that Thr83 is the phosphorylation site through which MST4 attenuates YAP activity. However, it is important to note that these experiments also showed that MST4 overexpression also suppressed YAPS127A-induced expression of CTGF and CYR61 expression (Fig. 3 B and Fig. S2 E), hinting at the independence of Thr83’s YAP-regulating effects from those mediated by the canonical Hippo cascade Ser127 target site.
YAP phosphorylation at Thr83 attenuates its nuclear import
Given our findings that MST4 limits YAP activity via Thr83 phosphorylation and such an effect is independent of the classic Hippo kinase cascade, we went on to investigate the mechanism through which Thr83 phosphorylation causes YAP inactivation. Interestingly, based on reported TEAD4-binding interface of YAP (Chen et al., 2010; Li et al., 2010), we noted that Thr83 is just positioned adjacent to this interface (Fig. 3 C). We then performed Co-IP assays in HEK293FT cells overexpressing YAP variants to evaluate their interaction with TEAD4. Interestingly, compared with YAPWT and YAPT83A, the YAPT83E mutant exhibited reduced association with TEAD4 (Fig. 3 D). Next, to determine whether the observed decreased TEAD4-YAPT83E association resulted either from reduced protein–protein interaction or from reduced nuclear import of YAP, we first performed an in vitro pull-down assay using purified recombinant TEAD4 proteins and indicated MBP-tagged YAP proteins. Results revealed that YAPT83E displayed comparable TEAD4-binding capacities as with YAPWT and YAPT83A (Fig. S2 F), suggesting that Thr83 phosphorylation of YAP does not affect its direct interaction with TEAD4. To test the other possibility, we next examined the cytoplasmic/nucleus distribution of indicated YAP mutants, and found that both YAPT83A and YAPS127A were dominantly enriched in the nucleus, whereas YAPT83E was mainly sequestered in the cytoplasm as compared with YAPWT (Fig. 3 E). Collectively, these data indicate that YAP phosphorylation at Thr83 impairs its nuclear import.
The cytoplasm-to-nucleus translocation of Yorkie, the Drosophila melanogaster homologue of YAP, is mediated by the classical importin α/β heterodimer complex (Wang et al., 2016). Distinct from Drosophila (which has only three importin α subunits), a total of seven importin α subunits (KPNA1–KPNA7) have been identified in mammalian cells (Bauer et al., 2015). To identify the specific subunits required for YAP nuclear import in mammalian cells, we conducted Co-IP experiments showing that YAP interacts with KPNA1, KPNA2, and KPNA7 (Fig. S2 G). Next, Co-IP assays between YAP mutants and KPNA1 (or KPNA7) were performed to evaluate whether pYAPT83 affects its binding ability to importins (Fig. 3 F and Fig. S2 H). Both YAPT83A and YAPS127A displayed higher binding affinities for KPNA1/KPNA7 than did YAPWT, whereas the phosphomimic YAPT83E had much weaker binding affinity for KPNA1/KPNA7 (Fig. 3 F). Consistently, overexpression of MST4 apparently suppressed the interaction of YAP with KPNA1, while depletion of MST4 promoted such interaction (Fig. 3 G). Overall, we concluded that MST4-mediated Thr83 phosphorylation of YAP impairs its binding to importin complex, leading to decreased nuclear translocation and activation of YAP.
Serum starvation induces concomitant phosphorylation of YAP by MST4 and LATS1/2
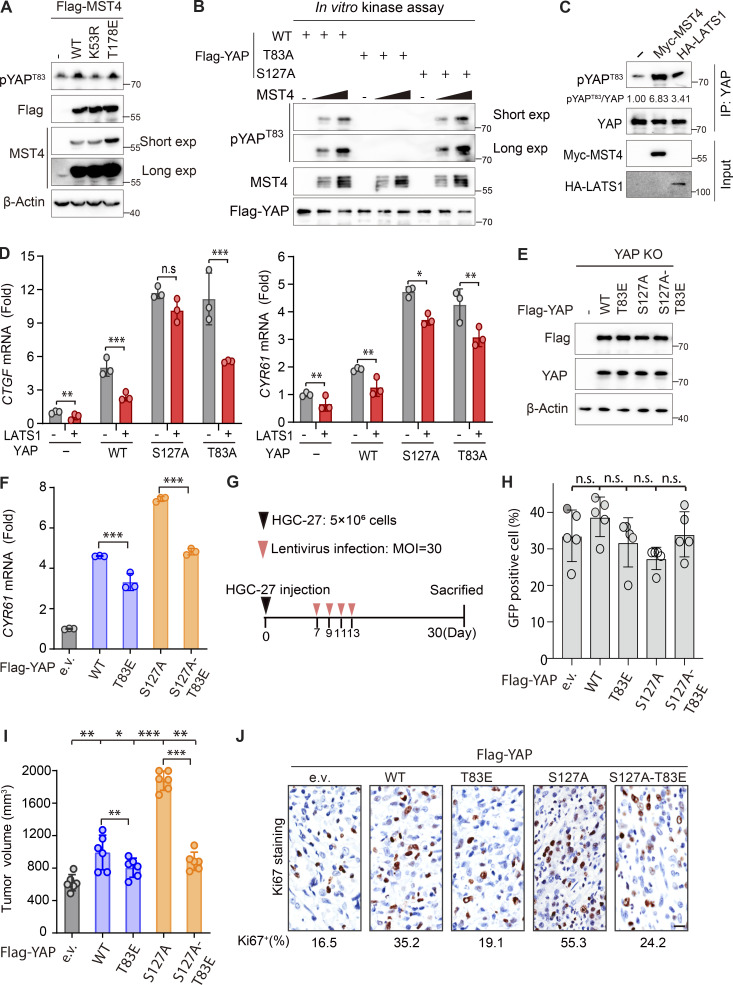
To better characterize YAP Thr83 phosphorylation in response to a stress stimulus (serum starvation), we raised a polyclonal antibody specifically recognizing phosphorylation of YAP Thr83 as its epitope (Fig. 4 A). And we confirmed that this antibody could recognize MST4-enforced YAPWT, but not YAPT83A mutant, phosphorylation (Fig. 4 A). Consistent with the requirement of MST4’s kinase activity, MST4WT and MST4T178E, but not MST4K53R, markedly increased pYAPT83 levels (Fig. S3 A). Using this antibody, we found that serum starvation treatment resulted in dramatic increases in YAP Thr83 phosphorylation levels (Fig. 4 B). Consistent with previous reports, this treatment also significantly increased YAP Ser127 phosphorylation levels (Fig. 4 B). As expected, this stress-induced YAP Thr83 phosphorylation was significantly reduced in MST4 KO cells (Fig. 4 C). Importantly, we found that phosphorylation of YAP at Ser127 was also greatly reduced in the MST4 KO cells (Fig. 4 C). Moreover, the phosphorylation levels at both sites were decreased in cells in which the canonical Hippo components LATS1/2 were knocked out (LATS1/2 DKO) in response to serum starvation (Fig. 4 D).
Figure 4.
Interplay between pYAPT83 and pYAPS127 in response to serum starvation. (A) Validation of the homemade pYAPT83 antibody. HEK293FT cells were cotransfected with Flag-tagged YAP (WT or T83A mutant) and Myc-MST4 for 48 h. Cell lysates were processed for Flag immunoprecipitation and were analyzed using indicated antibodies (n = 4). (B) Serum starvation triggers concomitant phosphorylation of YAP at Thr83 and Ser127. 293A cells cultured in medium with or without serum for 12 h were harvested and analyzed by Western blotting using indicated antibodies (n = 3, quantification values represent the mean from three repeats). (C) MST4 promotes YAP phosphorylation at Thr83 and Ser127 in response to serum starvation. 293A cells (WT and MST4 KO) were subjected to serum starvation for 12 h and analyzed by Western blotting using indicated antibodies (n = 3, quantification values represent the mean from three repeats). (D) LATS1/2 promotes YAP phosphorylation at Thr83 and Ser127 in response to serum starvation. 293A cells (WT and LATS1/2 KO) were subjected to serum starvation treatment and analyzed by Western blotting (n = 3, quantification values represent the mean from three repeats). (E) Cell lysates derived from HEK293FT cells expressing indicated Flag-tagged YAP proteins were incubated with Flag beads. Immunoprecipitates were subjected to Western blotting analysis using antibodies (n = 2, quantification values represent the mean from two repeats). (F and G) YAP Thr83 phosphorylation constrains YAPS127A-induced transcriptional activation and CTGF expression. 293A YAP KO cells reconstituted with indicated YAP constructs were subjected to either luciferase assay (F; n = 3, ***, P < 0.001, unpaired t test) or qPCR analysis of mRNA expression of CTGF (G; n = 3, ***, P < 0.001, unpaired t test). (H) YAP Thr83 phosphorylation limits YAPS127A-induced tumor formation in vivo. Nude mice injected with HGC-27 cells were further infected with lentivirus harboring indicated YAP mutants. Representative images showed the tumors induced by indicated YAP mutants (left panel). The tumor weights were measured and analyzed 3 wk after injection (three mice/group, n = 2 biological repeats, n.s., not significant, **, P < 0.01; ***, P < 0.001, unpaired t test; and one-way ANOVA with Dunnett’s post hoc analysis as compared with vector control). (I) Schematic illustration of the concomitant pYAPT83 and Ser127 in response to serum starvation. Data are presented as the mean ± SEM. See also Fig. S3.
Figure S3.
Interplay between pYAPT83 and pYAPS127 in response to serum starvation. (A) Phosphorylation of YAP Thr83 requires MST4’s kinase activity. Cell lysates derived from 293A cells transiently transfected with Flag-tagged MST4 constructs were subjected to Western blotting analysis using indicated antibodies (n = 3). (B) YAPS127A mutation does not affect MST4-mediated pYAPT83 in vitro. Purified YAP proteins (WT and mutants) were incubated with increased dose of MST4 protein at 30°C for 15 min in vitro. Samples were separated by SDS-PAGE and immunostaining by indicated antibodies (n = 3). (C) LATS1 overexpression promotes pYAPT83 levels. Cell lysates derived from HEK293FT cells transfected with vector, Myc-MST4, or HA-LATS1 were subjected to YAP immunoprecipitation assay and analyzed by Western blotting using indicated antibody (n = 2, quantification values represent the mean from two repeats). (D) LATS1 overexpression inhibits YAPT83A-induced gene expression. HEK293FT cells were transfected with indicated YAP constructs together with or without LATS1 for 48 h. Cells were processed for real-time PCR analysis of YAP target gene CTGF and CYR61 transcriptional levels (n = 3, n.s, not significant; *, P< 0.05; **, P< 0.01; ***, P< 0.001, unpaired t test). (E) Western blotting analysis of the expression of reconstituted YAP constructs in 293A YAP KO cells (n = 3). (F) YAP Thr83 phosphorylation constrains YAPS127A-induced CYR61 expression. 293A YAP KO cells reconstituted with indicated YAP constructs were subjected to qPCR analysis of mRNA expression of CYR61 (n = 3, ***, P < 0.001, unpaired t test). (G) Schematic illustration of the designed xenograft tumor formation assay. (H–J) YAP Thr83 phosphorylation limits YAPS127A-induced tumor formation. Lentiviral infection efficiency (H), average tumor volume (I), and Ki-67–positive cell rates (J) in each YAP mutant group were examined and quantified (three mice/group, n = 2, n.s., not significant, **, P < 0.01; ***, P < 0.001, unpaired t test and one-way ANOVA with Dunnett’s post hoc analysis as compared with vector group). Data are presented as the mean ± SEM. Related to Fig. 4.
These observations prompted us to speculate that both modifications may be essential for the sequestration of YAP in the cytoplasm in response to serum starvation. To test this hypothesis, we first excluded the possibility that YAPS127A may perturb MST4’s ability to phosphorylate YAPT83, as both YAPWT and YAPS127A can be efficiently phosphorylated by MST4 in vitro (Fig. S3 B). Next, we examined the phosphorylation of both sites in the precipitates of indicated YAP proteins (Fig. 4 E). As expected, YAPT83A and YAPS127A completely blocked the corresponding phosphorylation as compared with YAPWT (Fig. 4 E). Intriguingly, we found that pYAPS127 levels were remarkably attenuated in YAPT83A precipitates, and pYAPT83 levels were also reduced in YAPS127A precipitates (Fig. 4 E), which was entirely consistent with their subcellular localization patterns (Fig. 3 A). Conversely, we found that LATS1 overexpression, which resembles YAPS127E to sequester YAP in cytoplasm, enhanced the pYAPT83 levels (Fig. S3 C), and similarly suppressed YAPT83-induced CTGF and CYR61 expression (Fig. S3 D). Taken together, our results indicate that phosphorylation of one site (Thr83 or Ser127) can alter the subcellular localization of YAP and therefore enhance the phosphorylation of the other site by promoting the availability of YAP to the corresponding kinase, and that concomitant phosphorylation of both sites is required for full sequestration of YAP in the cytoplasm.
MST4 restricts YAP activity independent of the classic Hippo signaling
We next evaluated the biological importance of the MST4-mediated branch of YAP signaling in the absence of a classic Hippo signal. To this end, a YAPS127A-T83E mutant was generated that should in theory disrupt the canonical LATS1/2 kinases-mediated Ser127-dependent inhibitory effects but should mimic MST4-mediated attenuation of YAP activity. We then reintroduced YAPS127A-T83E and other YAP mutants into 293A YAP KO cells (Fig. S3 E), and examined their abilities to stimulate TEAD4-associated luciferase activity and to induce expression of YAP target genes CTGF and CYR61. First, we noted that compared with YAPWT, cells expressing the YAPT83E mutant had significantly decreased TEAD4-luciferase activity and transcription of CTGF and CYR61, while YAPS127A strongly stimulated both luciferase activity and target gene expression (Fig. 4, F and G; and Fig. S3 F). Consistent with our design, expression of the YAPS127A-T83E mutant significantly reduced luciferase activity and target genes’ induction compared with YAPS127A (Fig. 4, F and G; and Fig. S3 F), thereby demonstrating that YAP Thr83 phosphorylation attenuates YAP activation independent of canonical Hippo kinase cascade-mediated Ser127 phosphorylation.
We further investigated whether the YAP Thr83 phosphorylation can rescue the oncogenic functions of YAPS127A in vivo. To this end, nude mice were first injected subcutaneously with GC HGC-27 cells for 1 wk to incubate tumors, and were further infected with lentiviruses expressing the YAP mutants to further stimulate the tumor growth for 3 wk before harvest (Fig. S3 G). Although the infection efficiencies were comparable between groups (30–40%; Fig. S3 H), the YAPT83E mutant group showed decreased average tumor weights (797 mg vs. 1,084 mg) and volumes (804 mm3 vs. 992 mm3) as compared with YAPWT (Fig. 4 H and Fig. S3 I). The YAPS127A mutant, however, can further stimulate tumor growth compared with YAPWT (Fig. 4 H and Fig. S3 I). Strikingly, expression of the YAPS127A-T83E mutant efficiently reversed the tumor weights (from 1,559 to 814 mg) and volumes (from 1,887 to 883 mm3) compared with YAPS127A, and comparable to the YAPT83E group (Fig. 4 H and Fig. S3 I). These observations were further confirmed by immunohistochemistry (IHC) staining of the cell proliferation marker Ki67, which showed similar patterns to those of tumor weights and volumes (Fig. S3 J). Collectively, these results demonstrated that the MST4–YAP signaling is independent of the classic Hippo pathway (Fig. 4 I).
Recapitulation of the MST4–pYAPT83 signaling axis in GC
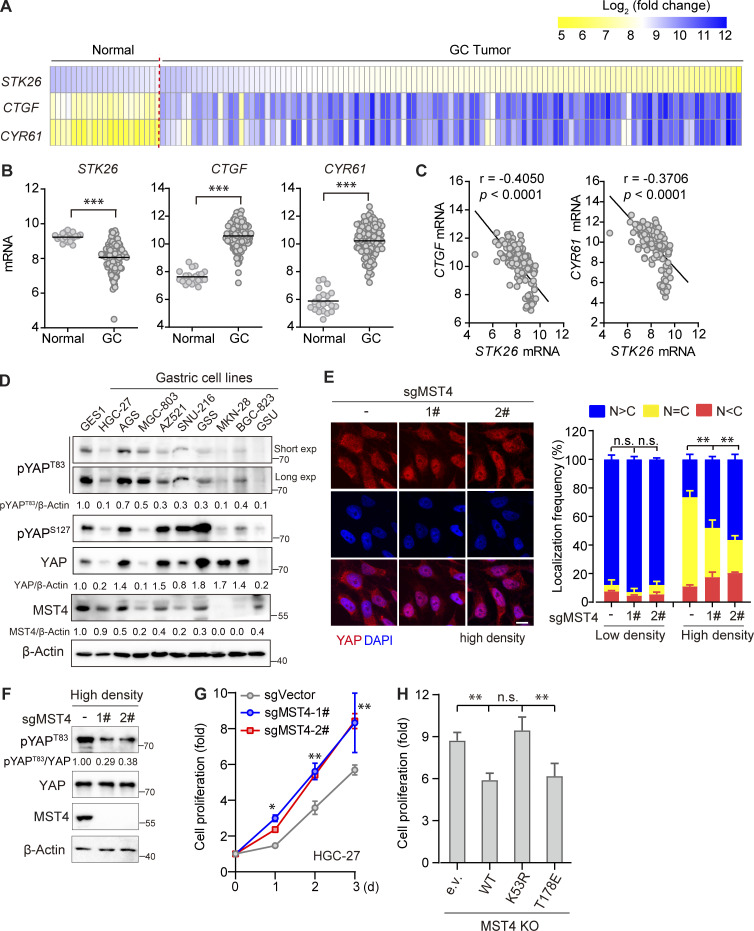
MST4 is known to exert multiple context-dependent functions in tumorigenesis (Chen et al., 2018; Madsen et al., 2015). Our finding that MST4 suppresses YAP activation implies that it may work as a tumor suppressor under certain conditions. Interestingly, analysis of microarray datasets publicly available revealed that expression of MST4 is significantly decreased in GC specimens compared with normal tissues (P < 0.001; Fig. 5, A and B). Moreover, the mRNA levels of YAP target genes CTGF and CYR61 are markedly elevated in patients with GC, and show very good negative correlation with the STK26 mRNA level (Fig. 5, A–C). Consistent with these observations, the expression levels of MST4 and pYAPT83 were both decreased in multiple GC cell lines as compared with normal gastric GES1 cells (Fig. 5 D).
Figure 5.
MST4 limits YAP activation and cell proliferation in GC cells. (A and B) Analysis of STK26, CTGF, and CYR61 mRNA expression in published GC dataset. Heatmap demonstration (A), and statistical analysis (B) of the mRNA expression levels of STK26 and YAP target genes CTGF and CYR61 between healthy (n = 21) and GC tissue samples (n = 111) based on previously published microarray database (GEO accession no. GSE54129, ***, P < 0.001, unpaired t test). (C) STK26 mRNA expression negatively correlated with CTGF and CYR61 mRNA expression. Pearson’s correlation analysis between STK26 mRNA and YAP target genes CTGF and CYR61 was performed by GraphPad Prism 8.0 software. (D) The MST4–YAPT83 axis protein expression was down-regulated in GC cancer cells compared with normal gastric cells. Western blotting analysis of MST4, pYAPT83, pYAPS127, and YAP protein levels in normal and different GC cell lines (n = 2, quantification values represent the mean from two repeats). (E) MST4 depletion increases YAP nuclear localization in response to high cell density. HGC-27 cells (WT and MST4 KO) were seeded at high (90%) cell density for 24 h before processing for IFA assay using anti-YAP antibodies (scale bar, 5 µm, n = 3, n.s, not significant; **, P< 0.01; one-way ANOVA with Dunnett’s post hoc analysis, compared with control gRNA). (F) MST4 depletion reduced pYAPT83 levels in HGC-27 cells. High cell density (90%) of HGC-27 cells (WT and MST4 KO) were harvested for immunoblotting analysis using indicated antibodies (n = 3, quantification values represent the mean from three repeats). (G) MST4 deficiency stimulates GC cell proliferation. Cell proliferation curves of HGC-27 cells (WT and MST4 KOs) over the indicated time were measured by CellTiter Luminescent-based assay (four replicates per cell line for three repeats, *, P< 0.05; **, P< 0.01; two-way ANOVA with Tukey’s post hoc test). (H) MST4 limits cell proliferation requires its kinase activity. Quantification of cell proliferation rates of HGC-27 KO cell reconstituted with indicated constructs on day 3 were measured as in G (n = 3, n.s, not significant; **, P< 0.01; one-way ANOVA with Dunnett’s post hoc analysis, compared with vector control). Data are presented as the mean ± SEM. See also Fig. S4.
To corroborate the MST4–pYAPT83 signaling axis in GC, we generated a MST4 KO line on the basis of HGC-27 cells (Fig. S4 A), and examined the effects of MST4 in modulating YAP localization in response to stress stimuli. In keeping with the results in 293A cells, we found YAP was efficiently localized into the nucleus, and its target genes CTGF and CYR61 were significantly induced in response to serum starvation in the HGC-27–derived MST4 KO cells (Fig. S4, B and C). Moreover, nuclear fractions of YAP were also significantly increased in MST4 KO cells under other conditions including higher cell confluence (Fig. 5 E and Fig. S4 D) and lower matrix stiffness (Fig. S4 E). Importantly, pYAPT83 levels were apparently decreased in MST4 KO cells under high-density conditions (Fig. 5 F).
Figure S4.
MST4 limits YAP activation and cell proliferation in GC cells. (A) Generation and validation of HGC-27 MST4 KO cells using CRISPR-Cas9 technology. (B) MST4 KO promotes YAP nuclear localization in response to serum starvation. HGC-27 cells (WT and MST4 KO) were subjected to serum starvation for 12 h and processed to IFA assay using anti-YAP antibody (scale bar, 5 µm; n = 3, n.s., not significant, **, P < 0.01, one-way ANOVA with Dunnett’s post hoc analysis). (C) MST4 KO promotes YAP target gene expression. HGC-27 cells (WT and KOs) were treated with serum-free medium for 12 h. mRNA was extracted, and transcriptional levels of CTGF and CYR61 were examined by real-time qPCR (n = 3, **, P < 0.01; ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with control gRNA). (D) HGC-27 cells (WT and MST4 KO) at low cell density (10%) were subjected to IFA assay using anti-YAP antibody (scale bar, 5 µm). (E) HGC-27 cells (WT and MST4 KO) were seeded at either soft (1.5 kPa) or high stiffness (28.0 kPa) plates for 24 h before processing for IFA assay using anti-YAP antibodies (scale bar, 5 µm; n = 3, n.s., not significant, *, P< 0.05; **, P < 0.01, one-way ANOVA with Dunnett’s post hoc analysis, compared with control gRNA). (F) Knockdown of MST4 stimulates GC cell growth. HGC-27 cells pretreated with indicated siRNAs were seeded at 1,000 cells/well density, and cell proliferation curves over the indicated time were measured by CellTiter Luminescent-based assay (four replicates per cell line for three repeats, ***, P< 0.001; two-way ANOVA with Tukey’s post hoc test). (G) Western blotting analysis of the reconstitution of MST4 constructs in HGC-27 MST4 KO cells used in Fig. 5 H. Data are presented as the mean ± SEM. n.c., non-targeting control. Related to Fig. 5.
We also examined the regulatory effect of MST4 on the proliferation of HGC-27 cells and found that depletion of MST4 by siRNA or CRISPR-Cas9 significantly promoted HGC-27 cell proliferation (Fig. 5 G and Fig. S4 F). Furthermore, reconstitution of the HGC-27 MST4 KO cells with MST4WT and MST4T178E, but not MST4K53R, attenuated the proliferation of these cells (Fig. 5 H and Fig. S4 G). Together with the observations in Fig. 4 H, these results recapitulated the MST4–pYAPT83 signaling axis in GC cells, suggesting that dysregulation of this axis may promote gastric tumorigenesis.
The MST4–pYAPT83 signaling axis suppresses gastric tumorigenesis in mice
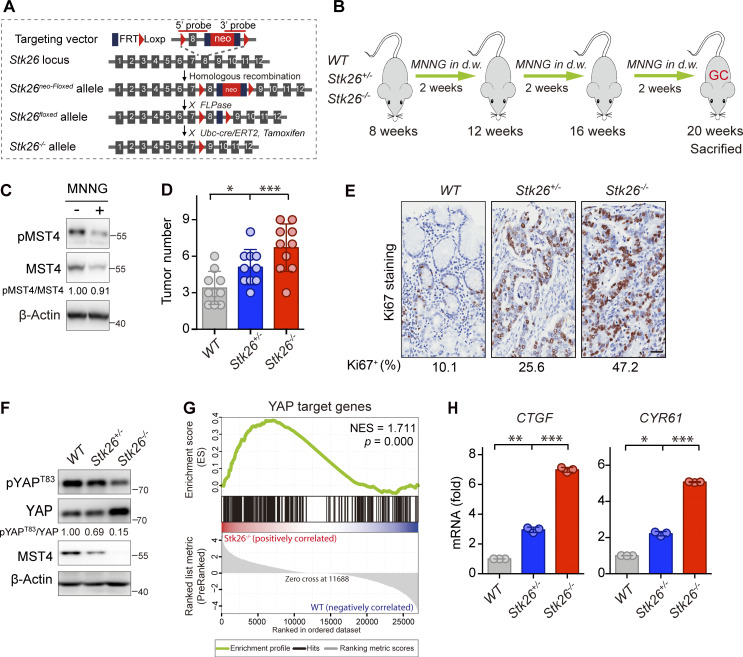
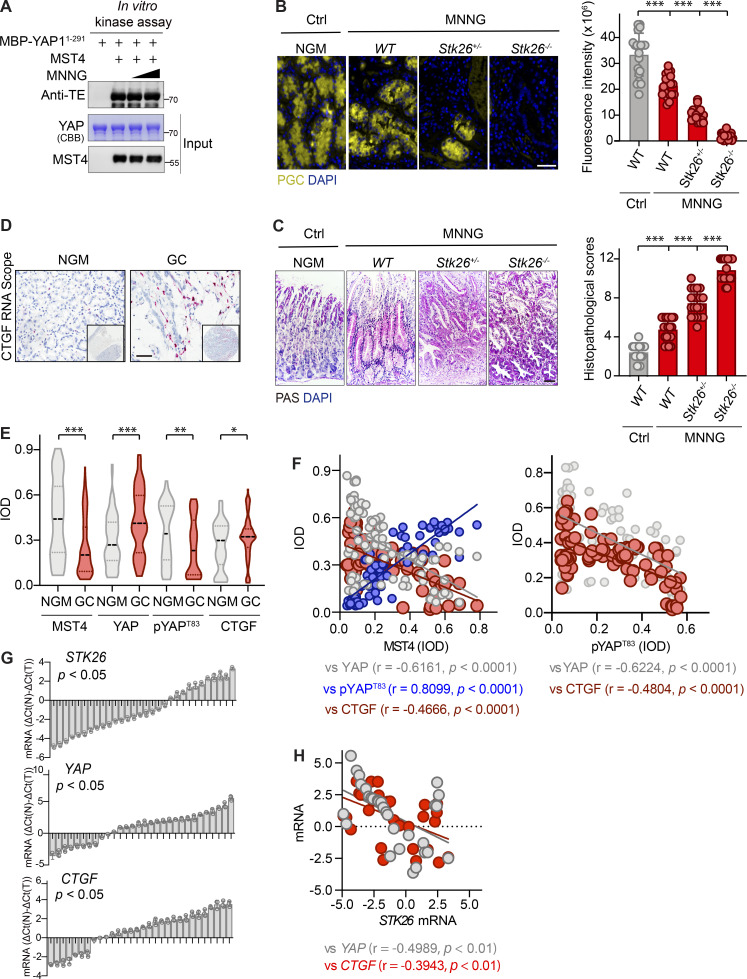
To assess in vivo the effect of the MST4–pYAPT83 signaling axis in gastric tumorigenesis, we next generated whole-body MST4 heterozygous and homozygous mice (Stk26+/− and Stk26−/−) by deletion of exon 8 (Fig. 6 A). For gastric tumor formation, 8-wk-old mice were treated with 100 mg/ml carcinogen N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) using a standard dosage timing regimen (Fig. 6 B). Interestingly, we observed an apparent decrease of MST4 protein level in WT mice treated with MNNG (Fig. 6 C), although in vitro assay showed that MNNG did not directly affect its kinase activity (Fig. S5 A). Supporting the notion that MST4 serves as a tumor suppressor for GC, we found the numbers of palpable tumors were significantly increased in Stk26+/− and Stk26−/− mice (Fig. 6 D). A similar increasing trend was also observed by IHC staining of the proliferation marker Ki67 between WT, Stk26+/−, and Stk26−/− mice (Fig. 6 E). Further histological validation of tumor tissues from each group revealed much lower pepsinogen C (PGC) signals, but increased periodic acid–Schiff (PAS) signals, in Stk26+/− and Stk26−/− mice when compared with WT mice (Fig. S5, B and C). Taken together, these results indicate that deficiency of MST4 promotes gastric tumorigenesis.
Figure 6.
MST4 suppresses gastric tumorigenesis and YAP activation in vivo. (A) Schematic illustration of the generation of Stk26+/− and Stk26−/− mice. (B) An experimental flow chart of the chemical MNNG treatment for induction of gastric tumorigenesis in mice model. 8-wk-old mice (WT, Stk26+/−, and Stk26−/−) were treated with drinking water containing 100 mg/ml MNNG followed a standard “three cycles” timing regimen. A MNNG “cycle” includes 2 wk of MNNG in drinking water (d.w.) followed by 2 wk of regular water. Mice were sacrificed at 20 wk old for subsequent multiple analysis. (C) MNNG treatment reduces MST4 protein expression in vivo. Gastric tissues derived from mice treated with or without MNNG were subjected to Western blotting analysis using indicated antibodies (n = 2, quantification values represent the mean from two repeats). (D) MST4 deficiency promotes GC development. Quantification of palpable gastric tumor numbers in WT, Stk26+/−, and Stk26−/− mice (10 mice/group, *, P < 0.05; ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with WT). (E) IHC staining of the proliferation marker Ki-67 in gastric tissues derived from WT, Stk26+/−, and Stk26−/− mice. Quantifications of Ki-67–positive cells (%) are displayed below (scale bar, 50 µm). (F) Levels of YAP Thr83 phosphorylation were reduced in GC tissue samples in Stk26−/− mice. Western blotting analysis of pYAPT83 and total YAP protein levels in gastric tumor tissues derived from WT, Stk26+/−, and Stk26−/− mice (two mice/group) using indicated antibodies (n = 3, quantification values represent the mean from three repeats). (G) YAP signaling pathway was activated in gastric tumors derived from Stk26−/− mice. GSEA analysis showing a significant positive enrichment of YAP target genes in gastric tumor tissues derived from WT (n = 2) and Stk26−/− (n = 2) mice by RNA-Seq. (H) CTGF and CYR61 mRNA expression were gradually up-regulated in Stk26+/− and Stk26−/− mice. Real-time qPCR analysis of transcriptional levels of YAP target genes CTGF and CYR61 in gastric tumor tissues derived from WT, Stk26+/−, and Stk26−/− mice (two mice/group, n = 3, *, P< 0.05; **, P< 0.01; ***, P< 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with WT). Data are presented as the mean ± SEM. See also Fig. S5.
Figure S5.
MST4 deficiency promotes GC and predicates poor clinical outcomes. (A) MNNG treatment did not affect MST4’s kinase activity in vitro. Purified MBP-YAP1-291 proteins were incubated with purified MST4 protein in the kinase assay buffer supplemented with 1 and 10 µg/μl of MNNG for 30 min at 30°C. Samples were subjected to Coomassie Brilliant Blue staining or Western blotting analysis using indicated antibodies (n = 3). (B and C) Histological validation of the MNNG-induced GC tumors derived from indicated groups. Gastric tissues derived from control (no MNNG treatment) WT mice and MNNG-treated WT, Stk26+/−, and Stk26−/− mice were subjected to IHC analysis using anti-PGC antibody (B, left panel, scale bar, 50 µm), and PAS staining kit (C, left panel, scale bar, 50 µm). Quantification of the fluorescence intensity of PGC signals (B, right panel), and histopathological scores of PAS staining (C, right panel) were performed by ImageJ/Fiji software (20 areas were analyzed for each group, ***, P < 0.001, one-way ANOVA with Dunnett’s post hoc analysis, compared with control WT). (D) Representative images showing the RNA-scope IHC staining of CTGF on tissue microarrays derived from 88 GC patients (scale bar, 100 µm). (E) Stereological analysis of IHC staining data and CTGF RNA-scope data from the GC microarray tissues by measuring IOD (*, P< 0.05; **, P< 0.01; ***, P< 0.001 by paired Student’s t test). (F) Linear correlation analysis of IOD values between MST4 and YAP (left panel, gray dots), MST4 and pYAPT83 (left panel, blue dots), MST4 and CTGF (left panel, red dots), pYAPT83and YAP (right panel, gray dots), and pYAPT83 and CTGF (right panel, red dots). (G) qPCR analysis of mRNA levels of STK26, YAP, and CTGF in 30 pairs of freshly prepared GC samples. Bar value [△Ct(N) − △Ct(T)] represents the difference of mRNA levels between normal tissue and tumor (n = 3, paired Student’s t test). (H) Relative mRNA levels of STK26 were compared with those of YAP (red dots) and CTGF (gray dots) by Pearson’s correlation analysis. Related to Fig. 6 and Fig. 7.
To further verify the relevance of YAP in MST4-regulated gastric tumorigenesis, we next examined YAP signaling in tumor tissues derived from Stk26+/− and Stk26−/− mice. Consistent with our in vitro mechanistic findings, pYAPT83 levels were gradually reduced in Stk26+/− and Stk26−/− mice, and we noted that the total YAP protein levels were significantly increased in an MST4 dose-dependent decrease manner (Fig. 6 F). Subsequent RNA sequencing (RNA-Seq) and gene set enrichment analysis (GSEA) of gastric tumor tissues from the WT and Stk26−/− mice revealed a significant positive enrichment of YAP target gene expression upon MST4 depletion (Fig. 6 G). These observations were further confirmed by quantitative PCR (qPCR) analysis showing that the transcription of CTGF and CYR61 were also gradually elevated upon depletion of MST4 (Fig. 6 H). Collectively, this evidence revealed that MST4 functions as a tumor suppressor in GC, deficiency of which would promote gastric tumorigenesis with diminished Thr83 phosphorylation and hyperactivation of YAP.
The MST4–pYAPT83 axis is negatively correlated to human GC
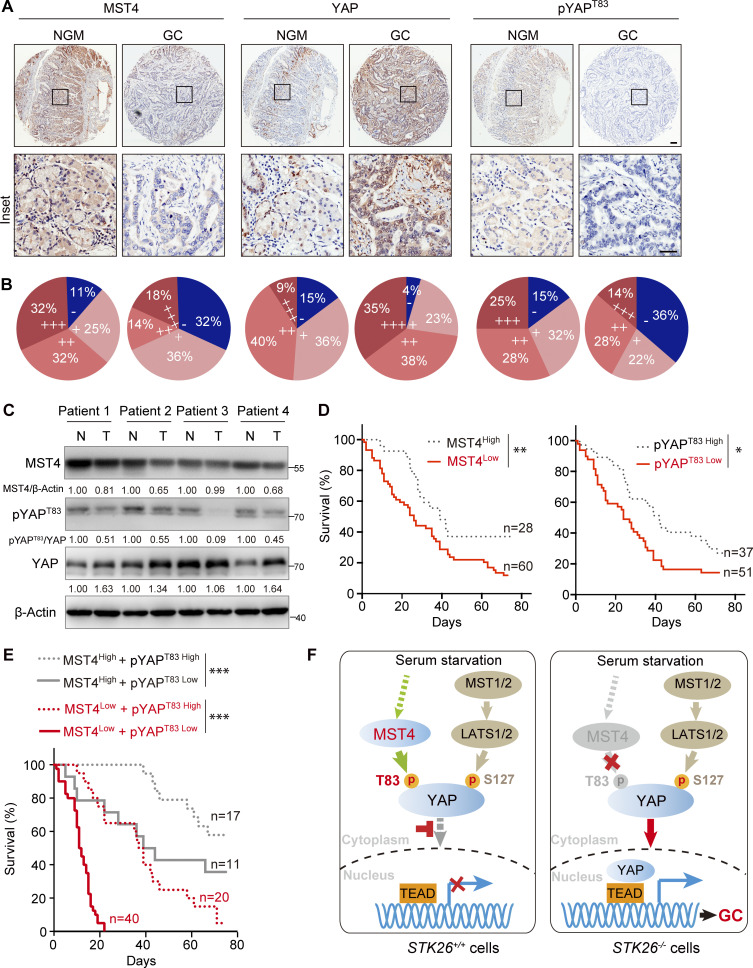
To further validate the pathological role of the MST4–pYAPT83 signaling axis in GC, we next examined the expression levels of MST4, pYAPT83, and YAP in an IHC analysis of a tissue microarray comprising 88 previously described GC specimens (Jiao et al., 2018). Strikingly, the protein levels of MST4, as well as pYAPT83, were observed to be down-regulated in 68.2% (60/88) and 58.0% (51/88) of gastric tumors, respectively (Fig. 7, A and B). Moreover, a significant positive correlation between MST4 and pYAPT83 levels was revealed (P = 0.0207; Table S1). On the contrary, protein levels of total YAP were up-regulated in 72.7% (64/88) of GC patients (Fig. 7, A and B), which showed a significant negative correlation with MST4 protein level (P = 0.0387; Table 1). Nevertheless, there was no statistically significant correlation between pYAPT83 and total YAP protein levels (P = 0.2253; Table S2). We also assessed the correlations between MST4 level, as well as pYAPT83 level, with the clinicopathological features of the 88 GC patients (Table 2 and Table S3). Although no significant correlations between MST4 protein expression and sex (P = 0.6598), age (P = 0.1394), or distant metastasis (P = 0.0837) were observed, we found significant negative correlations between low MST4 level and tumor size (P = 0.0003), lymph node metastasis (P = 0.0001), and tumor stages (P = 0.0246; Table 2). Analysis of pYAPT83 with these features revealed highly similar correlation patterns as with MST4 levels (Table S3).
Figure 7.
Pathological association of MST4–pYAPT83 axis in human GC. (A and B) The MST4–YAPT83 axis protein expression was down-regulated in human GC samples. (A) Representative images showing the IHC staining of MST4, YAP, and pYAPT83 on tissue microarrays derived from 88 GC patients. NGM, normal gastric mucosa (scale bar, 100 µm). (B) Staining protein levels of MST4, YAP, and pYAPT83 in normal and cancerous GC tissue indicating negative (−), weak (+), moderate (++), and strong (+++) expression levels. (C) The expression of MST4–pYAPT83 axis was down-regulated in four-pair freshly prepared GC samples. Western blotting analysis of MST4, YAP, and pYAPT83 protein levels between four-pair of adjacent normal (N) and tumor tissue (T) samples using indicted antibodies (n = 3, quantification values represent the mean from three repeats). (D) Lower MST4 and pYAPT83 expression predicates poor clinical outcomes. Kaplan–Meier survival analysis of patients between high and low levels MST4 (left panel) and pYAPT83 levels (right panel) from the 88 GC tissue microarray, respectively. *, P< 0.05, **, P< 0.01. (E) Kaplan–Meier survival analysis of patients with combined MST4/pYAPT83 at high or low levels from tissue microarray. ***, P< 0.001. (F) Schematic models illustrating both MST4-pYAPT83 and canonical MST1/2–LATS1/2–pYAPS127 axis are required for the inactivation of YAP and targeted gene expression in response to serum starvation (left panel, STK26+/+). Deficiency or down-regulation of the MST4–pYAPT83 axis activates YAP signaling pathway and contributes to the gastric tumorigenesis (right panel, STK26−/−). See also Fig. S5.
Table 1. MST4 are negatively correlated with those of YAP in GC.
| MST4 | Fisher’s exact test | ||||
|---|---|---|---|---|---|
| Low | High | Total | |||
| YAP | Low | 12 | 12 | 24 | 0.0387* |
| High | 48 | 16 | 64 | ||
| Total | 60 | 28 | 88 | ||
Asterisk indicates statistical significance. *, P < 0.05.
Table 2. Clinical significance of MST4 expression in GC.
| Groups | MST4 expression | n | Positive (%) | P value (Fisher’s test) | |||
|---|---|---|---|---|---|---|---|
| − | + | + + | + + + | ||||
| Sex | |||||||
| Male | 19 | 25 | 8 | 13 | 65 | 70.8 | 0.6598 |
| Female | 9 | 7 | 4 | 3 | 23 | 60.9 | |
| Age | |||||||
| <60 yr | 6 | 10 | 3 | 10 | 29 | 79.3 | 0.1394 |
| ≥60 yr | 22 | 22 | 9 | 6 | 59 | 62.7 | |
| Tumor size | |||||||
| pT1 + pT2 | 1 | 1 | 3 | 7 | 12 | 91.7 | 0.0003*** |
| pT3 + pT4 | 27 | 31 | 9 | 9 | 76 | 64.5 | |
| Lymph node metastasis | |||||||
| N0 + N1 | 4 | 8 | 10 | 14 | 36 | 88.9 | 0.0001*** |
| N2 + N3 | 24 | 24 | 2 | 2 | 52 | 53.8 | |
| Distant metastasis | |||||||
| M0 | 25 | 32 | 12 | 16 | 85 | 70.6 | 0.0837 |
| M1 | 3 | 0 | 0 | 0 | 3 | 0.0 | |
| Tumor stage | |||||||
| I + II | 9 | 8 | 8 | 12 | 37 | 75.7 | 0.0246* |
| III + IV | 19 | 24 | 4 | 4 | 51 | 62.7 | |
| Total | 28 | 32 | 12 | 16 | 88 | ||
Asterisks indicate statistical significance. *, P < 0.05; ***, P < 0.001.
In addition to the IHC analysis, a subsequent RNAscope in situ hybridization assay revealed that the mRNA level of CTGF was also increased in GC samples compared with adjacent normal mucosa (Fig. S5 D). We then stereologically quantified the integrated optical density (IOD) of immunostaining results from both IHC and RNAscope. We observed a significant down-regulation of MST4 (P < 0.001) and pYAPT83 (P < 0.01), as well as markedly increased expression of YAP (P < 0.001) and CTGF (P < 0.05), in GC samples compared with normal gastric mucosa (Fig. S5 E). Further linear regression analysis revealed that MST4 was positively correlated with pYAPT83 (P = 0.8099; P < 0.0001), but negatively correlated with YAP (P= −0.6161; P < 0.0001), and CTGF (P = −0.4666; P < 0.0001; Fig. S5 F). Similarly, negative correlation between pYAPT83 and YAP (P= −0.6224; P < 0.0001), as well as between pYAPT83 and CTGF (P= −0.4804; P < 0.0001), were also observed (Fig. S5 F).
Next, we examined the expression of MST4, pYAPT83, YAP, and CTGF in freshly prepared human GC samples. qPCR assays indicated that mRNA levels of STK26 were significantly down-regulated (17/30, P = 0.0301), whereas YAP (16/30, P = 0.0429) and CTGF (14/30, P = 0.0494) were up-regulated in GC samples (Fig. S5 G). Negative correlations between STK26 and YAP (r = −0.4989; P = 0.0041) as well as between STK26 and CTGF (r = −0.3943; P = 0.0065) were observed (Fig. S5 H). Consistently, immunoblotting analysis revealed that the protein levels of both MST4 and pYAPT83 were decreased, whereas the protein levels of total YAP were increased, in gastric tumors as compared with adjacent normal tissues (Fig. 7 C).
Lastly, Kaplan–Meier survival analysis of our microarray data revealed that low expression levels of MST4, as well as those of pYAPT83, indicate poor prognosis of GC patients (Fig. 7 D). Furthermore, patients classified as MST4High/pYAPT83High showed the best survival outcomes, whereas the MST4Low/pYAPT83Low status predicted the shortest survival (Fig. 7 E). Taken together, these results highlight the clinical relevance of the down-regulated MST4–pYAPT83 signaling axis for GC prognostics.
Discussion
The Hippo–YAP signaling pathway is well recognized for its pathological roles in tumorigenesis. Despite the fact that depletion of MST1/2 or LATS1/2 kinases promotes tumorigenesis in mice (Choi et al., 2018; Zhou et al., 2009), deletion or mutation of these genes is rarely identified in cancer, raising the question of why the downstream transcription coactivator YAP is frequently hyperactivated in most cancers. Emerging evidence suggests additional mechanisms exist to regulate YAP activation (Misra and Irvine, 2018). Here, we revealed that the MST4 kinase-mediated Thr83 phosphorylation of YAP represents such an additional mechanism of YAP inactivation. In response to serum starvation, Thr83 and Ser127 are concomitantly yet independently phosphorylated by MST4 and LATS1/2, respectively, to fully inhibit the nuclear translocation and activation of YAP (Fig. 7 F).
The Hippo–YAP pathway can respond to a wide range of stress signals, eventually leading to the manifestation of different states of YAP-dependent gene transcription. Recently, a burst of studies have identified various types of PTMs including ubiquitination, methylation, and O-GlcNAcylation to regulate YAP activation in Hippo-independent manners (Fang et al., 2018; Peng et al., 2017; Yao et al., 2018; Zhang et al., 2017, 2019). Together with these studies, our current work further demonstrates a complicated machinery for fine and tight control of YAP activity under various cellular contexts. Interestingly, some negative regulators of YAP, such as MST4 kinase characterized here, and the reported SET1A (Fang et al., 2018), RUNX3 (Qiao et al., 2016), and OTUD1 (Yao et al., 2018), were found to be down-regulated in different cancer specimens and to be associated with poor prognosis. Perhaps dysregulation of these YAP-related tumor suppressors could explain the hyperactivation of YAP in cancer cells without apparent change of the classic Hippo kinase cascade.
Among the MST family of kinases, MST4, MST3, and STK25 share almost identical three-dimensional structures (Thompson and Sahai, 2015). Intriguingly, during the preparation of this manuscript, STK25 was reported to promote LATS activation and therefore indirectly limit YAP activity (Lim et al., 2019). In that work, a siRNA-mediated depletion strategy was used to screen YAP regulators in response to F-actin disruption. Note that depletion of MST4, as well as MST3, also reduced YAP phosphorylation after dihydrocytochalasin B treatment in their primary screening. Thus, these observations support our current finding of the MST4–YAP signaling axis, and together with our current study, suggest that MST family of kinases can all function, either indirectly or directly, as negative regulators of YAP activation. At this stage, however, the specific physiological/pathological contexts for these MST kinases to exert such function remain unclear, as well as the possible redundancy. In this regard, we did observe functional redundancy between MST4 and MST3 at cellular levels (data not shown). Systematic investigation of MST kinases in vivo in mice models is required to eventually clarify such potential functional redundancy in limiting YAP activation and GC development. Another important aspect to further decipher the complex machinery regulating YAP activation, proteomics-based strategy, could be useful to probe the interaction network of MST kinases in response to different types of stress conditions.
Excessive nuclear translocation of YAP leads to its hyperactivation, which has been commonly observed to be associated with tumorigenesis in different human cancers (Harvey et al., 2013; Zheng and Pan, 2019). However, exactly how YAP translocates into the nucleus remains poorly understand, although it is generally thought that the nuclear import and export machineries are involved in this process. In Drosophila, it is suggested that importin α1 can interact with the N terminus of Yorkie, the YAP homologue, and drive its nuclear import (Wang et al., 2016). Meanwhile, quantitative analysis–based screening revealed mediators of YAP import/export machineries (Ege et al., 2018). PTMs, such as methylation, can also affect YAP nuclear export process (Fang et al., 2018). Consistent with these reports, here we found KPNA2 (importin α1), as well as KPNA1 (importin α5) and KPNA7 (importin α8), interact with YAP and mediate its nuclear import in mammals. Furthermore, we demonstrated that MST4-dediated Thr83 phosphorylation of YAP disrupts its interaction with importin(s), and thereby impairs its nuclear import. However, the detailed molecular mechanisms of the YAP-importin complex formation need to be further investigated, possibly from a structural perspective. In this regard, it is worth mentioning that the molecular requirements of TAZ translocation have been revealed recently (Kofler et al., 2018), showing a RhoA-regulated nuclear localization signal and TEAD-regulated nuclear export signal model of TAZ nucleocytoplasmic shuttling.
Given the strong positive correlation of YAP hyperactivation with poor prognosis of GC, growing efforts are being made to target the Hippo–YAP pathway for treating GC (Calses et al., 2019; Huang et al., 2020; Jiao et al., 2014, 2018; Kang et al., 2011; Tang et al., 2019a; Zanconato et al., 2016; Zhang et al., 2020). We previously observed that YAP was up-regulated in GC, and such up-regulation was positively correlated with tumor stages (Jiao et al., 2014, 2018). Consistently, YAP/TAZ was shown to be activated in LATS1 and LATS2 conditional KO mice to initiate gastric tumorigenesis via up-regulation of the downstream mediator MYC in vivo (Choi et al., 2018). YAP also appears to promote gastric metastasis via modulating the turnover of F- and G-actin dynamics (Qiao et al., 2017). Here, we found YAP was activated in tumor tissues derived from Stk26 KO mice, indicating that the MST4–YAP regulatory axis was essential to restrict YAP activity in vivo. However, currently it is not fully understood how YAP activation contributes to GC development (Zanconato et al., 2019). Characterization of YAP modulation in various specific cellular contexts is of importance to elucidate its biological function in GC development. In this regard, we used STK26 whole-body KO mice during modeling GC. Thus, the specific cell type in which the MST4–YAP signaling axis regulates GC still awaits further investigation. Meanwhile, given the heterogeneity and subtypes of GC (Cancer Genome Atlas Research Network, 2014; Cristescu et al., 2015), it is also intriguing to analyze the MST4–YAP signaling axis in a specific pathological setting in the future.
Our study demonstrated that the MST4–YAP signaling axis functions in parallel with the classic Hippo-YAP pathway. As such, we observed concomitant phosphorylation of YAP at both Thr83 and Ser127; changing the phosphorylation status of one site would indirectly alter the status of another due to the resultant nuclear translocation of YAP. Interestingly, we noticed that the MST4-mediated inhibitory effect on YAP is stronger in the YAPS127A context when compared with that in the YAPWT context both in vitro and in vivo. A possible interpretation is that the MST4–pYAPT83 axis became more prominent, and thus cells may be more sensitive to manipulation of this axis when the canonical Hippo signaling was disrupted (YAPS127A). At this point, the upstream signals of the MST4–YAP signaling axis remain unknown. Membrane proteins including G-protein–coupled receptors have been proposed to relay extracellular signals to the intracellular Hippo kinase cascade (Yu et al., 2012). Besides, the striatin-interacting phosphatase and kinase (STRIPAK) complex was recently revealed to integrate upstream signals to trigger Hippo signaling in cells (Chen et al., 2019; Gil-Ranedo et al., 2019; Tang et al., 2019b). In the future, it would be intriguing to investigate whether these proteins are involved in the upstream regulation of the MST4–YAP signaling, especially in a context of tumorigenesis.
In summary, our work identified an alternative MST4–YAP signaling axis in parallel with the classic Hippo–YAP signaling pathway, shedding new light on the coregulation of YAP activity via both Hippo-dependent and Hippo-independent functions. Importantly, we demonstrate dysregulation of this MST4–YAP axis promotes gastric tumorigenesis and prognosticates bad clinical outcomes of GC patients.
Materials and methods
Cell culture
Human HEK293FT, 293A, and GC cell lines including AGS, BGC-823, MGC-803, and HGC-27 cells were obtained from the cell library of the Chinese Academy of Sciences (Shanghai, China). GSS and GSU gastric cancer cells were purchased from Cell Bank of RIKEN BioResource Center (Tsukuba, Japan). MKN-28 and AZ-521 cells were obtained from Cell Bank of Japanese Collection of Research Bioresources (NIBIOHN). SNU-216 and GES-1 cells were from the Korean Cell Line Bank and Biofeng (Biofeng), respectively. 293A-LATS1/2 DKO and 293A-YAP KO cells were generously provided by F. Yu (Fudan University, Shanghai, China). All the GC cell lines were maintained in RPMI1640 medium (Invitrogen). HEK293FT and 293A cell lines, including the KO cell lines derived from these cell lines, were cultured in DMEM (Invitrogen). Cells were grown in culture medium supplemented with 10% FBS, 100 µg/ml penicillin, and 100 µg/ml streptomycin in 5% CO2, at 37°C with saturated humidity.
Plasmids
Bacteria-mediated YAP protein expression constructs, including the truncated and point mutations of YAP, were cloned into a modified pET-28a vector with an N-terminal MBP tag. Full-length MST4, however, was engineered into a pET-28a vector with a C-terminal uncleavable His6 tag. TEAD4 YAP-binding domain (TEAD4-YBD, residues 217–433 aa) was cloned into a modified pET-28a vector with an N-terminal tobbaco etch virus protease cleavable His6-SUMO tag. Mammalian expression plasmids, including the Flag-tagged MST1, MST2, MST3, MST4, STK25, and the YAP point mutants, were sub-cloned into a modified pCDNA-3.1 vector with an N-terminal 3xFlag tag. The SFB (S-tag, FLAG epitope tag, and streptavidin-binding peptide tag)-tagged KPNA1–7 were generously provided by M.S.Y. Huen (University of Hong Kong), and were transferred into a pCDNA-3.1 vector with an N-terminal hemagglutinin (HA) tag. For lentiviral-mediated overexpression constructs, the genes-of-interest were constructed into a pCDH1-MCS-CoGFP vector. All mutants were generated by site-directed mutagenesis and were verified by Sanger sequencing.
Transfection, lentivirus packaging, and infection
For RNAi-mediated depletion experiments, cells were transfected with two rounds of either nontargeting control or targeting siRNAs (Genepharma) using Lipofectamine RNAiMAX Transfection Reagent (13778150, Invitrogen) according to the manufacturer’s instructions. Sequences for targeting MST4 were 5′-GCCUGAUCCAAAGAAAGUATT-3′ (siMST4_#1) and 5′-AUUGUUACAAUAUAGGCACCATT-3′ (siMST4_#2). For plasmid-based transfection experiments, constructs were first incubated with polyethylenimine (23966–1, Polysciences) at a ratio of 1:3 for 15 min at room temperature before dropwise them to the culture medium. Lentiviral particles were produced by transient cotransfection of the lentiviral-based expression constructs together with packaging plasmids psPAX2 and pMD2.G at a ratio of 4:3:1 into HEK293FT cells. 48 h after transfection, the supernatants were filtered (0.45 µm) and were applied to recipient cell lines in the presence of 8 µg/ml polybrene (H9268, Sigma-Aldrich).
Generation of KO cells by use of CRISPR-Cas9 method
All the KO cell lines used in this study were generated using the CRISPR-Cas9 gene targeting approach (Sanjana et al., 2014). Briefly, sequences of two guide RNAs (gRNAs) targeting STK26 gene exon1 and exon2 were obtained from human GeCKO library (https://www.genscript.com/CRISPR-gRNA-library.html), and were sub-cloned into the pLentiCRISPRv2 vector (52961, Addgene). The sequences of the two gRNAs were as follows: MST4 gRNA1#: 5′-CTTGGACAGCCACCGGCGAG-3′; MST4 gRNA2#: 5′-CCAGTGCTGAACCACCGCCC-3′. Lentiviral particles were generated by cotransfection of lentiCRISPRv2-based gRNAs, PsPaX2, and pMD2.G plasmids at a ratio of 4:3:1 in 293FT cells. For generation of KOs, cells were infected with lentiviral particles harboring indicated gRNAs twice at 24-h intervals. KO cells were selected by 1 µg/ml puromycin for 1 wk before validation by Western blotting using MST4 antibody.
Immunoprecipitation and Western blotting
For whole-cell extracts, cells were lysed with 20 mM Tris·HCl, pH 8.0, 100 mM NaCl, 0.5% NP-40, and 1 mM EDTA (NETN buffer) supplemented with Micrococcal Nuclease (M0247S, NEB) on ice for 15 min. Cell lysates were boiled with 5× SDS loading buffer. Proteins were separated by SDS-PAGE, transferred to polyvinylidene fluoride membranes, and blotted with indicated antibodies. The following antibodies were used for immunoblotting: YAP (sc-101199, Santa Cruz, 1:500), MST4 (sc-376649, Santa Cruz, 1:1,000), Lamin A (sc-518013, Santa Cruz, 1:1,000), α-tubulin (sc-17787, Santa Cruz, 1:2,000), c-Myc (sc-40, Santa Cruz, 1:2,000), Flag (F3165, Sigma-Aldrich, 1:5,000), β-actin (A2228, Sigma-Aldrich, 1:5,000), pYAPS127 (13008, Cell Signaling Technology, 1:500), HA (3724, Cell Signaling Technology, 1:1,000), LATS1 (9153, Cell Signaling Technology, 1:500), TEAD4 (ab58310, Abcam, 1:1,000), MST1 (ab264127, Abcam, 1:1,000), MST2 (ab52641, Abcam, 1:1,000), MST4 (ab52491, Abcam, 1:1,000), anti-thiophosphate ester specific antibody (ab92570, Abcam, 1:4,000), and anti-MST4 + MST3 + STK25 (phospho T174 + T178 + T190) antibody (ab76579, Abcam, 1:500). The polyclonal antibody recognizing the phosphorylated YAP Thr83 residue (pYAPT83, 1:1,000) was generated by Shanghai Immune Biotech Co., Ltd. via immunizing rabbits with a YAP peptide spanning the phosphorylated Thr83 residue (“KTANVPQpTVPMRLRK”). The secondary HRP antibodies including goat anti-rabbit (31460, 1:4,000), and goat anti-mouse (31430, 1:4,000) were purchased from Thermo Fisher Scientific. Western blotting images were captured by the Mini Chemiluminescent Imaging and Analysis System (Beijing Sage Creation Science Co., Ltd.).
For Co-IP experiments, cells were lysed with NETN buffer for 15 min on ice, followed by centrifugation at 12,000 rpm for 10 min at 4°C. Supernatants were then transferred into new Eppendorf tubes and incubated with either Flag beads (L00425, GenScript) for 4 h or protein A/G plus agarose beads (sc-2003, Santa Cruz) conjugated with indicated antibodies overnight at 4°C with rotation. Beads were subsequently washed three times with NETN buffer and boiled with 1× SDS loading buffer. To avoid the noise of light/heavy chains, the secondary antibodies used for the immunoprecipitation assays including mouse anti-rabbit IgG light chain (A25022, 1:5,000) and goat anti-mouse IgG light chain (A25012, 1:5,000) were obtained from Abbkine.
Immunofluorescence staining assay (IFA)
Cells grown on coverslips were fixed with 3% paraformaldehyde for 15 min at room temperature followed by permeabilization in 0.5% Triton X-100 solution for 30 s. After blocking with 3% milk at room temperature for 20 min, cells were incubated sequentially with primary antibodies and secondary fluorophore-conjugated antibodies. DAPI (Sigma-Aldrich, 1:10,000) was used to stain nuclear DNA. The following antibodies were used for IFA assays: YAP (sc-101199, Santa Cruz, 1:100), Flag (F3165, Sigma-Aldrich, 1:1,000), and Alexa Fluor–conjugated secondary antibodies (Thermo Fisher Scientific, 1:400). All the images were captured using a 60× oil immersion lens on an Olympus FV1200 fluorescence microscope and were further processed by ImageJ software.
Luciferase reporter assay
HEK293FT or 293A cells were seeded in triplicates for each treatment in 48-well plates. 24 h later, cells were cotransfected with 5 × upstream activator sequence luciferase reporter, TEAD-luciferase reporter, and Renilla luciferase reporter, together with indicated plasmids. Cells were lysed in passive lysis buffer, and luciferase activities were detected using the Dual-luciferase Assay System (E1910, Promega) 36 h after transfection. Firefly luciferase activity levels were normalized to levels of Renilla luciferase activity in each well.
RNA extraction, reverse transcription, and qPCR
Total RNA from cultured cells were extracted using the RNA isolater Total RNA Extraction Reagent (R401-01-AA, Vazyme), and cDNA was reversed from RNA using the HiScript II Q Select RT SuperMix for qPCR (R223-01, Vazyme) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using SYBR Green reagents in a StepOnePlus system (Applied Biosystems). Primers used in this study were as follows: hCTGF forward: 5′-AAAAGTGCATCCGTACTCCCA-3′, hCTGF reverse: 5′-CCGTCGGTACATACTCCACAG-3′; hCYR61 forward: 5′-GGTCAAAGTTACCGGGCAGT-3′, hCYR61 reverse: 5′-GGAGGCATCGAATCCCAGC-3′; hGAPDH forward: 5′-GGCATCCTGGGCTACACTGA-3′, hGAPDH reverse: 5′-GAGTGGGTGTCGCTGTTGAA-3′; mCTGF: forward: 5′-GGACACCTAAAATCGCCAAGC-3′, mCTGF reverse: 5′-ACTTAGCCCTGTATGTCTTCACA-3′; mCYR61: forward: 5′-TAAGGTCTGCGCTAAACAACTC-3′, mCYR61 reverse: 5′-CAGATCCCTTTCAGAGCGGT-3′; mGAPDH: forward: 5′-AATGGATTTGGACGCATTGGT-3′, mGAPDH reverse, 5′-TTTGCACTGGTACGTGTTGAT-3′.
Cell proliferation assay
Cell proliferation assays were performed based on the protocol of the CellTiter-Lumi Plus Luminescent Cell viability Assay Kit (C0068M, Beyotime). Briefly, cells were seeded in triplicates into 96-well plates at a density of 3,000/well in 100 µl medium and were cultured for the indicated time. Cells were lysed by addition of an equal volume of CellTiter-Lumi Plus Reagent to the cultured medium for 2 min at room temperature on an orbital shaker. After that, around 130 µl supernatants were transferred into a new 96-well plate, and the luminescent signals were measured in a BioTek Synergy NEO microplate reader.
Protein purification
Proteins were expressed in Escherichia coli BL21 (DE3) CodonPlus cells. Protein expression was induced by 0.4 mM isopropyl β-D-thiogalactopyranoside in Terrific Broth medium, and cells were cultured for an additional 16 h at 18°C. Cells were then harvested by centrifugation and were resuspended in lysis buffer (20 mM Hepes, pH 7.5, 500 mM NaCl, 10% glycerol, and 1 mM DTT) for MBP-tagged proteins and lysis buffer plus 10 mM imidazole for His6- and His6-SUMO–tagged proteins. Cells were lysed by a high-pressure homogenizer at 800 bar, and cell debris was removed by centrifugation at 16,000 rpm for 40 min. For His6- and His6-SUMO–tagged proteins, the soluble fraction was loaded to Ni Sepharose preequilibrated beads with lysis buffer. The Ni Sepharose beads were washed sequentially with lysis buffer plus 20 mM imidazole, lysis buffer plus 40 mM imidazole, and eluted with lysis buffer plus 400 mM imidazole. For MBP-tagged proteins, the soluble fraction was loaded to Amylose Resin. The Amylose Resin was washed with lysis buffer and eluted with lysis buffer plus 20 mM maltose. Then protein samples were concentrated and loaded to a HiLoad 16/60 Superdex 75 or 200 pg column using 20 mM Hepes, pH 7.5, 100 mM NaCl, and 1 mM DTT as running buffer.
MBP pull-down assay
MBP-fused protein coupled on Amylose Resin (E8021L, NEB) were incubated with indicated prey proteins at 4°C for 3 h in buffers containing 20 mM Hepes, pH 7.5, 100 mM NaCl, and 1 mM DTT. After three washes, the proteins were eluted with the same buffer supplemented with 20 mM maltose. The input and output samples were separated by SDS-PAGE and detected by Coomassie Brilliant Blue staining or Western blotting using indicated antibodies.
In vitro kinase assays
For radioactive kinase assays, purified MST4 proteins were first incubated with ATP in kinase assay buffers (50 mM Tris, pH 7.5, 5 mM ATP, 0.1 mM EGTA, and 10 mM magnesium acetate) to fully activate kinase activity via auto-phosphorylation at 30°C for 30 min. Then, the auto-phosphorylated MST4 were further incubated with MBP-tagged YAP proteins in the kinase assay buffer supplemented with 0.1 mM [γ32P]-ATP at 30°C for another 30 min. Samples were separated by SDS-PAGE and analyzed by autoradiography.
ATP analogue ATPγS-based kinase assay was performed mainly based on the Kinase Reaction and Alkylation Protocol (Abcam) as described previously (Allen et al., 2007). Briefly, purified MST4 and MBP-tagged YAP proteins were coincubated in the reaction buffer containing 20 mM Hepes, pH 7.5, 100 mM NaCl, 1 mM ATPγS (ab138911, Abcam), and 10 mM MgCl2 for 30 min at 30°C. Reactions were followed by alkylation for 1 h at room temperature by addition of 2.5 mM p-nitrobenzyl mesylate (ab138910) to introduce thiophosphate esters on substrates. Samples were subjected for Western blotting analysis, and the phosphorylated proteins were assessed using the thiophosphate ester specific antibody (ab92570, Abcam).
For kinase assay using mammalian expressed proteins as substrates, YAP proteins (WT and mutants) were immunoprecipitated from HEK293FT cells transiently transfected with indicated constructs for 48 h using Flag beads. Proteins were incubated with purified MST4 proteins in buffers containing ATP for 30 min at 30°C with rotation. Samples were boiled with 2× SDS loading buffer and analyzed by Western blotting using the indicated antibodies.
MS
MS analysis was performed in the Mass Spectrometry Facility of the National Facility for Protein Science in Shanghai, Shanghai Science Research Center, Chinese Academy of Sciences (Shanghai, China). Briefly, 0.1 µg purified MST4 and 40 µg YAP (1–150 aa) proteins were incubated to induce phosphorylation in the buffer containing 50 mM Tris pH 7.5, 1 mM ATP, 10 mM MgAc2, and 0.1 mM EGTA for 30 min at 30°C. Proteins were precipitated with TCA solution (6.1 N). The pellet was subsequently dissolved in 8 M urea and 100 mM Tris, pH 8.5. Then, tris(2-carboxyethyl)phosphine (final concentration, 5 mM, Thermo Fisher Scientific) and iodoacetamide (final concentration, 10 mM, Sigma-Aldrich) were added for reduction and alkylation. The protein mixture was digested with trypsin (Promega). The reaction was stopped by formic acid, and the peptide mixture was desalted by monospin C18 column (Pierce). The peptide mixture was loaded onto a homemade 15-cm-long pulled-tip analytical column (Aqua, C18, 100 µm inner diameter × 360 µm outer dimension, 3 µm particle size, 125 Å pore diameter, Phenomenex) connected to an Easy-nLC 1000 nano HPLC (Thermo Fisher Scientific) for MS analysis. Survey full-scan MS spectra (from m/z 300–1,800) were acquired in the Orbitrap analyzer with resolution r = 70,000 at m/z 400. The top 20 MS/MS events were sequentially generated selected from the full MS spectrum at a 27% normalized collision energy. Acquired MS/MS data were analyzed against a Uniprot database by Sequest algorithm integrated in the Integrated Proteomics Pipeline software. Trypsin was defined as cleavage enzyme. Cysteine alkylation by iodoacetamide was specified as fixed modification with mass shift 57.02146. Serine, threonine, tyrosine phosphorylation was set as variable modification with mass shift 79.9663. A decoy database containing the reversed sequences of all the proteins was appended to the target database to accurately estimate peptide probabilities and false discovery rate, and the false discovery rate was set at 0.01.
Xenograft tumor formation assay
Healthy male BALB/c A-nu/nu mice (3–4 wk) were obtained from the Shanghai Experimental Animal Center and maintained in pathogen-free conditions in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Shanghai Institute of Biochemistry and Cell Biology (SIBCB; Shanghai, China). The approval ID for the use of animals was no. SIBCB-NAF-14-004-S329-023, issued by the Animal Core Facility of SIBCB.
For the tumor formation assay, mice were first injected with 5 × 106 HGC-27 cells/per mouse into the flank of the mice to induce tumor formation for 7 d. Once the tumor volume reached around 100 mm3, lentiviral particles harboring indicated YAP mutations (multiplicity of infection = 30) were further injected intratumorally into the nude mice on days 7, 9, 11, and 13, respectively. Tumor length and width were measured every 2 d to calculate tumor volume (= width2 × length × 0.523). 3 wk later, mice were scheduled for euthanasia before harvesting and measuring indicated tumor weight. Tumors were further subjected to fixation in 4% paraformaldehyde, followed by either examination of GFP signals in tumors to validate the infection efficiency, or by IHC staining with Ki67 antibodies (ab15580, Abcam, 1:100) to quantify the percentage of proliferating cells.
Generation of Stk26+/− and Stk26−/− mice and MNNG-induced GC mice model
Stk26Floxed mice were generated using Cre/LoxP recombination and the gene targeting and screening strategies outlined in Fig. 6 A. The experimental procedures used for the culture, transfection, and selection of E14K ES cells (129/Ola) have been previously described (Hao and Rajewsky, 2001). Chimeric mice were crossed to Flpe-deleter mice (Rodríguez et al., 2000) to generate Stk26Floxed mice. Ubc-cre/ERT2 mice were from Shanghai Model Organisms. Stk26Floxed mice were crossed with Ubc-cre/ERT2 mice to generate tamoxifen-inducible specific MST4 KO (Stk26−/−) mice. Mice were genotyped by PCR analysis of isolated tail DNA using the following primers: forward: 5′-TGAGCAGCAATAAGGAAGTATG-3′; and reverse: 5′-CTGGGCAGCCCTTGGACAATCC-3′. Cre activity was induced by intraperitoneal injection with tamoxifen suspended in soybean oil (75 mg/kg, Sigma-Aldrich) every 2 d. At 8 wk of age, mice were treated with 100 mg/ml MNNG in drinking water using a standard dosage timing regimen as shown in Fig. 6 B. Mice were sacrificed at 20 wk old and processed for subsequent tumor quantification, IHC staining, Western blotting, and RNA-Seq analysis.
RNA-Seq and GSEA analysis
Total RNA was extracted from gastric tumors derived from WT (n = 2) and STK26−/− (n = 2) mice using the Total RNA Extraction Reagent (R401-01-AA, Vazyme). RNA quality was assessed on a 2100 expert Bioanalyzer and sent for library preparation and sequencing on the Illumina Hisseq2000 platform by BGI genomics. GSEA software was used to characterize the differences in transcriptome profiles of genes from the STK26−/− mice compared with WT mice in specific signatures as previously reported (Zanconato et al., 2015).
In situ hybridization
mRNA expression was determined using in situ hybridization following the RNA-scope procedure (ACD). Briefly, tissue was pretreated with heat and protease before hybridization of the target probe CTGF (ACD). Preamplifier, amplifier, and alkaline phosphatase–labeled oligos were sequentially hybridized followed by the application of a chromogenic substrate to produce red punctate dots. Tissue was counter-stained with Mayer’s hematoxylin.
Tissue microarray and IHC staining
All samples collected and used in this study were derived from patients who signed an informed consent that was approved by the Ethics Committee of Taizhou Hospital of Zhejiang province. In total, tumor and adjecent normal gastric samples from 88 GC patients were collected, and tumor and normal tissue microarray sections were prepared by Shanghai Outdo Biotech Co. Ltd. For IHC, tissue microarray sections were incubated with anti-MST4 (1:100 dilution), anti-YAP antibody (1:100 dilution), or anti-pYAPT83 antibody (1:50 dilution). IHC stains were scored by two independent pathologists who were blinded to the clinical characteristics of the patients. The scoring system was based on the intensity and extent of staining: staining intensity was classified as 0 (negative), 1 (weak), 2 (moderate), or 3 (strong); staining extent was dependent on the percentage of positive cells (examined in 200 cells) and was classified into 0 (<5%), 1 (5–25%), 2 (26–50%), 3 (51–75%), or 4 (>75%). According to the staining intensity and staining extent scores, the IHC result was classified as 0–1, negative (–); 2–4, weakly positive (+); 5–8, moderately positive (++); and 9–12, strongly positive (+++).
The IOD of the immunostaining was also measured by using Image-Pro Plus Version 6.2 software (Media Cybernetics Inc.), and the cutoff value for the definition of subgroups was the median IOD. The IOD greater than median was considered as the high expression group, and the IOD median or less was considered as the low expression group.
Statistical analysis
Both cellular and animal studies tended to be underpowered. Estimation of sample size for planned comparisons of two independent means using a two-tailed test were undertaken using GraphPad Prism 8.0 statistical software. Data are expressed as mean ± SEM for continuous variables and as frequencies and proportions for categorical variables. Continuous data were compared using Student’s t tests (comparing two variables) or one-way ANOVA analysis (comparing multiple variables). For correlation, the Spearman rank correlation was used for continuous variables. Survival curves were calculated according to the Kaplan–Meier method; survival analysis was performed using the log-rank test. P < 0.05 was considered to indicate a significant difference. At least two biological replicates were used throughout the study.
Data availability
RNA sequencing raw data have been deposited in the Gene Expression Omnibus database under the accession no. GSE142644.
Online supplemental material
Fig. S1 shows the negative regulation of MST4 on YAP activation and the requirement of its kinase activity. Fig. S2 shows the identification and evaluation of the key residues required for MST4-mediated YAP inactivation. Fig. S3 shows the concomitant MST4-pYAPT83 and LATS1/2–pYAPS127 axis in response to stress both in vitro and in vivo. Fig. S4 shows the recapitulation of MST4-mediated YAP inactivation in GC cells. Fig. S5 shows the MST4–YAP axis is required for gastric tumorigenesis in vivo, and its dysregulation predicts poor prognosis in human GC samples. Table S1 shows the positive correlation between MST4 and pYAPT83 levels in the 88 GC patients’ tissue microarray. Table S2 shows the correlation of protein levels between YAP and pYAPT83 in the tissue microarray. Table S3 shows the correlations between pYAPT83 with the clinicopathological features of the 88 GC patients.
Supplementary Material
lists protein levels of MST4 and pYAP (T83) are positively correlated in GC.
describes the correlation of protein levels of total YAP and pYAP(T83) in GC.
describes the clinical significance of pYAP (T83) expression in GC.
Acknowledgments
We thank the staff, Ping Wu, and Chao Peng from the Mass Spectrometry Facility of the National Facility for Protein Science in Shanghai for MS data processing and analysis. We also thank the staff of the Core Facility for Cell Biology of SIBCB for technical support.
This project was supported by the National Key R&D Program of China (2017YFA0504504), Shanghai Pujiang Program (19PJ1408300), National Natural Science Foundation of China (81902806, 81725014, 31930026, 81822035, 81773212, 81972876, 81902389, 81802399, 81927801, and 81725008), ‘‘Strategic Priority Research Program’’ (XDB19020202, XDA12010315), Fundamental Research Funds for the Central Universities (22120190021, 22120190213, and 22120190137), and Shanghai Municipal Health Commission (2019LJ21).
Author contributions: Z. Zhou, S. Jiao, and L. An designed research; L. An, M. Chen, Y. Tang, Z. Cao, and J. Guan performed most cellular and in vitro experiments; P. Nie conducted the in vivo xenograft tumor formation assay; S. Jiao carried out the in vivo GC mouse model analysis and the clinical tissue analysis with input from C. Hou and H. Zhang; L. An, S. Jiao, W. Wang, and Y. Zhao analyzed the data; Z. Zhou, S. Jiao, L. An, M. Chen, and P. Nie wrote the manuscript; and Z. Zhou, S. Jiao, and H. Xu supervised the project.
References
- Allen, J.J., Li M., Brinkworth C.S., Paulson J.L., Wang D., Hübner A., Chou W.H., Davis R.J., Burlingame A.L., Messing R.O., et al. 2007. A semisynthetic epitope for kinase substrates. Nat. Methods. 4:511–516. 10.1038/nmeth1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, N.C., Doetsch P.W., and Corbett A.H.. 2015. Mechanisms Regulating Protein Localization. Traffic. 16:1039–1061. 10.1111/tra.12310 [DOI] [PubMed] [Google Scholar]
- Calses, P.C., Crawford J.J., Lill J.R., and Dey A.. 2019. Hippo Pathway in Cancer: Aberrant Regulation and Therapeutic Opportunities. Trends Cancer. 5:297–307. 10.1016/j.trecan.2019.04.001 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network . 2014. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 513:202–209. 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Chan S.W., Zhang X., Walsh M., Lim C.J., Hong W., and Song H.. 2010. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 24:290–300. 10.1101/gad.1865310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Sun Y., Wei Y., Zhang P., Rezaeian A.H., Teruya-Feldstein J., Gupta S., Liang H., Lin H.K., Hung M.C., and Ma L.. 2012. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat. Med. 18:1511–1517. 10.1038/nm.2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Zhang H., Shi Z., Li Y., Zhang X., Gao Z., Zhou L., Ma J., Xu Q., Guan J., et al. 2018. The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer. J. Biol. Chem. 293:14455–14469. 10.1074/jbc.RA118.003279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R., Xie R., Meng Z., Ma S., and Guan K.L.. 2019. STRIPAK integrates upstream signals to initiate the Hippo kinase cascade. Nat. Cell Biol. 21:1565–1577. 10.1038/s41556-019-0426-y [DOI] [PubMed] [Google Scholar]
- Cho, Y.S., Li S., Wang X., Zhu J., Zhuo S., Han Y., Yue T., Yang Y., and Jiang J.. 2020. CDK7 regulates organ size and tumor growth by safeguarding the Hippo pathway effector Yki/Yap/Taz in the nucleus. Genes Dev. 34:53–71. 10.1101/gad.333146.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W., Kim J., Park J., Lee D.H., Hwang D., Kim J.H., Ashktorab H., Smoot D., Kim S.Y., Choi C., et al. 2018. YAP/TAZ Initiates Gastric Tumorigenesis via Upregulation of MYC. Cancer Res. 78:3306–3320. [DOI] [PubMed] [Google Scholar]
- Cottini, F., Hideshima T., Xu C., Sattler M., Dori M., Agnelli L., ten Hacken E., Bertilaccio M.T., Antonini E., Neri A., et al. 2014. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat. Med. 20:599–606. 10.1038/nm.3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristescu, R., Lee J., Nebozhyn M., Kim K.M., Ting J.C., Wong S.S., Liu J., Yue Y.G., Wang J., Yu K., et al. 2015. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 21:449–456. 10.1038/nm.3850 [DOI] [PubMed] [Google Scholar]
- Ege, N., Dowbaj A.M., Jiang M., Howell M., Hooper S., Foster C., Jenkins R.P., and Sahai E.. 2018. Quantitative Analysis Reveals that Actin and Src-Family Kinases Regulate Nuclear YAP1 and Its Export. Cell Syst. 6:692–708.e13. 10.1016/j.cels.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, L., Teng H., Wang Y., Liao G., Weng L., Li Y., Wang X., Jin J., Jiao C., Chen L., et al. 2018. SET1A-Mediated Mono-Methylation at K342 Regulates YAP Activation by Blocking Its Nuclear Export and Promotes Tumorigenesis. Cancer Cell. 34:103–118.e9. 10.1016/j.ccell.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Gil-Ranedo, J., Gonzaga E., Jaworek K.J., Berger C., Bossing T., and Barros C.S.. 2019. STRIPAK Members Orchestrate Hippo and Insulin Receptor Signaling to Promote Neural Stem Cell Reactivation. Cell Rep. 27:2921–2933.e5. 10.1016/j.celrep.2019.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Z., and Rajewsky K.. 2001. Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J. Exp. Med. 194:1151–1164. 10.1084/jem.194.8.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, K.F., Zhang X., and Thomas D.M.. 2013. The Hippo pathway and human cancer. Nat. Rev. Cancer. 13:246–257. 10.1038/nrc3458 [DOI] [PubMed] [Google Scholar]
- Hong, A.W., Meng Z., Yuan H.X., Plouffe S.W., Moon S., Kim W., Jho E.H., and Guan K.L.. 2017. Osmotic stress-induced phosphorylation by NLK at Ser128 activates YAP. EMBO Rep. 18:72–86. 10.15252/embr.201642681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J.M., Nagatomo I., Suzuki E., Mizuno T., Kumagai T., Berezov A., Zhang H., Karlan B., Greene M.I., and Wang Q.. 2013. YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene. 32:2220–2229. 10.1038/onc.2012.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T., Kim C.K., Alvarez A.A., Pangeni R.P., Wan X., Song X., Shi T., Yang Y., Sastry N., Horbinski C.M., et al. 2017. MST4 Phosphorylation of ATG4B Regulates Autophagic Activity, Tumorigenicity, and Radioresistance in Glioblastoma. Cancer Cell. 32:840–855.e8. 10.1016/j.ccell.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Yuan W., Lai C., Zhong S., Yang C., Wang R., Mao L., Chen Z., and Chen Z.. 2020. EphA2-to-YAP pathway drives gastric cancer growth and therapy resistance. Int. J. Cancer. 146:1937–1949. 10.1002/ijc.32609 [DOI] [PubMed] [Google Scholar]
- Jiang, H., Wang W., Zhang Y., Yao W.W., Jiang J., Qin B., Yao W.Y., Liu F., Wu H., Ward T.L., et al. 2015. Cell Polarity Kinase MST4 Cooperates with cAMP-dependent Kinase to Orchestrate Histamine-stimulated Acid Secretion in Gastric Parietal Cells. J. Biol. Chem. 290:28272–28285. 10.1074/jbc.M115.668855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, S., Wang H., Shi Z., Dong A., Zhang W., Song X., He F., Wang Y., Zhang Z., Wang W., et al. 2014. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 25:166–180. 10.1016/j.ccr.2014.01.010 [DOI] [PubMed] [Google Scholar]
- Jiao, S., Zhang Z., Li C., Huang M., Shi Z., Wang Y., Song X., Liu H., Li C., Chen M., et al. 2015. The kinase MST4 limits inflammatory responses through direct phosphorylation of the adaptor TRAF6. Nat. Immunol. 16:246–257. 10.1038/ni.3097 [DOI] [PubMed] [Google Scholar]
- Jiao, S., Guan J., Chen M., Wang W., Li C., Wang Y., Cheng Y., and Zhou Z.. 2018. Targeting IRF3 as a YAP agonist therapy against gastric cancer. J. Exp. Med. 215:699–718. 10.1084/jem.20171116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, W., Tong J.H., Chan A.W., Lee T.L., Lung R.W., Leung P.P., So K.K., Wu K., Fan D., Yu J., et al. 2011. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin. Cancer Res. 17:2130–2139. 10.1158/1078-0432.CCR-10-2467 [DOI] [PubMed] [Google Scholar]
- Kofler, M., Speight P., Little D., Di Ciano-Oliveira C., Szászi K., and Kapus A.. 2018. Mediated nuclear import and export of TAZ and the underlying molecular requirements. Nat. Commun. 9:4966. 10.1038/s41467-018-07450-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz, L.M., Liu-Chittenden Y., Yin F., Zheng Y., Yu J., Huang B., Chen Q., Wu S., and Pan D.. 2013. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell. 25:388–401. 10.1016/j.devcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Zhao B., Wang P., Chen F., Dong Z., Yang H., Guan K.L., and Xu Y.. 2010. Structural insights into the YAP and TEAD complex. Genes Dev. 24:235–240. 10.1101/gad.1865810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S., Hermance N., Mudianto T., Mustaly H.M., Mauricio I.P.M., Vittoria M.A., Quinton R.J., Howell B.W., Cornils H., Manning A.L., and Ganem N.J.. 2019. Identification of the kinase STK25 as an upstream activator of LATS signaling. Nat. Commun. 10:1547. 10.1038/s41467-019-09597-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K.C., Park H.W., and Guan K.L.. 2018. Deregulation and Therapeutic Potential of the Hippo Pathway in Cancer. Annu. Rev. Cancer Biol. 2:59–79. 10.1146/annurev-cancerbio-030617-050202 [DOI] [Google Scholar]
- Liu, X., Yang N., Figel S.A., Wilson K.E., Morrison C.D., Gelman I.H., and Zhang J.. 2013. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene. 32:1266–1273. 10.1038/onc.2012.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen, C.D., Hooper S., Tozluoglu M., Bruckbauer A., Fletcher G., Erler J.T., Bates P.A., Thompson B., and Sahai E.. 2015. STRIPAK components determine mode of cancer cell migration and metastasis. Nat. Cell Biol. 17:68–80. 10.1038/ncb3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C.J., Lou H.J., Simpson C., van de Kooij B., Ha B.H., Fisher O.S., Pirman N.L., Boggon T.J., Rinehart J., Yaffe M.B., et al. 2019. Comprehensive profiling of the STE20 kinase family defines features essential for selective substrate targeting and signaling output. PLoS Biol. 17:e2006540. 10.1371/journal.pbio.2006540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, J.R., and Irvine K.D.. 2018. The Hippo Signaling Network and Its Biological Functions. Annu. Rev. Genet. 52:65–87. 10.1146/annurev-genet-120417-031621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, J.S., Meng Z., Kim Y.C., Park H.W., Hansen C.G., Kim S., Lim D.S., and Guan K.L.. 2015. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat. Cell Biol. 17:500–510. 10.1038/ncb3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, S., Kim W., Kim S., Kim Y., Song Y., Bilousov O., Kim J., Lee T., Cha B., Kim M., et al. 2017. Phosphorylation by NLK inhibits YAP-14-3-3-interactions and induces its nuclear localization. EMBO Rep. 18:61–71. 10.15252/embr.201642683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya, I.M., and Halder G.. 2019. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 20:211–226. 10.1038/s41580-018-0086-y [DOI] [PubMed] [Google Scholar]
- Oudhoff, M.J., Freeman S.A., Couzens A.L., Antignano F., Kuznetsova E., Min P.H., Northrop J.P., Lehnertz B., Barsyte-Lovejoy D., Vedadi M., et al. 2013. Control of the hippo pathway by Set7-dependent methylation of Yap. Dev. Cell. 26:188–194. 10.1016/j.devcel.2013.05.025 [DOI] [PubMed] [Google Scholar]
- Pan, D. 2010. The hippo signaling pathway in development and cancer. Dev. Cell. 19:491–505. 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, C., Zhu Y., Zhang W., Liao Q., Chen Y., Zhao X., Guo Q., Shen P., Zhen B., Qian X., et al. 2017. Regulation of the Hippo-YAP Pathway by Glucose Sensor O-GlcNAcylation. Mol. Cell. 68:591–604.e5. 10.1016/j.molcel.2017.10.010 [DOI] [PubMed] [Google Scholar]
- Qiao, Y., Lin S.J., Chen Y., Voon D.C., Zhu F., Chuang L.S., Wang T., Tan P., Lee S.C., Yeoh K.G., et al. 2016. RUNX3 is a novel negative regulator of oncogenic TEAD-YAP complex in gastric cancer. Oncogene. 35:2664–2674. 10.1038/onc.2015.338 [DOI] [PubMed] [Google Scholar]
- Qiao, Y., Chen J., Lim Y.B., Finch-Edmondson M.L., Seshachalam V.P., Qin L., Jiang T., Low B.C., Singh H., Lim C.T., and Sudol M.. 2017. YAP Regulates Actin Dynamics through ARHGAP29 and Promotes Metastasis. Cell Rep. 19:1495–1502. 10.1016/j.celrep.2017.04.075 [DOI] [PubMed] [Google Scholar]
- Rodríguez, C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., and Dymecki S.M.. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140. 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Sanjana, N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Ding Y., Zhan M., Li Y., Gao D., Wang G., Gao Y., Li Y., Wu S., Lu L., et al. 2019. Usp7 regulates Hippo pathway through deubiquitinating the transcriptional coactivator Yorkie. Nat. Commun. 10:411. 10.1038/s41467-019-08334-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, D.E., Dai Y., Lin L.W., Xu Y., Liu D.Z., Hong X.P., Jiang H.W., and Xu S.H.. 2019a. STUB1 suppresseses tumorigenesis and chemoresistance through antagonizing YAP1 signaling. Cancer Sci. 110:3145–3156. 10.1111/cas.14166 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tang, Y., Chen M., Zhou L., Ma J., Li Y., Zhang H., Shi Z., Xu Q., Zhang X., Gao Z., et al. 2019b. Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling. Cell Discov. 5:3. 10.1038/s41421-018-0077-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Klooster, J.P., Jansen M., Yuan J., Oorschot V., Begthel H., Di Giacomo V., Colland F., de Koning J., Maurice M.M., Hornbeck P., and Clevers H.. 2009. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev. Cell. 16:551–562. 10.1016/j.devcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Thompson, B.J., and Sahai E.. 2015. MST kinases in development and disease. J. Cell Biol. 210:871–882. 10.1083/jcb.201507005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro, A., Panciera T., and Piccolo S.. 2018. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 20:888–899. 10.1038/s41556-018-0142-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Huang J., Wang X., Yuan J., Li X., Feng L., Park J.I., and Chen J.. 2012. PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 26:1959–1971. 10.1101/gad.192955.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Xiao Z.D., Li X., Aziz K.E., Gan B., Johnson R.L., and Chen J.. 2015. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat. Cell Biol. 17:490–499. 10.1038/ncb3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Lu Y., Yin M.X., Wang C., Wu W., Li J., Wu W., Ge L., Hu L., Zhao Y., and Zhang L.. 2016. Importin α1 Mediates Yorkie Nuclear Import via an N-terminal Non-canonical Nuclear Localization Signal. J. Biol. Chem. 291:7926–7937. 10.1074/jbc.M115.700823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K.E., Li Y.W., Yang N., Shen H., Orillion A.R., and Zhang J.. 2014. PTPN14 forms a complex with Kibra and LATS1 proteins and negatively regulates the YAP oncogenic function. J. Biol. Chem. 289:23693–23700. 10.1074/jbc.M113.534701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, F., Zhou Z., Kim J., Hang Q., Xiao Z., Ton B.N., Chang L., Liu N., Zeng L., Wang W., et al. 2018. SKP2- and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity. Nat. Commun. 9:2269. 10.1038/s41467-018-04620-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F.X., Zhao B., Panupinthu N., Jewell J.L., Lian I., Wang L.H., Zhao J., Yuan H., Tumaneng K., Li H., et al. 2012. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 150:780–791. 10.1016/j.cell.2012.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F.X., Zhao B., and Guan K.L.. 2015. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 163:811–828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, X., Yao P.Y., Jiang J., Zhang Y., Su Z., Yao W., Wang X., Gui P., Mullen M., Henry C., et al. 2017. MST4 kinase phosphorylates ACAP4 protein to orchestrate apical membrane remodeling during gastric acid secretion. J. Biol. Chem. 292:16174–16187. 10.1074/jbc.M117.808212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato, F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M., and Piccolo S.. 2015. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17:1218–1227. 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato, F., Battilana G., Cordenonsi M., and Piccolo S.. 2016. YAP/TAZ as therapeutic targets in cancer. Curr. Opin. Pharmacol. 29:26–33. 10.1016/j.coph.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato, F., Cordenonsi M., and Piccolo S.. 2019. YAP and TAZ: a signalling hub of the tumour microenvironment. Nat. Rev. Cancer. 19:454–464. 10.1038/s41568-019-0168-y [DOI] [PubMed] [Google Scholar]
- Zhang, J., Smolen G.A., and Haber D.A.. 2008. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 68:2789–2794. 10.1158/0008-5472.CAN-07-6205 [DOI] [PubMed] [Google Scholar]
- Zhang, W., Gao Y., Li P., Shi Z., Guo T., Li F., Han X., Feng Y., Zheng C., Wang Z., et al. 2014. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 24:331–343. 10.1038/cr.2014.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Tang F., Terracciano L., Hynx D., Kohler R., Bichet S., Hess D., Cron P., Hemmings B.A., Hergovich A., and Schmitz-Rohmer D.. 2015. NDR functions as a physiological YAP1 kinase in the intestinal epithelium. Curr. Biol. 25:296–305. 10.1016/j.cub.2014.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Qiao Y., Wu Q., Chen Y., Zou S., Liu X., Zhu G., Zhao Y., Chen Y., Yu Y., et al. 2017. The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat. Commun. 8:15280. 10.1038/ncomms15280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Schaefer A., Wang Y., Hodge R.G., Blake D.R., Diehl J.N., Papageorge A.G., Stachler M.D., Liao J., Zhou J., et al. 2020. Gain-of-Function RHOA Mutations Promote Focal Adhesion Kinase Activation and Dependency in Diffuse Gastric Cancer. Cancer Discov. 10:288–305. 10.1158/2159-8290.CD-19-0811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Du J., Wang S., Shao L., Jin K., Li F., Wei B., Ding W., Fu P., van Dam H., et al. 2019. OTUB2 Promotes Cancer Metastasis via Hippo-Independent Activation of YAP and TAZ. Mol. Cell. 73:7–21.e7. 10.1016/j.molcel.2018.10.030 [DOI] [PubMed] [Google Scholar]
- Zhao, B., Wei X., Li W., Udan R.S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21:2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., Li L., Tumaneng K., Wang C.Y., and Guan K.L.. 2010. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24:72–85. 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., and Pan D.. 2019. The Hippo Signaling Pathway in Development and Disease. Dev. Cell. 50:264–282. 10.1016/j.devcel.2019.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D., Conrad C., Xia F., Park J.S., Payer B., Yin Y., Lauwers G.Y., Thasler W., Lee J.T., Avruch J., and Bardeesy N.. 2009. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 16:425–438. 10.1016/j.ccr.2009.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists protein levels of MST4 and pYAP (T83) are positively correlated in GC.
describes the correlation of protein levels of total YAP and pYAP(T83) in GC.
describes the clinical significance of pYAP (T83) expression in GC.
Data Availability Statement
RNA sequencing raw data have been deposited in the Gene Expression Omnibus database under the accession no. GSE142644.