Personalized medicine and circadian rhythms: Opportunities for modern society
Abstract
Circadian rhythms govern physiology and metabolism, leading to controlled homeostasis. We discuss the impact of circadian rhythms on society and the challenges for the imminent future of personalized medicine.
Circadian rhythms in everyday life
Life is dictated by cyclic activities aligned with the day–night rhythm. Circadian (from the Latin “circa diem”) rhythms pervade virtually all aspects of behavior and physiology. Light is a major zeitgeber (time giver) capable of influencing internal rhythms. Importantly, light-emitting devices are central to modern society. As such, we are continuously exposed to various sources of light that may impact negatively our circadian rhythms (Chang et al., 2015). There are ongoing social debates on topics that concern our circadian rhythms, like whether schools should adjust their schedules to better synchronize them to students’ daily rhythms (Smarr and Schirmer, 2018) or whether daylight savings should be abolished. Circadian rhythms regulate sleep/wake cycles, metabolism of nutrients, hormone secretion, immunological responses, and behavior, just to name a few things. Consequently, disruption of diurnal rhythms has been linked to various pathophysiological outcomes, such as obesity, metabolic syndrome, diabetes, and cancer.
The molecular clock
Virtually every cell in our body has a molecular clock. It consists in a group of transcription factors that generate 24-h oscillations by binding to thousands of sites throughout the genome and driving a system of feedback regulatory loops (Koike et al., 2012). Since the discovery of the molecular clock, an increasing number of studies have looked into 24-h gene expression rhythms. Studies in rodents show that 15% of all genes within a given tissue oscillate with minimal overlap between organs, meaning that approximately half of all expressed genes are rhythmic in at least one tissue (Zhang et al., 2014). More recently, profiling of the diurnal transcriptome of 64 tissues from baboons confirmed the extensiveness of circadian transcription (Mure et al., 2018). Furthermore, circadian oscillations are not limited to gene expression, but remarkably also encompass protein levels, posttranslational modifications, and metabolites. As our understanding of how circadian rhythms impact our physiology grows, awareness of their significance should be taken carefully into account by modern medicine and everyday lifestyle choices.
A time for every drug: The promise of chronopharmacology
At this time, when biomedical and translational research advocate for personalized or precision medicine, several considerations should be drawn from the numerous 24-h -omics datasets generated in recent years. One is of particular importance for clinical practice and drug development: a large proportion of drug targets display cyclic gene expression (Mure et al., 2018; Zhang et al., 2014). Yet, the time of day is contemplated rarely when administering medication. Likewise, a quick search of time-of-day dosing suggestions for Food and Drug Administration–approved drugs returns very few examples.
A problematic limitation is that, to date, most of the big data available for circadian gene expression have been derived from animal studies, as the time-resolution required for these studies is virtually impossible for humans. Notably, though, the CYCLOPS algorithm (cyclic ordering by periodic structure) was developed to enable the detection of rhythmic genes from human datasets lacking time-of-day sample collection information (Anafi et al., 2017). The CYCLOPS algorithm was used on a human RNA-sequencing dataset of 632 donors spanning 13 tissues, allowing the construction of a human database of circadian gene expression (Ruben et al., 2018). Similar to what was observed in murine models, 12% of the identified cyclic genes encode for drug targets, drug transporters, or metabolizing enzymes. This tells us that circadian timing very likely influences efficacy as well as toxicity of drugs and could explain, at least in part, the large percentage of failures in clinical trials. Among the drugs predicted to target cyclic genes, many were linked to the cardiovascular system, in keeping with the notion that many cardiovascular functions display 24-h rhythms and that adverse cardiovascular events happen more frequent in the early morning hours. Additionally, cardiotoxicity is one of the most frequent side effects and is often the reason for clinical trial discontinuation. Timing of drug administration to peak expression of its target could thus improve efficacy, and conversely, careful evaluation of rhythmic expression of “off targets” as well as its metabolism/clearance could help reduce toxicity and side effects. Underlining how these findings have high translational value, effectiveness of time-of-day dosing, also known as chronotherapy, has been demonstrated for some cardiovascular medications. A good example is the efficacy of the statin simvastatin, which is greatest when taken before bedtime. Antihypertensive drugs such as the Ca2+ channel blocker nifedipine and angiotensin II receptor antagonists also elicit greater benefits in the evening. Also, the MAPEC (Ambulatory Blood Pressure Monitoring and Cardiovascular Events) clinical study reported an improvement of cardiovascular risk following nighttime targeting of blood pressure (Rana et al., 2020).
Circadian dosing will certainly benefit other medical disciplines. Specifically, some promising data on anticancer treatments already exist, and a clinical trial evaluating time-of-day–dependent efficacy and toxicity of an anti-glioblastoma drug is currently recruiting patients (https://clinicaltrials.gov/ct2/show/NCT02781792). Also, vaccinations show time-of-day–dependent variations both in mice and humans (Long et al., 2016; Nobis et al., 2019). Thus, the time has come to start exploiting the growing amount of transcriptomic data now available and translate it to medical practice.
Environment matters
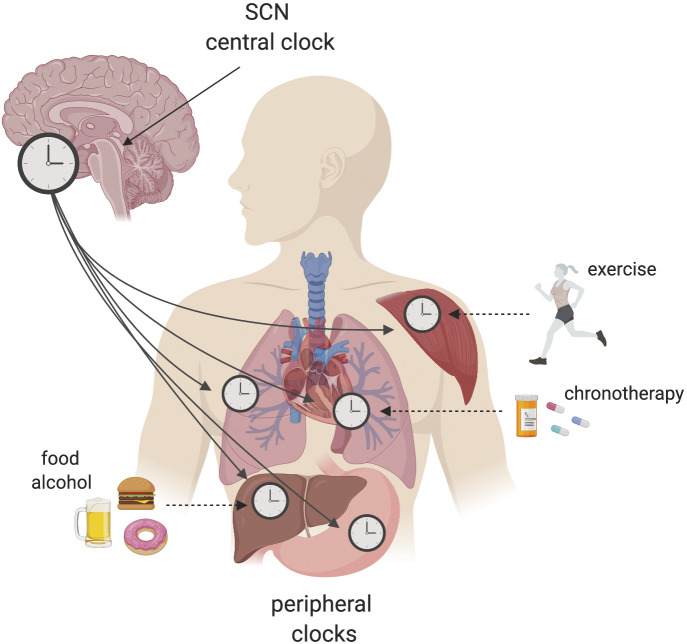
No two people are identical, and this is true for circadian rhythms too. When thinking about personalized medicine, translational potential, and clinical implementation, we need to carefully take into consideration interindividual variability. As a general rule, the human population may be classified into three main chronotypes: morning people, also called “morning larks”; people who prefer going to bed and waking up later, also called “night owls”; and people who fall in between. The picture isn’t that simple, though. We now know that an array of environmental factors, including dietary choices and exercise, can alter tissue-specific rhythms (Fig. 1). There is a time of day that is more appropriate for food intake and for exercise (Asher and Sassone-Corsi, 2015; Ezagouri et al., 2019; Sato et al., 2019). More than 60% of adults in the United States are overweight or obese due to overconsumption of foods with high caloric content. Studies in rodents have shown that high caloric content impacts our circadian rhythms in several ways: first of all, it modifies feeding behavior, leading to a spread-out consumption of calories throughout the day (Kohsaka et al., 2007); second, it affects circadian gene expression and metabolites in an array of tissues (Dyar et al., 2018; Eckel-Mahan et al., 2013). In the liver, it leads to activation of lipid-related transcription factors at a specific circadian time, promoting oscillation of transcriptional pathways involved in fatty acid synthesis, fatty acid oxidation, and glycerophospholipid metabolism. Paralleling gene transcription, a number of lipid metabolites also follow a similar pattern of gain of oscillation (Eckel-Mahan et al., 2013; Guan et al., 2018). A mouse study indicated that targeting PPAR-α—a major regulator of lipid metabolism—at its peak expression with an agonist used in the clinic for hyperlipidemia resulted in greater efficacy at lowering hepatic lipid accumulation when delivered (Guan et al., 2018). For this particular example, we can imagine that a normal weight patient would not profit from a time-of-day dosing protocol, while it would greatly benefit an overweight individual habitually consuming a diet rich in calories. Another very common habit of modern society is social drinking, with a yearly consumption of ∼6.13 liters per person and a percentage of the population consuming large amounts at a time (or “binge drinking”). A recent study showed how acute (binge) or chronic alcohol consumption differentially influence hepatic circadian gene expression rhythms (Gaucher et al., 2019).
Figure 1.
Interplay between the circadian clock network and environmental cues. The central clock regulator in the hypothalamic suprachiasmatic nucleus (SCN) connects with peripheral clocks to ensure timekeeping and physiology. The suprachiasmatic nucleus is entrained by light, while peripheral oscillators can be reset by environmental cues (zeitgebers, or time givers) such as food or exercise. A range of environmental factors can alter tissue-specific rhythms, leading to loss of temporal coherence among tissue clocks and increasing the risk of pathologies.
A major challenge for the upcoming years will be to precisely pinpoint the circadian phenotype of each individual. To do so, several steps should be implemented: self-assessment questionnaires alongside monitoring of physiological parameters such as light exposure, blood pressure, feeding behavior, and activity. Feasibility of the latter was recently proven in a small-scale study performed on healthy individuals (Skarke et al., 2017). Human datasets ought to be extended to include 24-h gene expression rhythms of diseased states known to be influenced by circadian disruption, and identification of circadian biomarkers could be sampled from serum or feces that both display robust rhythmicity of metabolites and microbiota composition, respectively. Finally, algorithms capable of integrating the above mentioned information ought to be developed for human studies.
Acknowledgments
We thank all members of the Sassone-Corsi laboratory for helpful discussion. The figure was created with Biorender.com.
Funding for C.M. Greco was provided by the National Cancer Institute of the US National Institutes of Health (NIH T32 2T32CA009054-36A1) and by the European Research Council (ERC MSCA-IF-2016 MetEpiClock 749869). Financial support for P. Sassone-Corsi was provided by the National Institute of Health, the Institut National de la Santé et de la Recherche Médicale, and a Novo Nordisk Challenge Grant.
References
- Anafi, R.C., et al. 2017. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1619320114 [DOI] [Google Scholar]
- Asher, G., and Sassone-Corsi P.. 2015. Cell. 10.1016/j.cell.2015.03.015 [DOI] [PubMed] [Google Scholar]
- Chang, A.M., et al. 2015. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1418490112 [DOI] [Google Scholar]
- Dyar, K.A., et al. 2018. Cell. 10.1016/j.cell.2018.08.042 [DOI] [Google Scholar]
- Eckel-Mahan, K.L., et al. 2013. Cell. 10.1016/j.cell.2013.11.034 [DOI] [Google Scholar]
- Ezagouri, S., et al. 2019. Cell Metab. 10.1016/j.cmet.2019.03.012 [DOI] [PubMed] [Google Scholar]
- Gaucher, J., et al. 2019. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1911189116 [DOI] [Google Scholar]
- Guan, D., et al. 2018. Cell. 10.1016/j.cell.2018.06.031 [DOI] [Google Scholar]
- Kohsaka, A., et al. 2007. Cell Metab. 10.1016/j.cmet.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Koike, N., et al. 2012. Science. 10.1126/science.1226339 [DOI] [Google Scholar]
- Long, J.E., et al. 2016. Vaccine. 10.1016/j.vaccine.2016.04.032 [DOI] [Google Scholar]
- Mure, L.S., et al. 2018. Science. 10.1126/science.aao0318 [DOI] [Google Scholar]
- Nobis, C.C., et al. 2019. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1905080116 [DOI] [Google Scholar]
- Rana, S., et al. 2020. Circ. Res. 10.1161/CIRCRESAHA.119.313349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben, M.D., et al. 2018. Sci. Transl. Med. 10.1126/scitranslmed.aat8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S., et al. 2019. Cell Metab. 10.1016/j.cmet.2019.03.013 [DOI] [PubMed] [Google Scholar]
- Skarke, C., et al. 2017. Sci. Rep. 10.1038/s41598-017-17362-6 [DOI] [Google Scholar]
- Smarr, B.L., and Schirmer A.E.. 2018. Sci. Rep. 10.1038/s41598-018-23044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R., et al. 2014. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1408886111 [DOI] [Google Scholar]