This study demonstrates that loss of TIF1γ in HSC aggravates fibrosis and that TIF1γ, identified as a novel target, has potential in the development of new therapeutic approaches to inhibiting or treating liver fibrosis.
Abstract
Transforming growth factor β (TGFβ) is a crucial factor in fibrosis, and transcriptional intermediary factor 1γ (TIF1γ) is a negative regulator of the TGFβ pathway; however, its role in liver fibrosis is unknown. In this study, mesenchymal stem cells derived from human embryonic stem cells (hE-MSCs) that secrete hepatocyte growth factor (HGF) were used to observe the repair of thioacetamide (TAA)-induced liver fibrosis. Our results showed that TIF1γ was significantly decreased in LX2 cells when exposed to TGFβ1. Such decrease of TIF1γ was significantly prevented by co-culture with hE-MSCs. Interaction of TIF1γ with SMAD2/3 and binding to the promoter of the α-smooth muscle gene (αSMA) suppressed αSMA expression. Phosphorylation of cAMP response element–binding protein (CREB) and binding on the TIF1γ promoter region induced TIF1γ expression. Furthermore, hepatic stellate cell–specific TIF1γ-knockout mice showed aggravation of liver fibrosis. In conclusion, loss of TIF1γ aggravates fibrosis, suggesting that a strategy to maintain TIF1γ during liver injury would be a promising therapeutic approach to prevent or reverse liver fibrosis.
Introduction
Fibrosis is the pivotal stage of liver scarring and can progress to cirrhosis and eventually cause liver failure, for which the only effective therapy is liver transplantation (Bataller and Brenner, 2005). However, the shortage of the available donated organs and long-term postsurgical immunosuppression have forced researchers to look for alternative therapeutic strategies (Burra et al., 2012). Recent technological developments have enabled the elucidation of the cellular mechanisms of liver fibrosis as well as therapeutic approaches to liver-oriented cell therapy. Regarding the underlying mechanism, activation of hepatic stellate cells (HSCs) plays a pivotal role in extracellular matrix production during liver fibrosis. Although the activation and transdifferentiation of HSCs to myofibroblasts are regarded as key pathogenic mechanisms of fibrogenesis, the key factors that play a role in the activation of HSCs remain to be fully elucidated.
The stimulation of hepatocyte regeneration by human mesenchymal stem cells (hMSCs) has been shown to be a therapeutic strategy to alleviate end-stage liver disease. However, its clinical potential is still a matter of debate (Baertschiger et al., 2009; Shi et al., 2011; Terai et al., 2005; Wang et al., 2012). In our previous study, we successfully derived hMSCs from human embryonic stem cells (hE-MSCs), and we demonstrated that hE-MSCs could be consistently produced, maintained, and expanded (Lee et al., 2010, 2012). In this study, hE-MSCs, which secrete hepatocyte growth factor (HGF) as the most abundant growth factor, were used as a strategic tool to screen for a target for repairing liver injury in vivo and in vitro. From these experiments, we identified transcriptional intermediary factor 1 gamma (TIF1γ) among six candidate proteins as a negative regulator of TGFβ signaling in HSCs. TIF1γ, also known as tripartite motif–containing 33, has been revealed to act as a ubiquitin E3 ligase (Xue et al., 2015). Notably, it serves as a transcriptional corepressor by interacting with SMAD family member 2/3 (Hesling et al., 2011).
In this study, our findings on the potential suppressive role of TIF1γ in liver fibrosis were corroborated by experiments in vitro using LX2 HSCs and in vivo using site- and time-specific Tif1γ gene targeting in transgenic (TG) mice. To know the role of TIF1γ in liver fibrosis, a transgene construct consisting of modified Cas9 conjugated to an estrogen receptor (ERT2) that is activated by tamoxifen (TMX), but not estradiol (Metzger and Chambon, 2001), and under the control of the lecithin retinol acyltransferase (Lrat) promoter to specifically knock out Tif1γ in HSCs was developed and used (Mederacke et al., 2013).
Results
Transplantation of hE-MSCs prevents thioacetamide (TAA)-induced liver fibrosis in nude mice
We previously found that hE-MSCs abundantly secrete HGF (Lee et al., 2018). Because HGF is an important growth factor in the liver (Böhm et al., 2010), we hypothesized that hE-MSCs might be efficacious in liver therapy.
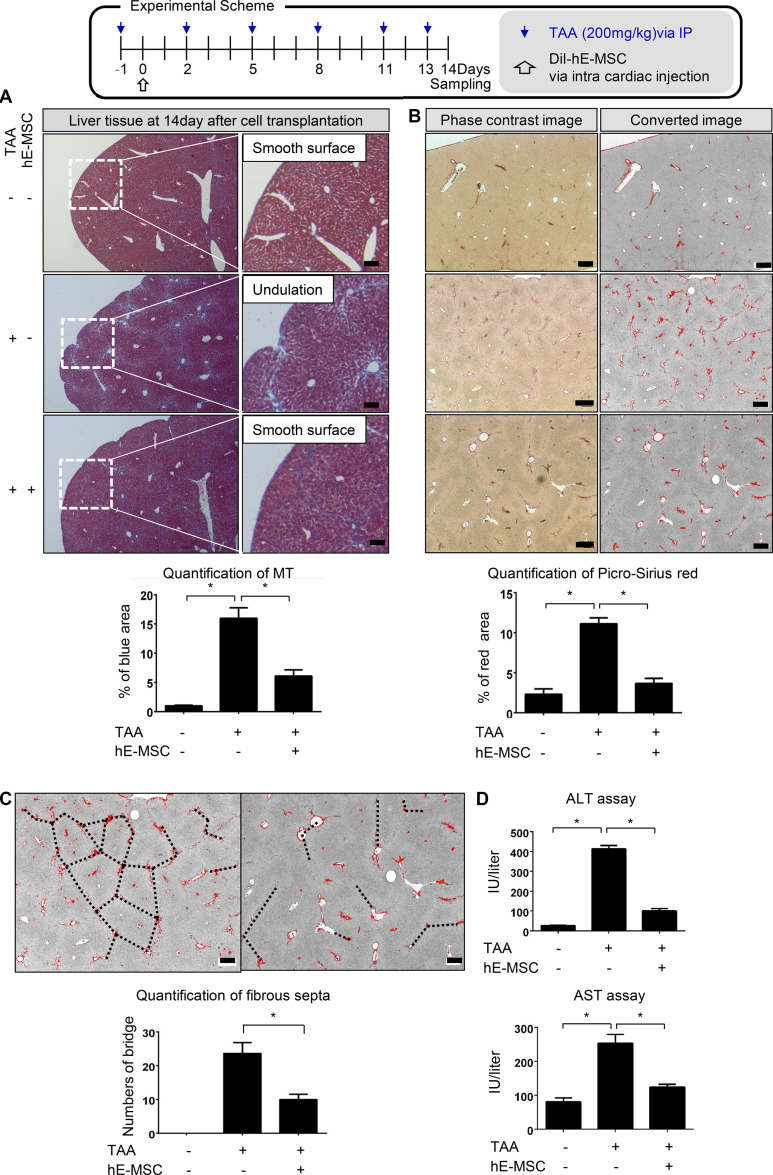
Histological analysis of collagen fibers using Masson’s trichrome staining revealed that transplantation of hE-MSCs reduced liver surface undulations and the fibrotic area at day 14 of TAA-induced liver injury (0.99 ± 0.18% in control [no treatment] vs. 16.0 ± 4.4% in TAA treatment vs. 6.1 ± 3.1% in TAA/hE-MSC treatment, n = 5; Fig. 1 A). Collagen deposits in 14-d tissues were visualized and quantified using Picro-Sirius red staining, which detects collagen types I and III (Fig. 1 B). The area of collagen deposit by TAA injury was significantly reduced by transplantation of hE-MSCs (2.3 ± 1.1% in control vs. 11.1 ± 1.2% in TAA treatment vs. 3.7 ± 1.0% in TAA/hE-MSC treatment, n = 5; Fig. 1 B). Fibrous septa were evaluated in liver tissues of 14-d TAA–treated mice with or without hE-MSC transplantation. Fibrosis stage was assessed based on staining with Picro-Sirius red, according to two histological classifications, METAVIR (stages I–IV) and Ishak (stages I–V; Bataller and Brenner, 2005; Standish et al., 2006). Fibrotic expansion with occasional bridging/septa (Ishak stage 3/METAVIR stage F2) was detected in TAA-treated mice (Fig. 1 C). However, hE-MSC transplantation prevented the formation of bridging/septa (Ishak stage 2/METAVIR stage F2; 23.7 ± 5.5 in TAA treatment vs. 10 ± 2.6 in TAA/hE-MSC treatment, n = 5; Fig. 1 C), which correlated well with a decrease in fibrous area. Injection of TAA into the peritoneum three times a week significantly elevated the levels of liver enzymes aspartate aminotransferase (AST) and alanine aminotransferase (ALT), leading to liver fibrosis. However, one systemic administration of hE-MSCs during the period of TAA injections prevented this elevation of liver enzyme (ALT: 26 ± 2.8 IU/liter in control vs. 413 ± 24.0 IU/liter in TAA treatment vs. 101 ± 15.6 IU/liter in TAA/hE-MSC treatment; AST: 80.5 ± 16.3 IU/liter in control vs. 253 ± 36.8 IU/liter in TAA treatment vs. 124 ± 11.3 IU/liter in TAA/hE-MSC treatment; Fig. 1 D).
Figure 1.
hE-MSC transplantation represses liver fibrosis after injury in mice. (A) Experimental scheme of hE-MSC transplantation into mice having a liver injury by TAA administration. Quantification of liver fibrosis by MT staining in three groups (normal, TAA, and TAA/hE-MSCs; mice, n = 5 in each group, compiled from two independent experiments). Quantification of the fibrotic area is presented as the blue portion (%) in total area, including red and blue portions. *, P < 0.05. Scale bars, 200 µm. IP, immunoprecipitation. (B) Picro-Sirius red staining of fibrotic areas in the livers at 14 d after hE-MSC transplantation. Quantification of liver fibrosis by Picro-Sirius red staining in three groups (normal, TAA, and TAA/hE-MSCs; mice, n = 5 in each group, compiled from two independent experiments). *, P < 0.05. Scale bars, 200 µm. (C) Measurement of fibrous septa using Picro-Sirius red staining. Portal-to-portal fibrous bridging/septa were counted per 2.7 × 3.4 mm2 (TAA vs. TAA/hE-MSCs; n = 5). Scale bars, 200 µm. Data are expressed as the mean ± SD. *, P < 0.05. (D) Serum levels of hepatotoxicity indicators AST and ALT at 14 d (normal, TAA, and TAA/hE-MSCs; mice, n = 5 in each group, compiled from two independent experiments). Statistical analysis by one-way ANOVA (A, B, and D) or unpaired Student's t test (C).
TIF1γ suppresses the activation of human LX2 HSCs
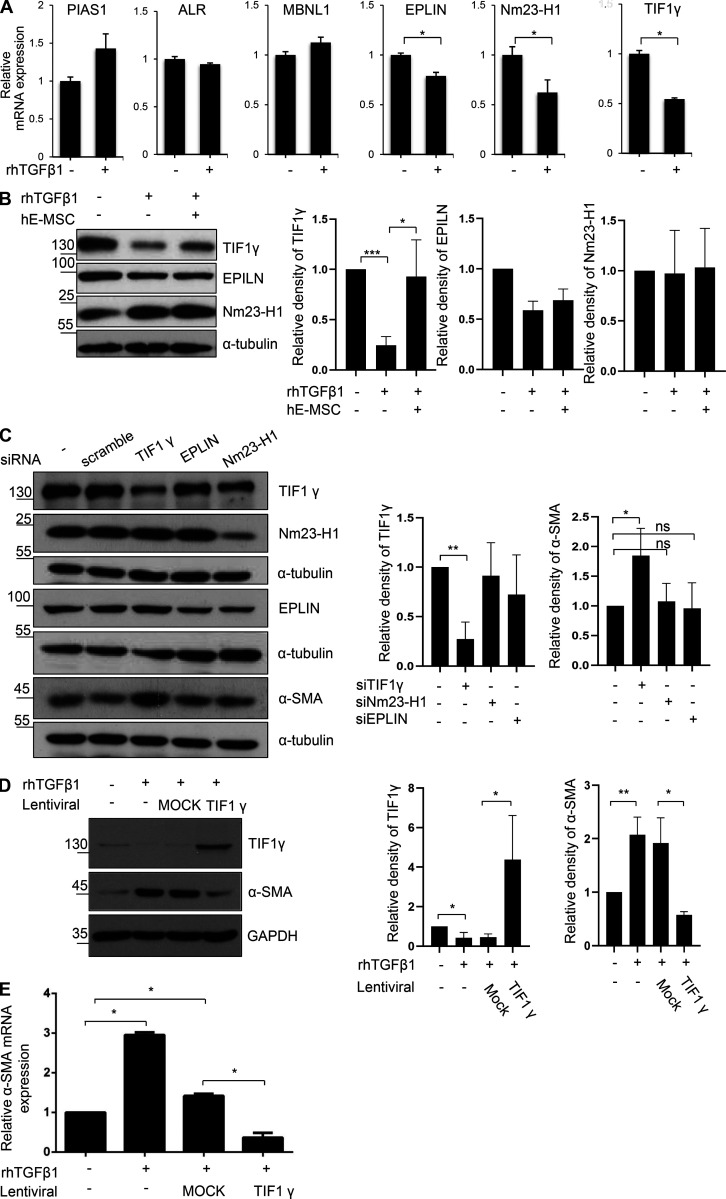
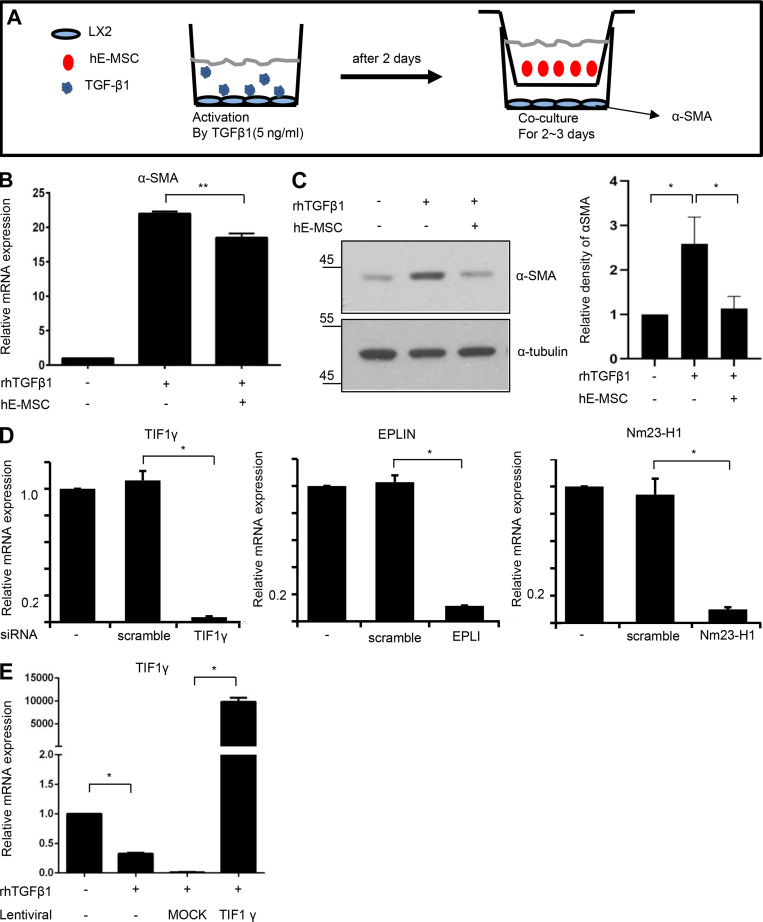
To evaluate how hE-MSC transplantation suppressed liver fibrosis in vitro, we searched for candidate antifibrotic factors that were declined in TGFβ1-activated LX2 as human HSCs. It has been reported that fibrotic events are associated with a TGFβ1 signaling pathway in activated HSCs; thus, we selected and tested six proteins (Dayoub et al., 2011; Hesling et al., 2011; Netherton and Bonni, 2010; Vajda et al., 2009; Zhang et al., 2011; Zhao et al., 2013; Table S1). Among the six candidates, we identified three proteins whose mRNA expression decreased after exposure to TGFβ1 in LX2 cells: epithelial protein lost in neoplasm (EPLIN), a cytoskeleton-associated protein that inhibits actin filament depolymerization; Nm23-H1, nucleoside diphosphate kinase A, a metastasis suppressor; and TIF1γ (Fig. 2 A). To validate the involvement of each of these three proteins in the effect of hE-MSCs on fibrosis, we co-cultured hE-MSCs with TGFβ1-activated LX2 cells (Fig. S1 A). When LX2 cells were exposed to TGFβ1, they were induced to myofibroblast-like cells through increased expression of α-smooth muscle actin (αSMA; Fig. S1, B and C). This change was significantly prevented by co-culture with hE-MSCs (Fig. S1, B and C). In these experiments, only TIF1γ was down-regulated by TGFβ1 and up-regulated in response to hE-MSCs in LX2 cells, whereas EPLIN and Nm23-H1 showed no changes at the protein level (Fig. 2 B). Finally, TIF1γ was functionally validated in loss- and gain-of-function experiments. When TIF1γ expression in LX2 cells was knocked down by specific siRNA, we observed αSMA up-regulation, whereas knockdown of EPLIN or Nm23-H1 did not affect αSMA expression (Fig. 2 C and Fig. S1 D). TIF1γ overexpression reduced the αSMA level in TGFβ1-treated LX2 cells, suggesting that TIF1γ is a novel candidate for anti-fibrosis treatment (Fig. 2, D and E; and Fig. S1 E).
Figure 2.
TIF1γ is a repressor for myofibroblastic activation of human hepatic stellate LX2 cells. (A) RT-qPCR analysis of six genes after TGFβ1 treatment. *, P < 0.05 (unpaired Student's t test). Mean values ± SD were calculated from three independent experiments, with biological triplicates in each experiment. (B) Representative Western blot of TIF1γ, EPLIN, and Nm23-H1 expression in TGFβ1-treated LX2 cells with or without hE-MSCs (quantified by ImageJ). Reproducible results from three independent experiments are shown. Mean values ± SD were calculated from three independent experiments, with biological triplicates in each experiment. *, P < 0.05; ***, P < 0.0001. (C) Representative Western blot of TIF1γ, EPLIN, and Nm23-H1, and αSMA expression in LX2 cells transfected with siEPLIN, siNm23-H1, or siTIF1γ for gene knockdown (quantified by ImageJ). Reproducible results from three independent experiments are shown. Mean values ± SD were calculated from three independent experiments, with biological triple in each experiment. *, P < 0.05; **, P < 0.01. (D) Representative Western blot of TIF1γ and αSMA expression in TGFβ1-treated LX2 cells transduced by lentiviral vector CMV-TIF1γ (quantified by ImageJ). Results represent the mean ± SD of three independent biological experiments. *, P < 0.05; **, P < 0.01. (E) RT-qPCR analysis of αSMA gene in TGFβ1-treated LX2 cells transduced by lentiviral vector CMV-TIF1γ. *, P < 0.05. Results represent the mean ± SD of three independent experiments, with biological triplicates in each experiments. Statistical analysis was performed by one-way ANOVA. ns, not significant.
Figure S1.
The process for selection of TIF1γ using in vitro cell system. (A) Experimental scheme for validation of anti-fibrosis potential of hE-MSCs in vitro. (B) RT-qPCR analysis of αSMA expression. Results represent the mean ± SD of three independent biological experiments. **, P < 0.01. (C) Western blot analysis of αSMA protein expression. Quantified by ImageJ. Results represent the mean ± SD of three independent biological experiments. *, P < 0.05. (D) RT-qPCR analysis for the validation of knockdown by siRNA. Results represent the mean ± SD of three independent experiments. *, P < 0.05. (E) RT-qPCR analysis for validation of TIF1γ overexpression by lentiviral vector. Results represent the mean ± SD of three independent experiments. *, P < 0.05.
HGF from hE-MSCs increases TIF1γ that interacts with SMAD2/3, leading to suppression of αSMA
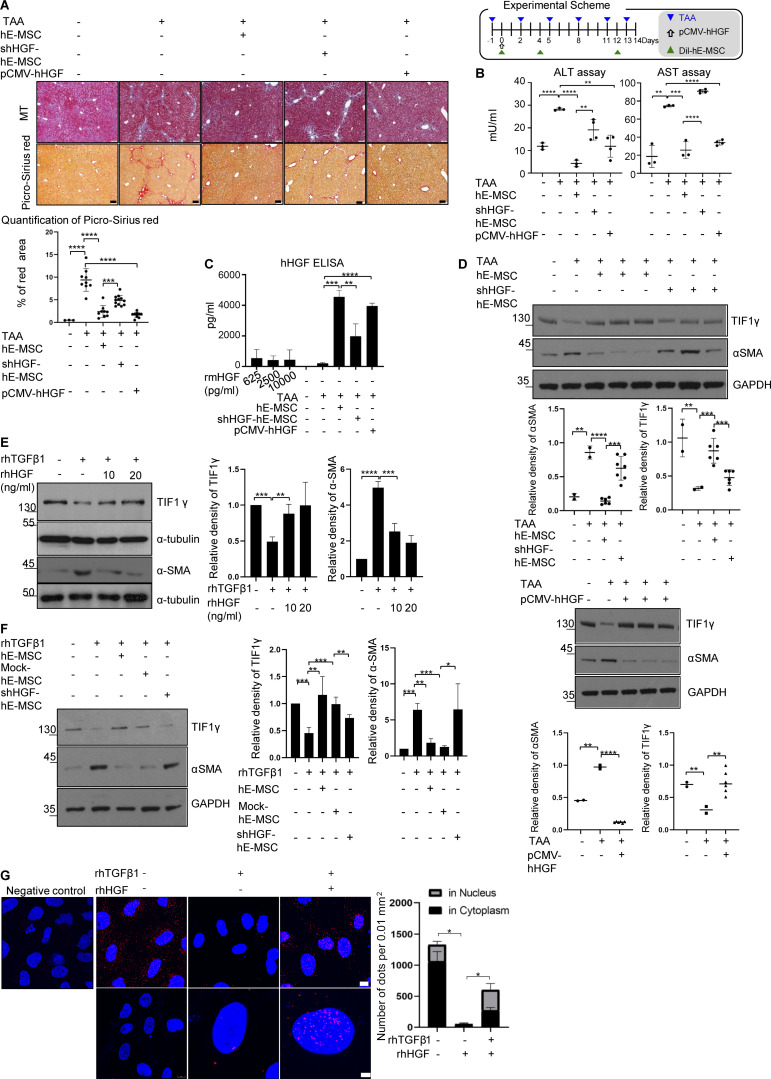
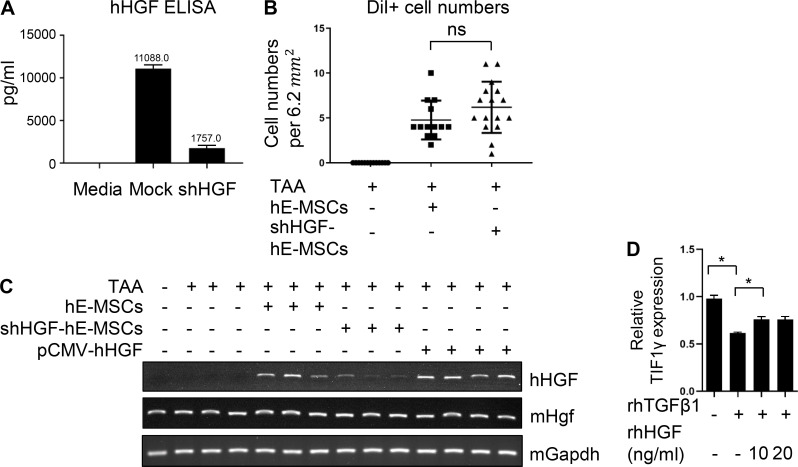
To further investigate the importance of hMSC-derived HGF, we conducted in vivo experiments using hE-MSCs transduced with small hairpin RNA against HGF (Fig. 3 A). We also evaluated human HGF (hHGF) ELISA results (Fig. S2 A). hE-MSCs were labeled with a fluorescent dye, DiI, for in vivo tracking as well as cell count in liver tissues. There was no significant difference in the DiI-labeled cells between hE-MSCs and shRNA for HGF (shHGF)–transduced hE-MSCs group (Fig. S2 B). However, the shHGF-transduced hE-MSCs group showed a significant increase in fibrous area (2.38 ± 1.34% in TAA/hE-MSC vs. 4.86 ± 1.10% in TAA/shHGF hE-MSC treatment). In pCMV-hHGF vector treatment, which was used to show the effect of HGF in vivo, fibrous area was reduced compared to the control group (1.65 ± 0.62% in pCMV-HGF vs. 9.39 ± 2.52% in TAA treatment; Fig. 3 A). ALT and AST levels also correlated with fibrous area (ALT: 11.87 ± 1.47 mU/ml in control vs. 28.19 ± 0.56 mU/ml in TAA treatment vs. 4.28 ± 1.45 mU/ml in TAA/hE-MSC treatment vs. 19.13 ± 4.55 mU/ml in TAA/shHGF hE-MSC treatment vs. 11.89 ± 4.84 mU/ml in TAA/pCMV-hHGF treatment; AST: 18.62 ± 12.27 mU/ml in control vs. 74.60 ± 1.07 mU/ml in TAA treatment vs. 25.66 ± 9.47 mU/ml in TAA/hE-MSC treatment vs. 90.86 ± 2.01 mU/ml in TAA/shHGF hE-MSC treatment vs. 33.96 ± 2.77 mU/ml in TAA/pCMV-hHGF treatment; Fig. 3 B). We performed RT-PCR using liver tissue from each group to detect whether hHGF was produced as a result of hE-MSCs in vivo. Expression of hHGF mRNA was detected in liver tissues of mice treated with hE-MSCs or pCMV-hHGF. Its level was low in the liver of mice treated with shHGF-transduced hE-MSCs (Fig. S2 C and Table S2). To verify the observed expression of hHGF in liver of mice treated with hE-MSCs or pCMV-hHGF in vivo, we measured serum hHGF levels in each group (Fig. 3 C). ELISA results verified that hHGF was indeed detected in blood of mice treated with hE-MSCs and pCMV-hHGF. Its serum level was low in mice treated with shHGF-transduced hE-MSCs, which was correlated with the mRNA expression pattern of hHGF in mice liver. Western blotting analysis of liver tissues showed that TIF1γ expression was decreased by TAA and restored by hE-MSCs or pCMV-hHGF in mouse liver tissue (Fig. 3 D). However, in the liver of mice treated with shHGF-transduced hE-MSCs, TIF1γ expression was not restored (Fig 3 D). The expression pattern of αSMA was opposite compared to TIF1 (Fig 3 D).
Figure 3.
Induction of TIF1γ by HGF and complex formation with SMAD. (A) Experimental scheme of hE-MSC or shHGF hE-MSCs or pCMV-hHGF vector transplantation into mice with a liver injury by TAA administration. Representative histochemical staining of MT and Picro-Sirius red staining in each group (each panel from an independent individual). Quantification of liver fibrosis by Picro-Sirius red staining in five groups (normal, TAA and TAA/hE-MSCs, TAA/shHGF hE-MSCs, and TAA/pCMV-hHGF; n = 3, 5, 5, 5, and 5, respectively) is presented as the red portion (%) in total area. One or two fields in each sample were measured using ImageJ. Mean values ± SD were calculated from measured fields of each sample in each group. Scale bars, 10 µm. ***, P < 0.001; ****, P < 0.0001. (B) Serum levels of hepatotoxicity indicators AST and ALT at 14 d (normal, TAA and TAA/hE-MSCs, TAA/shHGF hE-MSCs, TAA/pCMV-hHGF. n = 3, 3, 3, 4, and 4, respectively). Mean values ± SD were calculated from three or four samples with technical duplicates in each sample. **, P < 0.01. ***, P < 0.001; ****, P < 0.0001. (C) ELISA of hHGF in mouse serum (normal, TAA and TAA/hE-MSCs, TAA/shHGF hE-MSCs, TAA/pCMV-hHGF; n = 3, 3, 3, 4, and 4, respectively). Recombinant mHGF was used as a negative control (625, 2,500, and 10,000 pg/ml) and did not show reactivity with hHGF antibody. hHGF antibody (AF-294-NA; R&D Systems) was used in ELISA reactive with hHGF, but not mHGF. Mean values ± SD were calculated from three or four samples with technical duplicates in each sample. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Representative Western blot of TIF1γ and αSMA expression in five groups (normal, TAA and TAA/hE-MSCs, TAA/shHGF hE-MSCs, and TAA/pCMV-hHGF). Each lane represents one independent individual. Reproducible results from two independent Western blots are shown (quantified by ImageJ). Mean values ± SD were calculated from two or six bands in two independent Western blots. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (E) Representative Western blot for TIF1γ and αSMA in LX2 cells treated with rhTGF β1 (5 ng/ml) and/or rhHGF (10 or 20 ng/ml; quantified by ImageJ). Results represent the mean ± SD of two independent biological experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (F) Representative Western blot of TIF1γ and αSMA in LX2 co-cultured with shHGF- or mock-transfected hE-MSCs (quantified by ImageJ). Results represent the mean ± SD of two independent biological experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (G) Representative PLA to confirm interaction of TIF1γ and SMAD2/3 in LX2 cells. Rabbit TIF1γ antibody and mouse SMAD2/3 antibody were used. Anti-rabbit-plus and anti-mouse-minus probes bound to primary antibodies. Ligase was used to ligate the probes and polymerase used to amplify the specific sequence that could hybridize to the fluorescent probe to visualize interactions as dots. Red dots represent endogenous interactions between TIF1γ and SMAD2/3. The graph shows the total number of dots in the nucleus (gray) and cytoplasm (black). Dots were counted using ImageJ in four random fields per group. Mean values ± SD were calculated from four fields in two independent experiments, with biological duplicates in each experiments. *, P < 0.05. The number of total dots was compared by one-way ANOVA. Scale bars represent 10 µm (upper panel) and 2 µm (bottom panel).
Figure S2.
Validation for the expression of hHGF by hE-MSC or shHGF or pCMV-hHGF in vivo and in vitro. (A) Validation of shHGF hE-MSCs using ELISA of hHGF. Results represent the mean ± SD of two independent experiments. (B) Counting of DiI-labeled hE-MSCs in liver tissue. Three or four randomized fields (6.2 mm2) were counted using immunofluorescence (TAA/hE-MSCs and TAA/shHGF hE-MSCs; n = 5 and 5, respectively; three TAA individual tissues used negative control). (C) RT-PCR of hHGF in mouse liver tissues. Using specific hHGF primers or mHgf primers, mRNA of hHGF or mHgf in mouse liver was detected. Each lanes were presented each individual mouse liver tissue. (D) RT-qPCR of TIF1γ in human HSCs and LX2 cells treated with hTGFβ1 (5 ng/ml) and/or hHGF (10 or 20 ng/ml). Results represent the mean ± SD of two independent experiments. *, P < 0.05. ns, not significant.
To validate the effect of HGF from hE-MSCs on TIF1γ expression in human HSCs, LX2 cells were incubated with recombinant human HGF (rhHGF). TGFβ1 significantly suppressed TIF1γ and increased αSMA in LX2 cells, which was significantly reversed or prevented by rhHGF (Fig. 3 E and Fig. S2 D). Next, to confirm that HGF secreted from hE-MSCs is responsible for the maintenance of TIF1γ expression in HSCs, we knocked down HGF in hE-MSCs using HGF-specific small hairpin RNA, and we co-cultured the cells with TGFβ1-treated LX2 cells. While co-culture of LX2 cells with hE-MSCs decreased the induction of αSMA by TGFβ1 in LX2 cells, this effect was suppressed when HGF was knocked down, suggesting that HGF from hE-MSCs is strictly required for their anti-fibrosis effect on LX2 cells or HSCs (Fig. 3 F).
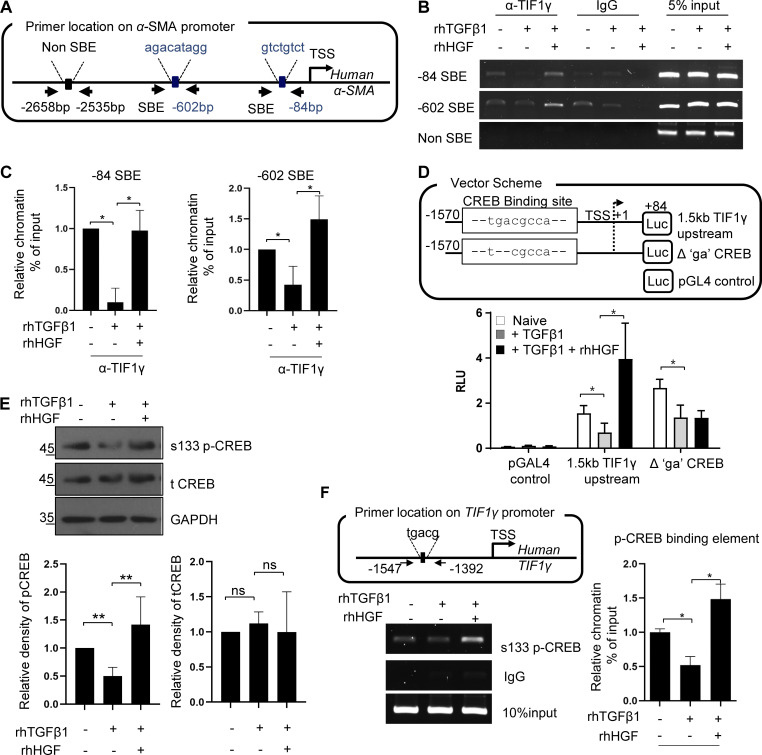
Although it is known that the SMAD2/3 complex is involved in the regulation of αSMA expression by TGFβ1 (Pardali et al., 2017), it is not known how αSMA is transcriptionally regulated by TGFβ1 in HSCs. We assumed that TIF1γ and SMAD2/3 interactions would be altered by TGFβ treatment and HGF treatment in LX2 cells. First, using a proximity ligation assay (PLA), we attempted to determine the cellular localization of TIF1γ and SMAD2/3 and their interaction. Interactions between TIF1γ and SMAD2/3 were sporadically observed and scattered throughout the nucleus and cytoplasm under basal conditions and drastically reduced by TGFβ1 treatment (total number of dots: 1,329.5 ± 143 vs. 52.7 ± 33/0.01 mm2, respectively; Fig. 3 G). When HGF was added, interactions between TIF1γ and SMAD2/3 were significantly increased and found to be concentrated in nucleus (total number of dots: 603 ± 62.8/0.01 mm2; in nucleus: 330.7 ± 102.2/0.01 mm2; Fig. 3 G). Next, we performed chromatin immunoprecipitation (ChIP) using TIF1γ antibody to identify which part of the αSMA promoter interacts with TIF1γ. The sequence of the SMAD-binding element (SBE) was used for the identification of a TIF1γ-binding site in the αSMA promoter (Fig. 4 A). GPminer (Lee et al., 2012) did not predict a TIF1γ-binding site in the αSMA promoter, but it did predict SBEs. PLA results suggested that SMAD2/3 forms a complex with TIF1γ; therefore, the SBE could be the TIF1γ-binding site. In ChIP data, the PCR product of –84SBE and –602SBE was increased in the HGF compared with the TGFβ1 treatment group (Fig. 4 B; quantified in Fig. 4 C). PLA and ChIP data indicated that binding of TIF1γ with SMAD2/3 on the αSMA promoter was augmented by HGF, resulting in decreased expression of αSMA, whereas it was reversed by TGFβ1.
Figure 4.
Interaction of TIF1γ with SMAD and binding to the promoter of the αSMA suppressed αSMA expression, and binding of pCREB on the TIF1γ promoter region induced TIF1γ expression. (A) Diagram showing the location of primers in αSMA promoter that can amplify each SBE. TSS, transcription start site. (B) Representative ChIP analysis of the αSMA promoter region with TIF1γ antibody and specific primers. The SBE sequence was analyzed using GPminer v. 2.0. TIF1γ antibody was used to pull down the protein–DNA complex, and the precipitated DNA fragments were analyzed with specific primers (indicated in the diagram) to confirm the binding sites. (C) Quantification of ChIP data. Results represent the mean ± SD of three independent biological experiments. *, P < 0.05. Statistically analyzed by one-way ANOVA. (D) Identification of CREB-binding sites in the putative promoter region (∼1.5 kb) of TIF1γ. Luciferase assay of a CREB-site point-mutated fragment (deletion of GA in TGACGCCA), evaluated using TGFβ1 and/or HGF. Results represent the mean ± SD of three independent biological experiments. *, P < 0.05 (one-way ANOVA). RLU, renilla luciferase unit. (E) Representative Western blot for total CREB and pCREBs133 in LX2 cells treated with TGFβ1 and/or HGF. Results represent the mean ± SD of three independent experiments. **, P < 0.01. (F) ChIP using primers for the CREB-binding site in the TIF1γ promoter, with the pCREBs133 antibody. Results represent the mean ± SD of three independent experiments. *, P < 0.05 (one-way ANOVA). ns, not significant.
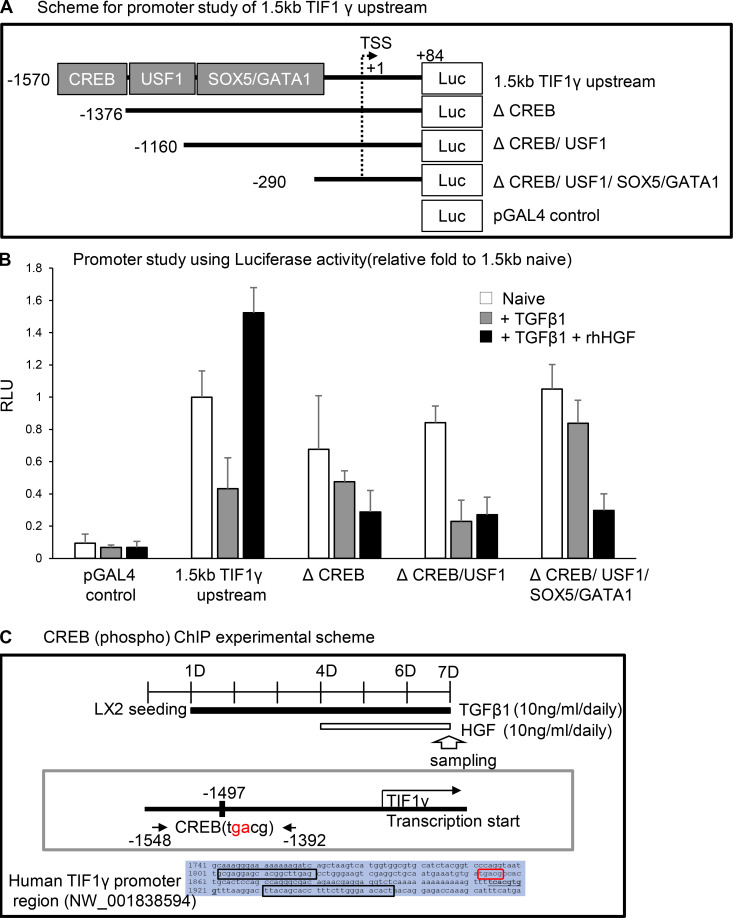
Mechanism of TIF1γ up-regulation by HGF: Phosphorylated cAMP response element–binding protein (pCREB) binds on the TIF1γ promoter
Next, to identify the promoter region involved in transcriptional activation of TIF1γ by HGF, we screened the ∼1.5-kb putative promoter region for transcription factor–binding sites. We identified binding sites for several transcription factors, including CREB, USF1, SOX5, and GATA1 (Fig. S3 A). Among them, the site for CREB was regulated by HGF, which was confirmed by deletion and point mutation (TGACG→T–CG) in a promoter study (Fig. 4 D and Fig. S3, A and B). Western blot and pCREBs133 ChIP assays revealed that Ser133-phosphorylated CREB (pCREBs133) induced transcription of TFI1-γ by HGF (Fig. 4, E and F). Ser133 phosphorylation is known as a marker for CREB activation, and pCREBs133 was reduced by TGFβ1 treatment and increased by HGF (Fig. 4 E). In a ChIP assay, binding of pCREBs133 to the CREB site of the TIF1γ promoter was increased by HGF, even in the presence of TGFβ1 (Fig. 4 F and Fig. S3 C).
Figure S3.
The screening and analysis of transcription factor on TIF1γ promoter. (A) Scheme of the promoter study of TIF1γ. (B) Promoter study by measuring luciferase reporter activity. Reproducible results from two independent experiments are shown. RLU, renilla luciferase unit. (C) Experimental setup for CREB (phospho) ChIP. To detect the CREB-binding site at residue 1,497, PCR was performed with CREB 1,548 5′-GCGAGGAGCACGGCTTGAG-3′ (forward) and CREB 1,392 5′-AGTGTTCCCAAGAAAGGTGCTGTAA-3′ (reverse) primers after chromatin pull-down with antibody against phosphorylated (Ser133) CREB.
TIF1γ is down-regulated in liver fibrosis
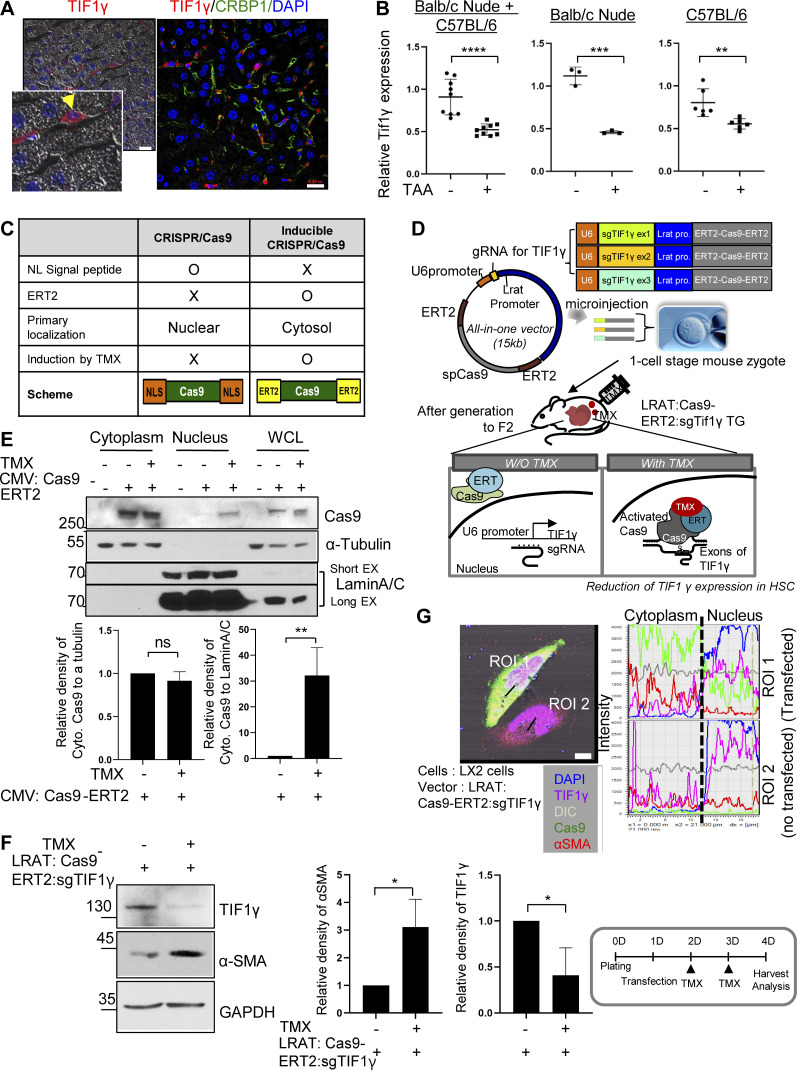
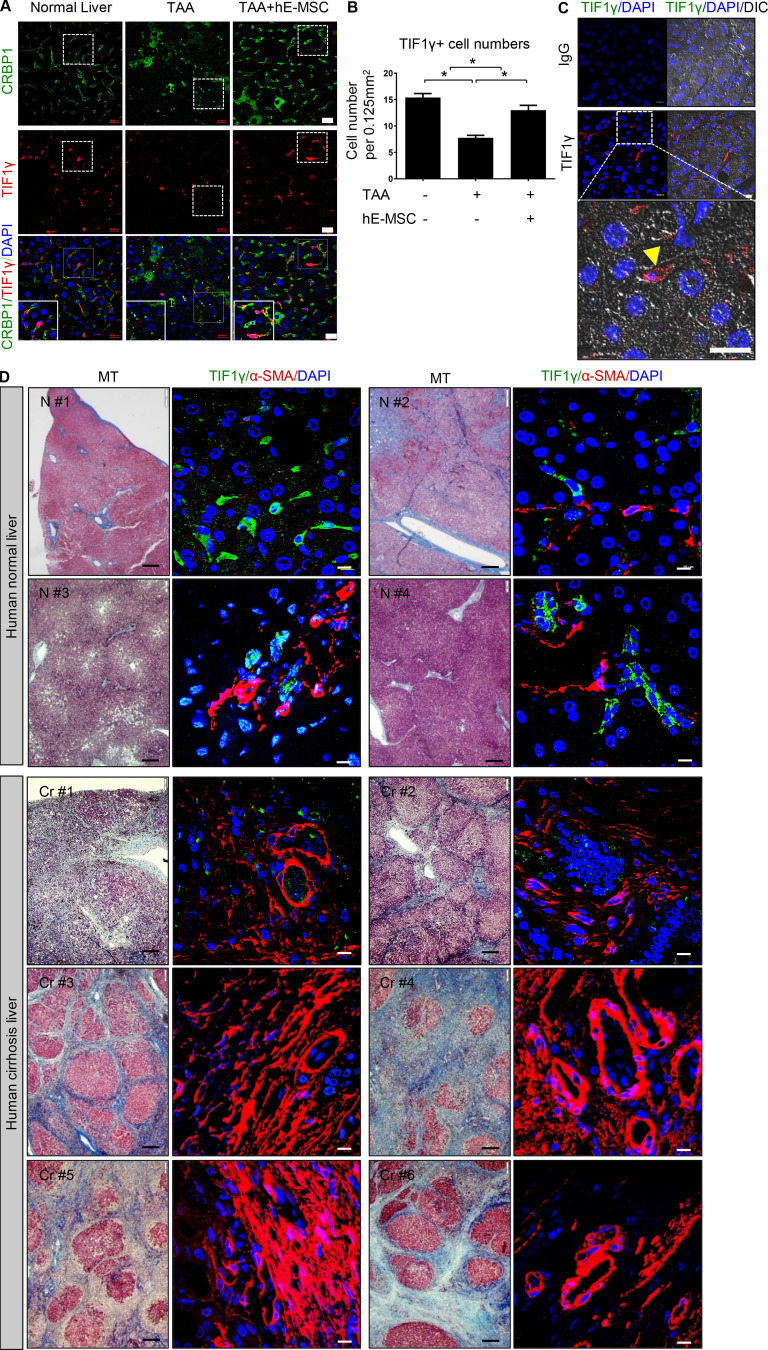
We detected TIF1γ-positive cells in mouse livers by immunohistochemistry and observed TIF1γ-positive cells in the space of Disse (perisinusoidal space; Fig. S4 A). Next, we isolated primary HSCs from normal and TAA-treated mouse liver tissues. Primary HSCs from TAA-treated liver tissue were shown to have decreased Tif1γ mRNA expression levels compared with HSCs from normal liver (Fig. S4 B). Interestingly, the magnitude of the decrease was much greater in BALB/c-nude mice than in C57BL/6N mice. As per a previously published report, liver injury in BALB/c mice results in severe fibrosis, whereas C57BL/6 mice develop comparatively minimal fibrosis (Shi et al., 1997).
Figure S4.
The expression of TIF1γ in normal mouse liver or HSC, and validation for development of TG mice with tamoxifen-inducible and HSC-specific knockout of Tif1γ. (A) TIF1γ expression in mouse liver analyzed by immunofluorescence. TIF1γ-positive cells were located in the space of Disse. Costaining of TIF1γ and CRBP1 (HSC marker) in mouse normal liver. Scale bars, 20 µm. The yellow arrowhead indicates the HSC in the space of Disse. (B) RT-qPCR of Tif1γ of primary HSC from normal and TAA-treated mouse liver tissues (BABL/c nude, n = 2; C57BL/6N, n = 4; technical triplicate). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Dot shows replication. Statistical analysis was performed using one-way ANOVA. (C) Comparison between CRISPR/Cas9 and inducible Cas9. (D) Vectors and experimental setup used for the establishment of TMX-inducible Tif1γ–knockout TG mice. (E) Validation of working Cas9-ERT2 system using 293T cells transfected with the CMV-promoter-Cas9-ERT2 construct. Lamin A/C was used as a nuclear faction control and α-tubulin as a cytosolic faction control. Results represent the mean ± SD of three independent experiments. **, P < 0.01. (F) Representative Western blot validation of effective silencing by the Cas9-ERT2:sgTIF1γRNA system in LX2 cells transfected with Lrat:Cas9-ERT2: sgTIF1γRNA construct. Results represent the mean ± SD of three independent experiments. *, P < 0.05. (G) Immunofluorescence imaging analysis of the Cas9-ERT2/sgTIF1γ system in LX2 cells. Scale bar, 7.5 µm. Comparison of a transfected (region of interest [ROI] 1) and a nontransfected (ROI 2) cell after TMX. Reproducible results from two independent experiments are shown.
Additionally, in normal livers, most of the TIF1γ-positive cells were stained with the HSC marker CRBP1 (Van Rossen et al., 2009; Fig. 5 A and Fig. S4 A). In damaged livers of 14-d TAA–treated nude mice, TIF1γ-positive cells were significantly reduced in number (Fig. 5 B). However, transplantation of hE-MSCs significantly prevented this reduction in TIF1γ-positive cells, resulting in the maintenance of normal live architecture and cells double positive for TIF1γ and CRBP1 (15.4 ± 1.7 cells in control [no treatment] vs. 8.6 ± 1.9 cells in TAA treatment vs. 13 ± 1.6 cells in TAA/hE-MSC treatment, n ≥ 4; Fig. 5, A and B). These data indicated that TIF1γ is a potential anti-fibrosis factor expressed in HSCs that is down-regulated by profibrotic signals such as TAA or TGFβ1 and up-regulated by anti-fibrosis therapy such as hE-MSC transplantation.
Figure 5.
TIF1γ expression in liver analyzed by immunofluorescence. (A) TIF1γ and CRBP1 staining in normal, TAA-treated, and hE-MSC–transplanted mouse livers after TAA treatment. Scale bars, 20 µm. (B) Quantification of TIF1γ-positive cells in the livers at 14 d (normal, TAA, and TAA/hE-MSCs; n = 3 in each group). TIF1γ-positive cells per 0.125 mm2 were counted using immunofluorescence images (n ≥ 4 for each slide). Mean values ± SD were calculated from counted fields of each sample in each group. (C) Immunofluorescence of TIF1γ in human normal liver. TIF1γ-positive cells (arrowhead) were located in the space of Disse. Scale bar, 10 µm. (D) TIF1γ and αSMA double staining in human normal (n = 4) and cirrhotic (n = 6) liver tissues. Human liver tissue was purchased from SuperBioChip Laboratories. Black scale bars, 20 µm; white scale bars, 10 µm.
To test whether our findings in the mouse model can be extrapolated to humans, we conducted TIF1γ immunohistochemistry in human normal and cirrhotic livers with a liver fibrosis grade of ISHAK6/METAVIR-F4 (Bataller and Brenner, 2005; Standish et al., 2006). Similar to the mouse model, we observed that TIF1γ was expressed in the space of Disse of normal liver (Fig. 5, C and D) and decreased in human cirrhotic livers in parallel to an increase in αSMA (Fig. 5 D).
Generation of TG mice with inducible, HSC-specific knockout of Tif1γ: Knockout of Tif1γ accelerates liver fibrosis in mice
To elucidate whether loss of TIF1γ acts as an accelerator in fibrosis in vivo, we generated TG mice using the Cas9-ERT2 genome editing system, which is currently widely used to target genes of interest in animals (Staahl et al., 2017). We generated a Cas9-ERT2 fusion system for induction by TMX (Fig. S4 C). ERT2 is known as a nuclear receptor that is transported into the nucleus by TMX (Indra et al., 1999). To induce knock out Tif1γ specifically in HSCs, we selected individual sites of exons 1, 2, and 3 in TIF1γ for constructing guide RNAs. Moreover, we used the ∼5-kb lecithin retinal acyltransferase (LRAT) putative promoter region for exclusive expression in HSCs, because LRAT is reported to be exclusively expressed in HSCs of the mouse liver (Mederacke et al., 2013). We generated three constructs targeting each exon of the TIF1γ gene: Lrat:Cas9-ERT2: sgTif1γexon1, LRAT:Cas9-ERT2: sgTif1γexon2, and LRAT:Cas9-ERT2: sgTif1γexon3 (Fig. S4 D). Before generating TG mice by introducing the three constructs concomitantly into embryos to knock out Tif1γ in HSCs by TMX treatment, this Cas9-ERT2 system was validated in vitro using 293T and LX2 cells. In CMV promoter-Cas9-ERT2–transfected 293T cells, Cas9 was detected in the nuclear fraction in addition to the cytosol upon TMX treatment, whereas Cas9 was not detected in the nuclear fraction and was detected only in the cytosol without TMX treatment (Fig. S4 E), demonstrating that TMX treatment induces translocation of Cas9 into the nucleus.
In LRAT:Cas9-ERT2: sgTIF1γ-transfected LX2 cells, TMX treatment decreased TIF1γ and increased αSMA (Fig. S4 F). Multicolor immunofluorescence microscopic imaging (Fig. S4 G) showed that TMX treatment induced the translocalization of cytosolic Cas9 protein into the nucleus, leading to a reduction in nuclear TIF1γ and induction of αSMA in the cytosol in LRAT:Cas9-ERT2: sgTIF1γ-transfected LX2 cells as compared with nontransfected cells.
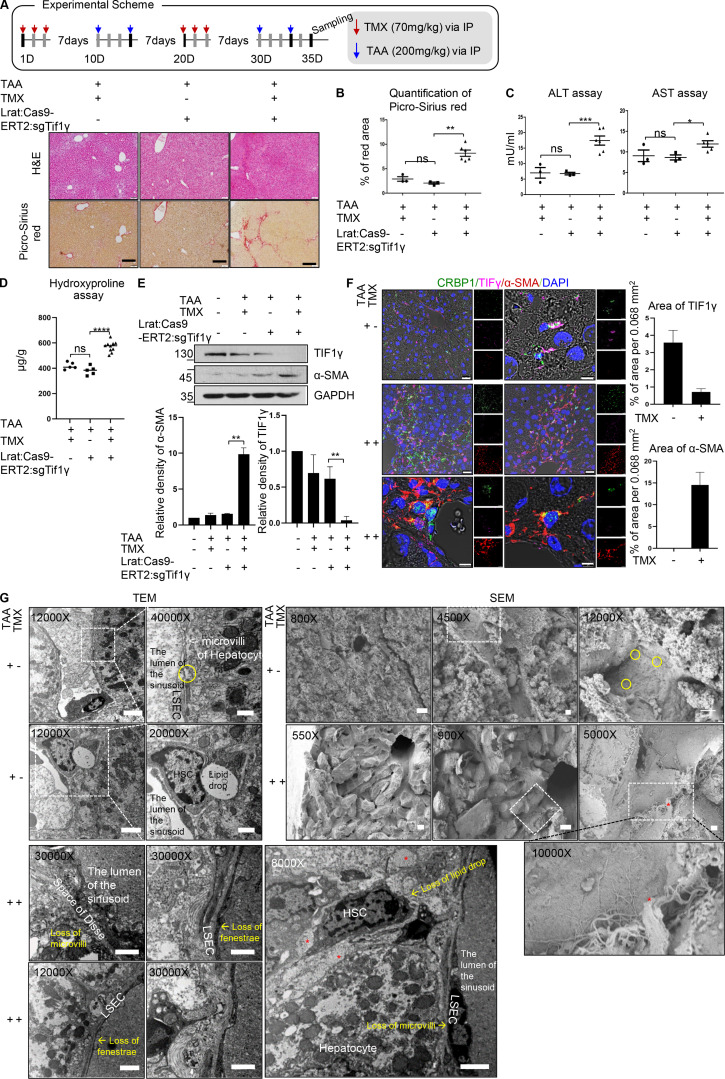
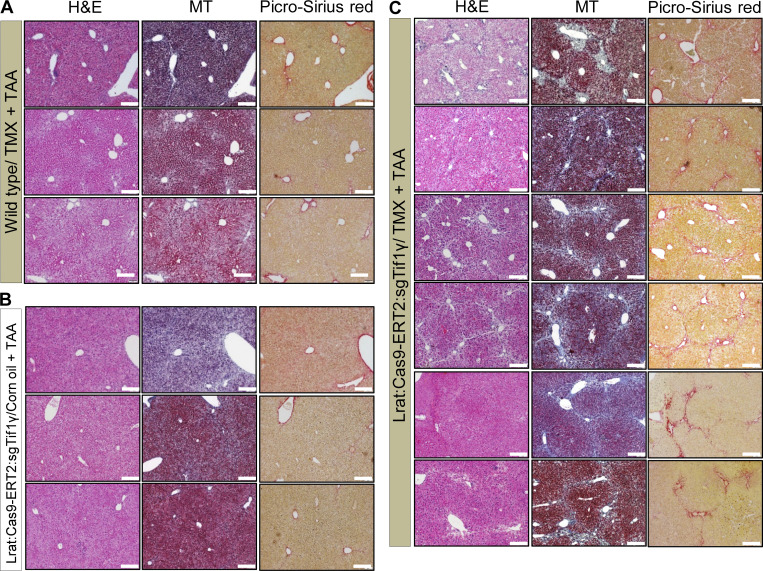
The validated DNA constructs were injected in mouse embryos to target Tif1γ by guide RNAs and to generate Lrat:Cas9-ERT2: sgTif1γ-TG mice. We treated the mice with TAA only twice to prevent severe fibrosis. Liver injury with TAA and TMX induction aggravated liver fibrosis in TG mice as compared with TMX/TAA-treated wild-type mice and corn oil/TAA–treated TG mice. TMX/TAA-treated wild-type mice, as well as corn oil/TAA-treated TG mice, showed a nearly normal liver phenotype, as indicated by Picro-Sirius red staining (2.9 ± 0.8% in TMX/TAA-treated wild-type mice vs. 2.1 ± 0.4% in corn oil/TAA-treated TG mice vs. 8.1 ± 1.6% in TMX/TAA-treated TG; Fig. 6, A and B; and Fig. S5, A–C). In the liver function test, TMX/TAA-treated TG mice showed significantly worse ALT, AST, and hydroxyproline profiles than TMX/TAA-treated wild-type mice and corn oil/TAA-treated TG mice (ALT: 7.0 ± 2.9 mU/ml in TMX/TAA-treated wild-type mice vs. 6.8 ± 0.7 mU/ml in corn oil/TAA-treated TG mice vs. 17.4 ± 3.6 mU/ml in TMX/TAA-treated TG; AST: 9.1 ± 2.4 mU/ml in TMX/TAA-treated wild-type mice vs. 8.6 ± 1.0 mU/ml in corn oil/TAA-treated TG mice vs. 11.6.4 ± 1.8 mU/ml in TMX/TAA-treated TG mice; hydroxyproline: 419.16 ± 27.65 μg/g in TMX/TAA-treated wild-type mice vs. 381.26 ± 34.63 μg/g in corn oil/TAA-treated TG mice vs. 570.54 ± 42.35 μg/g in TMX/TAA-treated TG mice; Fig. 6, C and D).
Figure 6.
Confirmation of TG mice with TMX-inducible and HSC-specific knockout of Tif1γ. (A) Observation of fibrosis and bridging structures by Picro-Sirius red staining upon knockdown of TIF1γ. Representative histochemical staining of Picro-Sirius red and H&E in each group. There were three groups: TMX/TAA in wild-type (n = 3), corn oil/TAA in TG (n = 3), and TMX/TAA in TG (n = 6). Two experiments were performed. Scale bars, 20 µm. IP, immunoprecipitation; D, day. (B) Quantification of fibrosis using Picro-Sirius red staining images. Fibrosis region was measured using ImageJ (TMX/TAA in wild-type, n = 3; corn oil/TAA in TG, n = 3; TMX/TAA in TG, n = 6). Data are expressed as the mean ± SD. Differences between groups were analyzed by one-way ANOVA (lane 1 and 2, n = 3; lane 3, n = 6).**, P < 0.01. (C) Serum levels of AST and ALT at 35 d in the experimental scheme. AST was measured through absorbance of 450 nm, and ALT was detected through 570 nm (TMX/TAA in wild-type, n = 3; corn oil/TAA in TG, n = 3; TMX/TAA in TG, n = 6; two experiments). Data are expressed as the mean ± SD. Differences between groups were analyzed by one-way ANOVA (lane 1 and 2, n = 3; lane 3, n = 6). *, P < 0.05; ***, P < 0.001. (D) Hydroxyproline assay from liver tissues at 35 d in the experimental scheme. The hydrolyzed hydroxyproline from homogenized liver tissue was detected at 560-nm absorbance using GloMax (Promega) system (TMX/TAA in wild-type, n = 3; corn oil/TAA in TG, n = 3; TMX/TAA in TG, n = 6; each sample measured one or two times; one experiment). Differences between groups were analyzed by one-way ANOVA (lane 1 and 2, n = 5; lane 3, n = 10).****, P < 0.0001. (E and F) Representative Western blot analysis (E) andimmunofluorescence of TIF1γ and αSMA protein in livers at 35 d (F). CRBP1 used as a HSC marker in immunofluorescence (quantified by ImageJ). Results represent the mean ± SD of two independent biological experiments. Scale bars represent 25 µm for the 400× image (left, lower magnification) and 2 µm for the 2,000× image (right, higher magnification). **, P < 0.01. (G) Representative transmission and SEM images of TG mice liver after corn oil/TAA treatment at 35 d. The circles indicate fenestrae of LSECs. Asterisks indicate collagen deposits in the periphery of HSCs. Scale bars represent 2 µm for 12,000× magnification, 1 µm for 20,000× and 30,000× magnification, and 500 nm for 40,000× magnification in transmission electron microscopy images. Scale bars represent 10 µm for 550×, 800×, and 900× magnification and 1 µm for 4,500×, 5,000×, and 12,000× magnification. Reproducible results from two independent experiments are shown. ns, not significant.
Figure S5.
The observation of TG mice with tamoxifen-inducible and HSC-specific knockout of Tif1γ. (A) Wild-type mouse livers after TAA injury induced with TMX were stained with HE, MT, and Picro-Sirius red, demonstrating mild fibrosis. Scale bars, 20 µm. (B) Lrat:Cas9-ERT2:sgTif1γ-TG mouse livers after TAA injury induced with vehicle corn oil were stained with HE, MT, and Picro-Sirius red, demonstrating mild fibrosis, as observed in wild-type mice. Scale bars, 20 µm. (C) Lrat:Cas9-ERT2:sgTif1γ TG mouse livers after TAA injury induced with TMX were stained with HE, MT, and Picro-Sirius red. Scale bars, 20 µm. Supplementary data of Fig. 6 A, duplicated for clarity.
Western blotting and immunofluorescence results validated TIF1γ knockout and an increase in αSMA expression in TMX/TAA-treated TG mice (Fig. 6, E and F). Limited changes in control (TMX/TAA-treated wild-type mice and corn oil/TAA–treated TG mice) as compared with normal wild-type mice were observed, indicating only a slight effect of the TAA treatment. However, TMX/TAA-treated TG mice showed lower TIF1γ and higher αSMA expression than control mice, which corresponded with the Picro-Sirius red, ALT, AST, and hydroxyproline results.
Structural changes within the fibrosis were analyzed by transmission electron microscopy (TEM) and scanning electron microscopy (SEM). We observed four important pathological findings in the livers of TMX/TAA-treated TG mice compared with corn oil/TAA-treated littermates: (1) loss of fenestrae of liver sinusoidal endothelial cells (LSECs), (2) loss of microvilli on hepatocytes, (3) loss of lipid droplets in HSCs, and (4) collagen fibrils surrounding HSCs (Fig. 6 G). SEM indicated the loss of fenestrae of LSECs, the TEM indicated the emergence of collagen fibrils (Fig. 6 G). Hepatic fibrosis showed several features in previous studies (Braet and Wisse, 2002; Elpek, 2014): (1) conversion of normal sinusoidal architecture to defenestrated capillarization of the sinusoidal endothelium, (2) transformation of fat-storing HSCs into collagen-secreting myofibroblasts, and (3) loss of hepatocyte microvilli.
Together, these findings in TG mice indicated that knockout of TIF1γ in HSCs accelerated liver fibrosis in response to injury.
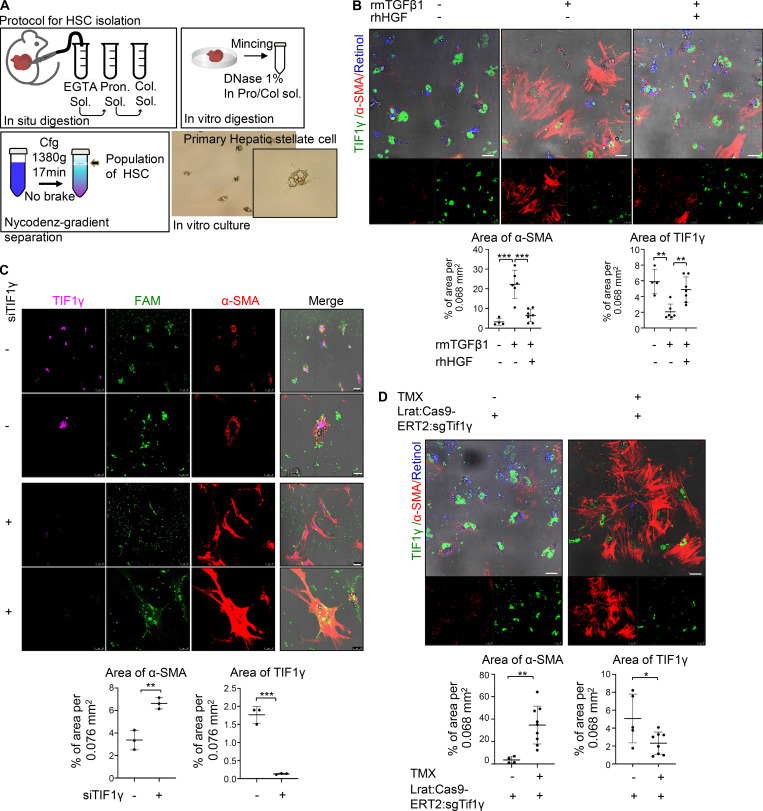
Validation of anti-fibrosis action of TIF1γ using primary HSCs
We isolated HSCs from mice (Fig. 7 A) using a previously reported method (Mederacke et al., 2015) to confirm whether the phenomenon observed in mice in vivo could be reproduced in primary HSCs. Isolated HSCs from normal mouse liver were plated on a cell-culture dish and treated with mTGFβ1 and hHGF. In accordance with the LX2 in vitro and in vivo results, Tif1γ was decreased by Tgfβ1 and increased by HGF (Fig. 7 B). Also, we confirmed that HSCs display a fibrotic character due to TIF1γ reduction (Fig. 7 C), as αSma was rapidly increased by siTif1γ, morphological changes indicating fibrosis were observed, and retinol was lost. Finally, we confirmed that HSCs prepared from Lrat:Cas9-ERT2: sgTif1γ TG mice also showed reduced Tif1γ upon treatment with TMX in vitro. Similar to the results in vivo, knockout of Tif1γ by TMX treatment induced the activation of HSCs, including an increase in αSMA (Fig. 7 D). In conclusion, these data confirm that in primary HSCs as well as LX2 cells, αSMA levels are regulated by TIF1γ.
Figure 7.
Validation of TIF1γ effect in primary HSC. (A) Schematic drawing illustrating the procedure of preparation of primary HSCs. (B) Representative immunofluorescence staining of primary HSCs prepared from a C57BL6/N wild-type mouse. Reproducible results from two independent experiments are shown. The percent TIF1γ or αSMA-positive area per 0.068 mm2 was quantified using ImageJ. Scale bars, 25 µm. **, P < 0.01; ***, P < 0.001. (C) Representative immunofluorescence staining of wild-type HSCs transfected with TIF1γ siRNA for 3 d. Reproducible results from two independent experiments are shown. Percent TIF1γ or αSMA-positive area per 0.076 mm2 was quantified using ImageJ program. Scale bars represent 25 µm (low magnification) and 10 µm (high magnification). **, P < 0.01; ***, P < 0.001. (D) Representative immunofluorescence staining of HSCs from TG mouse. HSCs treated with 10 nM TMX for 4 d after HSC isolation from TG liver. Reproducible results from two independent experiments are shown. Percent TIF1γ or αSMA-positive area per 0.068 mm2 was quantified using ImageJ. Scale bars, 25 µm. *, P < 0.05; **, P < 0.01.
Discussion
Stem cell therapy is a promising treatment option for liver fibrosis, which often requires liver transplantation. However, human adult stem cells, such as bone marrow–derived MSCs, do not always show sufficient efficacy in clinical trials (Mansilla et al., 2011; Ogden and Mickliem, 1976; Rando, 2006), and the efficacy depends on donor age, underlying diseases, and individual variations.
In our previous study, we successfully established a standard method to derive a large amount of hE-MSCs from a single preparation of hESCs, which can be generated and stored according to HLA type and thus can be used as an off-the-shelf source of allogeneic stem cells (Lee et al., 2010). Therefore, we suggest that hE-MSCs are an ideal source of stem cells for regenerative medicine, as they avoid some limitations of adult stem cells. In the current study, we found that transplantation of hE-MSCs in mouse fibrotic liver slowed down fibrosis and accelerated functional recovery of the injured liver. Normal serum levels of AST and ALT indicated that the effect of hE-MSCs was sustained systemically as well as at the target site, suggesting that hE-MSCs administered by intracardiac injection could reach and act on the injured organ. Moreover, we revealed that the administered hE-MSCs were homed to the damaged liver, where they released HGF, which induces TIF1γ in HSCs and prevents them from becoming myofibroblast-like cells, which are principal producers of extracellular matrix during liver fibrosis. The mechanisms underlying stem cell–based therapies are still under investigation; nevertheless, our results suggest that the mechanism of action of hE-MSCs in the prevention of liver fibrosis may be the secretion of paracrine factors that block the molecular mechanism for progression of fibrosis.
The pathophysiologic role of TGFβ1 in the liver is well known; TGFβ1 is crucial from initial liver injury through inflammation and fibrosis to cirrhosis and cancer (Fabregat et al., 2016). In liver fibrosis, TGFβ1 induces the activation and transformation of HSCs to αSMA-expressing myofibroblasts. TGFβ1 signaling leads to phosphorylation of the signal mediators SMAD2/3 and interaction with SMAD4 (Massagué et al., 2005). The SMAD complex can bind to DNA through the SBE and activate αSMA expression (Dennler et al., 1998; Meng et al., 2016). Therefore, targeting of TGFβ1 signaling in HSCs might be a useful therapy to prevent liver fibrosis.
TIF1γ has been suggested to inhibit TGFβ1 signaling through competition with SMAD4 for binding to activated SMAD2/3 in the differentiation of embryos and stem cells and epithelial-to-mesenchymal transition of mammary epithelial cells (He et al., 2006; Heldin and Moustakas, 2006; Hesling et al., 2011; Massagué and Xi, 2012). This study revealed the interaction TIF1γ with SMAD2/3 and the binding of this complex to the αSMA promoter in the human HSC cell line, LX2. A PLA showed that TIF1γ interacts with SMAD2/3 in the cytoplasm as well as in the nucleus in naive LX2 cells. The interaction was decreased by TGF-β1, whereas HGF effectively restored the interaction in the nucleus. This suggests that the interaction can occur in the cytosol and nucleus without a stimulator, and in response to the positive stimulator HGF, the complex translocates to the nucleus to exert its function. Next, we performed ChIP using TIF1γ antibody to know whether the TIF1γ–SMAD2/3 complex can bind the αSMA promoter. Structural–functional domains of TIF1γ have been reported (Heldin and Moustakas, 2006; Venturini et al., 1999); however, a DNA-binding sequence for binding to the promoter as a transcription factor has not been reported. Therefore, the SBE sequence was used to detect the binding of the TIF1γ–SMAD2/3 complex to the αSMA promoter after pull-down using TIF1γ antibody. The ChIP data showed that TIF1γ binds with the SMAD2/3 complex to repress αSMA expression.
For successful repression of αSMA by TIF1γ, the mechanism regulating TIF1γ levels needs to be known. Among several transcription factor–binding sites in the TIF1γ promoter region, mutation of the CREB-binding site eliminated the response to HGF, and binding of activated CREB to the TIF1γ promoter was enhanced by HGF.
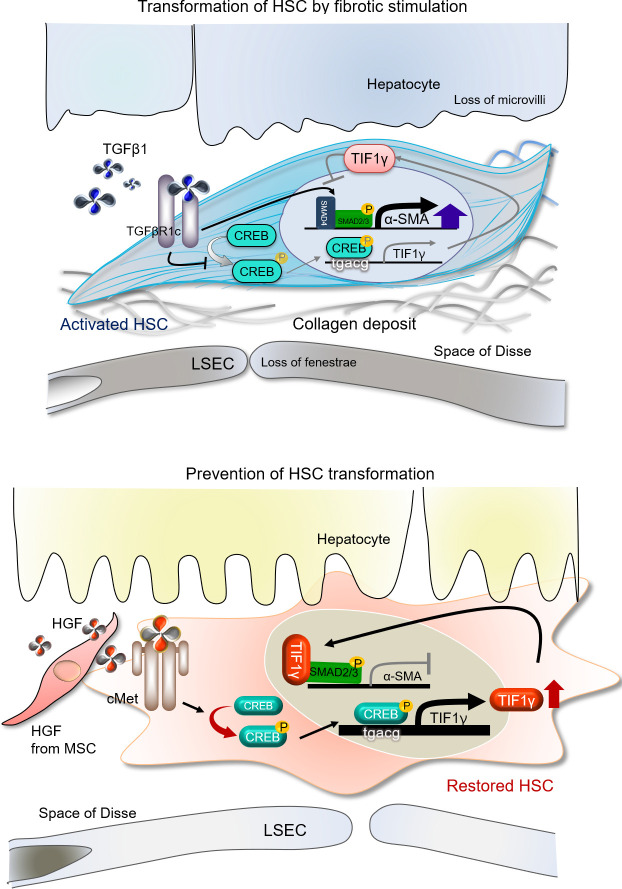
In conclusion, we suggest the following mechanisms of TIF1γ in HSCs during fibrotic stimulation and restoration (Fig. 8): suppression of CREB phosphorylation reduces TIF1γ expression and the interaction with SMAD2/3, whereas up-regulation of TIF1γ enhances the interaction with SMAD2/3 and inhibits αSMA expression. Experiments in TG mice with inducible, HSC-specific knockout of Tif1γ demonstrated that TIF1γ has potential as a novel therapeutic approach for the prevention of liver fibrosis. In addition to cell therapy using hE-MSCs, gene therapy would be feasible to directly increase or activate TIF1γ. For this purpose, we developed the construct, which induces the expression of this gene in the inflamed liver undergoing fibrosis, such as, TGFβ1-promotor–driven TIF1γ. This construct could be selectively delivered to liver stellate cells using retinol–liposome conjugate, because liver stellate cells uptake and store retinol (Sato et al., 2008).
Figure 8.
Role of TIF1γ during HSC transformation and restoration. TIF1γ expression in HSCs is decreased by TGFβ1 and increased in the presence of HGF. Phosphorylation of CREB and binding of pCREB on the TIF1γ promoter region induced TIF1γ expression. Interaction of TIF1γ with SMAD2/3 and binding of the complex to the promoter of αSMA suppressed αSMA expression. Therefore, TIF1γ has potential in the development of new therapeutic approaches to inhibiting or treating liver fibrosis.
Materials and methods
Study design
All animal study protocols were approved by the Institutional Animal Care and Use Committee (17–0040-S1A0) of Seoul National University Hospital, Korea. Male BALB/c-nude mice >12 wk (Orientbio) weighing 20–25 g were used for experiments. Mice were administered an intraperitoneal injection of 200 mg/kg TAA (Sigma-Aldrich) to induce liver fibrosis or PBS as a control three times a week for 1–3 wk. We used TAA instead of carbon tetrachloride (CCl4), which is commonly used to establish animal models of liver fibrosis, because in South Korea, CCl4 has been banned from usage under the Montreal Protocol. TAA-treated mice were randomly assigned into two groups receiving hE-MSCs or PBS once via intracardiac injection. Alternatively, TAA-treated mice were intraperitoneally injected with pCMV-hHGF (18 μg/head, HG10463-UT pCMV3-HG; Sino Biological) vector at 0, 4, and 12 d. This study was approved by the Institutional Review Board of Seoul National University Hospital, Korea (no. 1410-093-619).
LX2 culture
The human HSC line LX2 was a generous gift from Dr. Friedman (Mount Sinai School of Medicine, New York, NY). LX2 cells were grown in high-glucose DMEM supplemented with GlutaMax (Gibco), 2% FBS, and 1% (vol/vol) penicillin/streptomycin (LX2 complete medium; Gibco) at 37°C in a humidified incubator with 5% CO2. Mycoplasma contamination risk of cells used in this study was assessed via MycoQsearch Mycoplasma Real-Time PCR Detection Kit (CellSafe).
Loss- and gain-of-function analyses
Loss of function was analyzed in LX2 cells after siRNA-mediated knockdown of TIF1γ (sc-63127; Santa Cruz Biotechnology), EPLIN (sc-60593; Santa Cruz Biotechnology), or Nm23-H1 (sc-29414; Santa Cruz Biotechnology). LX2 cells were transfected with siRNA in DMEM GlutaMax without FBS using Metafectene-Pro (Biontex) for 7 h. Then, the medium was changed to complete fresh medium, and cells were incubated for 1–4 d without a change of medium. Lentiviral vector of shTIF1γ (catalog no. TL300849; OriGene) was transfected in LX2 for 24 h. Then, the medium was changed to complete fresh medium, and cells were incubated with hTGFβ1 (5 ng/ml) for 3 d.
Quantitative RT-PCR (RT-qPCR) analysis
Total RNA was isolated from cultured cells using the QIAshredder and RNeasy Plus Mini kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized from 1 µg RNA using the PrimeScript first-strand cDNA Synthesis Kit (Takara). qPCR was performed using the Power SYBR Green PCR master mix (Applied Biosystems) in an ABI PRISM-7500 sequence detection system (Applied Biosystems). GAPDH was used as an internal control to calculate relative changes in gene expression. Primers were designed using the Primer3 software (Whitehead Institute/MIT Center for Genome Research) and synthesized by Bioneer. The following primers were used: GAPDH, forward: 5′-CAACGAATTTGGCTACAGCA-3′, reverse: 5′-TGTGAGGAGGGGAGATTCA-3′; αSMA, forward: 5′-GGCAAGTGATCACCATCGGA-3′, reverse: 5′-TCTCCTTCTGCATTCGGTCG-3′; TIF1γ, forward 5′-CTCCGGGATCATCAGGTTTA-3′, reverse: 5′-ACTGCTCAACATGCAAGCAC-3′; Nm23-H1, forward 5′-GCCTGGTGAAATACATGCAC-3′, reverse: 5′-AGTTCCTCAGGGTGAAACCA-3′; EPLIN, forward 5′-CTGCGTGGAATGTCAGAAGA-3′, reverse: 5′-TTTTGCTTGCCCATAGATCC-3′; PIAS1, forward 5′-CATCGCCATTACTCCCTGTT-3′, reverse: 5′-AAGCGCTGACTGTTGTCTGA-3′; ALR, forward 5′-CCTGTGAGGAGTGTGCTGAA-3′, reverse: 5′-TCCACTTTTGAGCAGTCGAA-3′; and MBNL1, forward 5′-CAGCCGCCTTTAATCCCTAT-3′, reverse: 5′-TGTCAGCAGGATGAGCAAAC-3′.
hE-MSC culture
hE-MSCs were obtained as previously described (Lee et al., 2010). In brief, SNUhES3 hESCs (Institute of Reproductive Medicine and Population, Medical Research Center, Seoul National University Hospital, Seoul, Korea) were cultured in culture dishes without fibroblast growth factor-2 to establish embryonic bodies at 37°C with 5% CO2 for 14 d. The embryonic bodies were attached to gelatin-coated dishes for 16 d in low-glucose DMEM (Invitrogen) with 10% FBS (Invitrogen), and the derived cells were expanded in EGM-2MV medium (Lonza). The expanded hE-MSCs were tested for differentiation into adipocytes, osteocytes, myocytes, and chondrocytes under the appropriate conditions to evaluate their differentiation potential. For in vitro and in vivo experiments, hE-MSCs of passages 13–14 were used.
To knock down HGF, hE-MSCs were transduced with shHGF lentivirus (infectious units of virus, 5 × 105) specific to HGF (catalog no. TL312467V; OriGene), and knockdown was verified via ELISA.
Mycoplasma contamination risk of cells used in this study was assessed using the MycoQsearch Mycoplasma Real-Time PCR Detection Kit (CellSafe).
Co-culture
LX2 cells were plated on 10-cm dishes (2 × 105 cells/ml, Nunc; Thermo Fisher Scientific) and incubated at 37°C with 5% CO2 for 2–3 d. When cultures reached ∼50% confluence, the medium was changed to fresh medium containing 0.5% FBS and the cells were treated with 5 ng/ml hTGFβ1 (R&D Systems) every other day for 4 d. The medium was changed at every treatment with the cytokine. LX2 cells pretreated with hTGFβ1 were co-cultured with 8 × 105 hE-MSCs per dish in a transwell insert (0.4-µm pore size; Corning) in complete fresh medium containing 0.5% FBS and 5 ng/ml hTGFβ1, and samples were harvested after co-culture. Alternatively, hE-MSCs treated with small hairpin RNA specific to HGF were co-cultured with LX2 cells activated with hTGFβ1, after which 10 ng/ml or 20 ng/ml rhHGF (R&D Systems) was added.
Western blot assay
Cells or tissue samples were lysed in protein lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% deoxycholate, 1% NP-40, and 0.1% SDS with protease inhibitor cocktail; Roche). Total protein extracts (25–30 µg) were boiled for 5 min at 95°C, separated by SDS-PAGE, and transferred to polyvinylidene fluoride membranes (Merck) using a Bio-Rad transfer unit. Membranes were blocked with 5% skim milk diluted in Tris-buffered saline containing 0.1% Tween-20 and incubated with antibodies against TIF1γ (1:1,000, ab84455, human TRIM33 aa 1,077–1,127 [C terminal]; Abcam), αSMA (1:3,000, C6198, N-terminal synthetic decapeptide of αSMA; Sigma-Aldrich), EPLIN (1:500, ab50196, synthetic peptide: GVLAASMEAK ASSQQEKEDK PAETKKLRIA WPPPTELGSS GSALEEGIKM, corresponding to amino acids 502–551 of human EPLIN; Abcam), and anti-Nm23-H1 (1:1,000, sc-465, purified nm23-H1 of human origin; Santa Cruz Biotechnology); anti-α-tubulin antibody (1:5,000, T6199, the C-terminal end of the α-tubulin isoform [aa 426–430]; Sigma-Aldrich) or anti-GAPDH antibody (1:3,000, ab9485, full-length native protein [purified] corresponding to human GAPDH; Abcam) were used to detect internal control proteins. Membranes were washed and incubated with HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and washed, and immunoreactive bands were detected using Luminata Classicco (Merck).
Luciferase assay
LX2 cells were seeded in 12-well plates (Nunc; Thermo Fisher Scientific) and transfected with 100 ng firefly and 10 ng Renilla constructs (Promega) using FuGENE HD (Promega) for 6 h. Luciferase activity was measured following to the manufacturer’s protocols using a GLOMAX 20/20 luminometer (Promega).
ELISA
The secretion of HGF in cell culture supernatants was analyzed by ELISA using the hHGF ELISA kit (Cusabio Biotech) according to the manufacturer’s protocols. The absorbance at 450 nm was measured using a Multiskan GO Microplate Spectrophotometer (Thermo Fisher Scientific).
hHGF in the serum was detected using hHGF ELISA kit (SHG00B; R&D Systems) in accordance with the manufacturer’s instructions. The serum was prepared using serum collection tube (microtainer, 365967; BD) and diluted 1:2 with calibrator diluent RD59 buffer provided from the kit. Briefly, the samples, standard of recombinant hHGF, and positive control were added to the precoated 96-well, washed and treated with the detection antibody and HRP-conjugated antibody. The visualization of the captured hHGF was accomplished with 3,3′,5,5′-tetramethylbenzidine substrate solution. Yellow intensity was detected using the Glomax (Promega) system at 450 nm (normalized at 570 nm). Calculations were performed automatically (https://www.hycultbiotech.com/elisa_calculationsheet).
Subcellular protein fractionation
293T cells (ATCC) were collected in a tube after trypsinization. Nuclear-cytoplasmic buffer (0.5% digitonin prepared in PBS containing a phosphatase and protease inhibitor cocktails; genDEPOT) was added at fourfold the packed cell volume and pipetted carefully. After centrifugation, the supernatant was retained as the cytoplasmic fraction, and the pellet was resuspended in RIPA buffer (Thermo Fisher Scientific). Lysate in RIPA was retained as the nuclear fraction. α-Tubulin (anti-α-tubulin antibody (1:5,000, T6199, the C-terminal end of the α-tubulin isoform [aa 426–430]; Sigma-Aldrich) was assayed as a cytoplasmic fraction–specific marker, and lamin A/C (catalog no. 2032S, endogenous levels of total full-length lamin A; Cell Signaling) was used as a nuclear fraction–specific marker.
PLA
All procedures refer to the method provided by the Duolink kit (DUO92101-1KT; Sigma-Aldrich). LX2 cells were treated with 4% paraformaldehyde (163–20145-P01; Wako) for 15 mi and permeablized with 0.5% Triton X-100 (catalog no. T8787; Sigma-Aldrich) in PBS. A blocking step was performed with blocking solution provided from this kit for 1 h, and then primary antibodies were treated with anti-TIF1γ (1:100, ab47062, synthetic peptide corresponding to human TRIM33 [aa 600–700] conjugated to keyhole limpet hemocyanin; Abcam) and anti-Smad2/3 (1:50, sc-6032, recombinant Smad2 of human origin; Santa Cruz Biotechnology) at 4°C for 16 h. Samples washed with wash buffer A three times and then treated with Duolink PLA probe plus and minus for 1 h at 37°C. The ligation and amplification steps were performed to visualize signal. Images were acquired using a confocal microscope (LSM710; Carl Zeiss) and analyzed with ImageJ (National Institutes of Health) software.
ChIP assay
LX2 cells were treated with 1% formaldehyde (F8775; Sigma-Aldrich) for cross-linking for 10 min, and the reaction was quenched using 125 mM glycine (G8898; Sigma-Aldrich) for 5 min. Cells were then lysed using ChIP buffer (50 mM, NaCl 150 mM Tris-HCl, pH 8.0, 0.5% Triton X-100, 1 mM EDTA, 0.1% sodium deoxycholate, and 0.1% SDS including 1× Xpert protease inhibitor cocktail; genDEPOT) and sonicated to shear the DNA to 200–500-bp fragments using a BIORUPTOR sonicator (Diagenode). The target antibody was added, and the protein A/G agarose bead (Abcam) was added to pull down the target DNA. Beads were washed three times sequentially with different wash buffers (low-salt buffer: 20 mM Tris-HCl, 150 mM NaCl, 0.5% Triton X-100, and 0.1 mM EDTA; high-salt buffer: same as low-salt buffer except for 500 mM NaCl; lithium-chloride buffer: same as low-salt buffer except for 250 mM LiCl instead of NaCl). The samples were heated to 65°C for 4 h to remove the cross-links. The DNA fragments were recovered using a PCR purification kit (catalog no. 28106; Qiagen) and analyzed using qPCR and semiquantitative PCR.
The following primers were used for ChIP on the human αSMA promoter: −84 SBE forward, 5′-GAGAGTTTTGTGCTGAGGTCCC-3′; −84 SBE reverse, 5′-CCTGCTCTCCTCCCACTTGC-3′; −602 SBE forward, 5′-AAGGATGGTCCCTACTTATGCTG-3′; −602 SBE reverse, 5′-GGGAGGTGAGTGGAAATAGGAA-3′; non-SBE forward, 5′-AAAGGTGGGAAATGGAAAGG-3′; non-SBE reverse, 5′-TCTGCTGGTGCCGAAAAAT-3′.
Antibodies (epitope)
Mouse monoclonal [7A9-3A3] a-CRISPR-Cas9 (ab191468, recombinant fragment corresponding to S. pyogenes CRISPR-Cas9 [N terminal]; Abcam), rabbit polyclonal a-HGF (ab83760, synthetic peptide corresponding to a region within the N-terminal sequence 108–157 [VKKEFGHEFD LYENKDYIRN CIIGKGRSYK GTVSITKSGI KCQPWSSMIP] of human HGF, NP_001010932; Abcam), mouse monoclonal [24612.111] a-HGF (ab10678, rhHGF expressed in the insect cell line Sf 21; Abcam), mouse monoclonal a-CRBP1 (sc-271208, 1–135 aa of CRBP1 of human origin; Santa Cruz Biotechnology), rabbit polyclonal a-CRBP1 (sc-30106; Santa Cruz Biotechnology), rabbit polyclonal a-TIF1γ (ab47062, synthetic peptide corresponding to human TRIM33 [aa 600–700] conjugated to keyhole limpet hemocyanin; Abcam), and mouse monoclonal Cas9 (ab191468; Abcam, Recombinant fragment corresponding to S. pyogenes CRISPR-Cas9 [N terminal]) were used for immunofluorescence. Rat monoclonal [YOL1/34] a-α tubulin (sc-53030, raised against full-length purified α tubulin of Saccharomyces cerevisiae origin; Santa Cruz Biotechnology), rabbit polyclonal a-EPLIN (ab50196, a region within synthetic peptide [GVLAASMEAK ASSQQEKEDK PAETKKLRIA WPPPTELGSS GSALEEGIKM] corresponding to aa 502–551 of human EPLIN; Abcam), mouse monoclonal a-Nm23-H1 (sc-56928, Santa Cruz Biotechnology, raised against full-length nm23-H1 of human origin), rabbit polyclonal a-αSMA (ab5694, Abcam, raised against a synthetic peptide corresponding to N-terminus of actin from human smooth muscle), rabbit polyclonal a-TIF1γ (ab84455, Abcam, Synthetic peptide within Human TRIM33 aa 1,077–1,127 [C terminal]), rabbit polyclonal a-GAPDH (ab9485, full-length native protein [purified] corresponding to human GAPDH; Abcam), and mouse monoclonal Cas9 (ab191468, recombinant fragment corresponding to S. pyogenes CRISPR-Cas9 [N terminal]; Abcam) were used for immunoblot assays. Rabbit monoclonal a-CREB (9197s, recombinant protein specific to the amino terminus of human CREB-1 protein; Cell Signaling) and rabbit monoclonal a-S133 P-CREB (9198s, synthetic phosphopeptide corresponding to residues surrounding Ser133 of human CREB; Cell Signaling) were used for ChIP assay and Western blot analysis.
Immunohistochemistry
After blood collection, mouse livers were perfused with cold PBS and removed. The livers were fixed in a 10% neutral formalin solution, embedded in paraffin, and cut into serial sections (4–5 µm thick). Paraffin sections were stained with H&E (ab24588; Abcam), Masson's trichrome (MT; CS-MTRI; IHC World), or Picro-Sirius red (365548; Sigma-Aldrich) using standard protocols. MT and Picro-Sirius red staining was used to detect collagen to visualize connective tissues. Images were obtained using a Leica light microscope. To evaluate the therapeutic effect of hE-MSCs, the percentage of the fibrotic liver area was estimated by quantitative image analysis of MT- and Picro-Sirius red–stained sections using the SABIA (Metoosoft) and ImageJ (National Institutes of Health) software packages. The degree of liver fibrosis was represented according to the METAVIR scale or Ishak stage, which grade fibrosis from F0 (no fibrosis) to F4 (cirrhosis) and from 0 (no fibrosis) to 6 (cirrhosis), respectively.
Human liver tissues purchased from SuperBioChip Laboratories and paraffin-embedded tissue sections were deparaffinized in xylene and rehydrated in graded alcohol. After sections were subjected to heat-mediated antigen retrieval with citrate buffer (DAKO), nonspecific binding sites were blocked with 1% bovine serum albumin in PBS containing 0.01% Triton X-100. Depending on the antibody used, permeabilization was optionally conducted with 0.1% Triton X-100 in PBS for 10 min before blocking. Then, the tissue sections were incubated with the following primary antibodies overnight at 4°C: anti-TIF1γ (1:1,000; Abcam), anti-cellular retinol-binding protein 1 (CRBP1, 1:100; Santa Cruz Biotechnology), anti-αSMA (1:800; Sigma-Aldrich), anti-hepatocyte (hepatocyte paraffin-1 [Hep Par-1]; 1:300; DAKO), anti-Cas9 (1:50; Abcam), or anti-HGF (1:100; Abcam). After washing, the sections were incubated with secondary Alexa Fluor–conjugated antibodies (Invitrogen) for 2 h at room temperature, washed with PBS, and mounted in fluorescence mounting medium with 4′,6-diamidino-2-phenylindole (IHC World). Images were acquired using a confocal microscope (LSM710; Carl Zeiss).
Construction of TMX-inducible TIF1γ-knockout vector
pCAG-ERT2CreERT2 (catalog no. 13777) and pX330-U6-Chimeric_BB-CBh-hSpCas9 (catalog no. 42230) plasmids were purchased from Addgene, and murine Tif1γ CRISPR/Cas9 knockout plasmid (sc-430111) was purchased from Santa Cruz Biotechnology. The murine Lrat promoter sequence was predicted in GPminer described above (Lee et al., 2012), and a region (−5,500 to +72 bp) with predicted high activity was selected. The Lrat promoter was obtained by PCR (forward: 5′-GACATTGATTATTGACTAGTCCTTAAAGAGAGGCATCCGGGGTC-3′; reverse: 5′-GTTCTTCTCCTTTGCTAGCCATGACGCTCACGCTAAAGAGCTTGAAG-3′) using normal murine DNA (C57BL/6N). To analyze Lrat promoter activity, the CMV promoter of pcDNA DEST47 was replaced with the Lrat promoter. To generate the Lrat promoter–dependent and TMX-induced TIF1γ-knockout construct, the CAG promoter and Cre of pCAG-ERT2CreERT2 were replaced with the Lrat promoter and SpCas9. Next, three guide RNAs (gRNAs) for Tif1γ with the U6 promoter (gRNA 1, 5′-GGTGCGGCTGGGCCCGACGA-3′; gRNA 2, 5′-CTACATTCTTGACGACATAC-3′; gRNA 3, 5′-GAAGATAATGCAAGTGCAGT-3′) were inserted into the plasmids. pLrat:Cas9-ERT2: sgTif1γ constructs were confirmed by Sanger sequencing.
Generation of TMX-inducible TIF1γ-knockout mice
TG mice expressing pLrat:Cas9-ERT2: sgTif1γ were generated and interbred in pathogen-free conditions at Macrogen (Seoul, Korea). All manipulations were conducted with the approval of Macrogen Institutional Animal Care and Use Committee. To prepare embryos, C57BL/6N female mice of 5–8 wk of age were intraperitoneally injected at 48-h intervals with pregnant mare serum gonadotropin (7.5 IU) and human chorionic gonadotropin (5 IU) for superovulation. After the injections, female mice were mated with C57BL/6N stud male mice. Female mice with vaginal plugs were sacrificed, and fertilized embryos were harvested. The three pLrat:Cas9-ERT2: sgTif1γ DNAs were linearized, and the same concentrations of these constructs were microinjected into one cell of each embryo using standard microinjection procedures (Macrogen). Briefly, 4 ng/µl of a mixture of the three constructs was injected directly into the pronucleus of the zygote using a micromanipulator, and microinjected embryos were incubated at 37°C for 1–2 h. 14–16 injected one-cell-stage embryos were transplanted by surgical methods into the oviducts of pseudopregnant recipient mice (Institute of Cancer Research). Founders were identified by PCR using tail genomic DNA and primers specific to Cas9-ERT2 (forward: 5′-TGCTACAGAACAGTTGCAGCC-3′; reverse: 5′-ACCTTGTACTCGTCGGTGATC-3′) and Tif1γ gRNAs (U6 forward: 5′-GTCGACGAGGGCCTATTTCCCATGATT-3′; gRNA1 reverse: 5′-TCGTCGGGCCCAGCCGCACC-3′; gRNA2 reverse: 5′-GTATGTCGTCAAGAATGTAG-3′; and gRNA3 reverse: 5′-ACTGCACTTGCATTATCTTC-3′). After generation to F2, male 12-wk-old mice were used for experiments.
Serum assays
Blood samples were drawn from the hearts of anesthetized mice at 14 d after transplantation. Serum was separated by centrifugation at 3,000 rpm for 15 min and stored at −80°C until analysis. To test liver function after TAA treatment, ALT and AST activity was measured using an automatic chemistry analyzer (Hitachi 7070) according to the manufacturer’s instructions. Alternatively, AST (ab105135; Abcam) and ALT (ab105134; Abcam) assays were performed using the colorimetric method in TG mice. The substrates, glutamate and pyruvate, were used for generating standard curves to measure enzyme amounts. AST and ALT activities were determined following the manufacturer’s procedure and were expressed as milliunits per milliliter.
Hydroxyproline assay
Hydroxyproline from collagen was detected using a hydroxyproline assay kit (ab222941; Abcam) according to the manufacturer’s instructions. Briefly, 10 mg liver tissue was homogenized with distilled water and boiled, including the same volume of 10 N NaOH, for 1 h and neutralized using 10 N HCl. The precipitant was centrifuged and collected. 33 μg/10 μl samples were then dried on a 65°C hotplate. Measurement at 560 nm absorbance was obtained using the GloMax (Promega) system, and the following formula was used for calculation: hydrolyzed hydroxyproline concentration = B (amount of hydroxyproline)/V (sample volume) × D (dilution factor).
Electron microscopy
Liver samples for electron microscopy were fixed with 2.5% glutaraldehyde and cut into ∼1-mm3 pieces for transmission electron microscopy. Briefly, after dehydration, thin sections were stained with saturated uranyl acetate and lead citrate and observed under a JEM-1400 Plus transmission electron microscope. For SEM, each sample was ion-sputter coated and observed with a Hitachi S-4700 scanning electron microscope.
Primary HSC isolation and culture
Primary HSCs were cultured following the protocol reported by Mederacke et al. (2015). For in situ digestion, mice were sequentially perfused with EGTA, pronase, and collagenase solutions. Liver tissues were separated and digested in situ with 1% DNase for 25 min. After passage through a strainer, the cells were washed and aspirated with Gey's Balanced Salt Solution two times. HSCs were separated by centrifugation at 1,380 ×g on Nycodenz density gradient for 17 min without a brake. HSCs were cultured in DMEM containing 10% FBS.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 6 software (GraphPad Software). Data are expressed as the mean ± SD. Differences between groups were analyzed by the unpaired t test or one-way ANOVA. P values <0.05 were considered statistically significant.
Online supplemental material
Fig. S1 shows the process for selection of TIF1γ using an in vitro cell system. Fig. S2 shows the expression of hHGF by hE-MSC or shHGF or pCMV-hHGF in vivo and in vitro. Fig. S3 shows the screening and analysis of transcription factor on the Tif1γ promoter. Fig. S4 shows the expression of Tif1γ in normal mouse liver or HSCs and validation for the development of TG mice with TMX-inducible and HSC-specific knockout of Tif1γ. Fig. S5 shows the observation of TG mice with TMX-inducible and HSC-specific knockout of Tif1γ. Table S1 shows six anti-fibrosis factor candidates. Table S2 shows primers for mRNA of hHGF or mouse HGF (mHGF) in mouse liver.
Supplementary Material
shows six anti-fibrosis factor candidates.
shows primers for mRNA of hHGF or mHGF in mouse liver.
Acknowledgments
We thank Dr. Friedman for gifting LX2 cells.
This study was supported by the Korea Health Industry Development Institute (Korea Health Technology R&D Project Strategic Center of Cell and Bio Therapy grant HI17C2085 and Korea Research-Driven Hospital grant HI14C1277), funded by the Ministry of Health & Welfare, Republic of Korea. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions: Study concept and design: E.J. Lee and H. Kim. Acquisition, analysis, or interpretation of data: all authors. Drafting of the manuscript: E.J. Lee, I. Hwang, and H. Kim. Critical revision of the manuscript for important intellectual content: all authors. Obtained funding: H. Kim. Technical support: E.J. Lee, I. Hwang, J.Y. Lee, J.N. Park, K.C. Kim, I. Kim, D. Moon, S. Lee, D.W. Jun, H.S. Kim, and S. Park. E.J. Lee, I. Hwang, J.N. Park, and K.C. Kim.
References
- Baertschiger, R.M., Serre-Beinier V., Morel P., Bosco D., Peyrou M., Clément S., Sgroi A., Kaelin A., Buhler L.H., and Gonelle-Gispert C.. 2009. Fibrogenic potential of human multipotent mesenchymal stromal cells in injured liver. PLoS One. 4:e6657. 10.1371/journal.pone.0006657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataller, R., and Brenner D.A.. 2005. Liver fibrosis. J. Clin. Invest. 115:209–218. 10.1172/JCI24282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm, F., Köhler U.A., Speicher T., and Werner S.. 2010. Regulation of liver regeneration by growth factors and cytokines. EMBO Mol. Med. 2:294–305. 10.1002/emmm.201000085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braet, F., and Wisse E.. 2002. Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: a review. Comp. Hepatol. 1:1. 10.1186/1476-5926-1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burra, P., Arcidiacono D., Bizzaro D., Chioato T., Di Liddo R., Banerjee A., Cappon A., Bo P., Conconi M.T., Parnigotto P.P., et al. 2012. Systemic administration of a novel human umbilical cord mesenchymal stem cells population accelerates the resolution of acute liver injury. BMC Gastroenterol. 12:88. 10.1186/1471-230X-12-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayoub, R., Wagner H., Bataille F., Stöltzing O., Spruss T., Buechler C., Schlitt H.J., and Weiss T.S.. 2011. Liver regeneration associated protein (ALR) exhibits antimetastatic potential in hepatocellular carcinoma. Mol. Med. 17:221–228. 10.2119/molmed.2010.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler, S., Itoh S., Vivien D., ten Dijke P., Huet S., and Gauthier J.M.. 1998. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091–3100. 10.1093/emboj/17.11.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elpek, G.O. 2014. Cellular and molecular mechanisms in the pathogenesis of liver fibrosis: An update. World J. Gastroenterol. 20:7260–7276. 10.3748/wjg.v20.i23.7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat, I., Moreno-Càceres J., Sánchez A., Dooley S., Dewidar B., Giannelli G., and Ten Dijke P.. IT-LIVER Consortium . 2016. TGF-β signalling and liver disease. FEBS J. 283:2219–2232. 10.1111/febs.13665 [DOI] [PubMed] [Google Scholar]
- He, W., Dorn D.C., Erdjument-Bromage H., Tempst P., Moore M.A., and Massagué J.. 2006. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 125:929–941. 10.1016/j.cell.2006.03.045 [DOI] [PubMed] [Google Scholar]
- Heldin, C.H., and Moustakas A.. 2006. A new twist in Smad signaling. Dev. Cell. 10:685–686. 10.1016/j.devcel.2006.05.006 [DOI] [PubMed] [Google Scholar]
- Hesling, C., Fattet L., Teyre G., Jury D., Gonzalo P., Lopez J., Vanbelle C., Morel A.P., Gillet G., Mikaelian I., and Rimokh R.. 2011. Antagonistic regulation of EMT by TIF1γ and Smad4 in mammary epithelial cells. EMBO Rep. 12:665–672. 10.1038/embor.2011.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra, A.K., Warot X., Brocard J., Bornert J.M., Xiao J.H., Chambon P., and Metzger D.. 1999. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 27:4324–4327. 10.1093/nar/27.22.4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E.J., Lee H.N., Kang H.J., Kim K.H., Hur J., Cho H.J., Lee J., Chung H.M., Cho J., Cho M.Y., et al. 2010. Novel embryoid body-based method to derive mesenchymal stem cells from human embryonic stem cells. Tissue Eng. Part A. 16:705–715. 10.1089/ten.tea.2008.0596 [DOI] [PubMed] [Google Scholar]
- Lee, E.J., Xu L.J., Kim G.H., Kang S.K., Lee S.W., Park S.H., Kim S., Choi T.H., and Kim H.S.. 2012. Regeneration of peripheral nerves by transplanted sphere of human mesenchymal stem cells derived from embryonic stem cells. Biomaterials. 33:7039–7046. 10.1016/j.biomaterials.2012.07.053 [DOI] [PubMed] [Google Scholar]
- Lee, E.J., Hwang I., Lee J.Y., Park J.N., Kim K.C., Kim G.H., Kang C.M., Kim I., Lee S.Y., and Kim H.S.. 2018. Hepatocyte Growth Factor Improves the Therapeutic Efficacy of Human Bone Marrow Mesenchymal Stem Cells via RAD51. Mol. Ther. 26:845–859. 10.1016/j.ymthe.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.-Y., Chang W.-C., Hsu J.B.-K., Chang T.-H., and Shien D.-M.. 2012. GPMiner: an integrated system for mining combinatorial cis-regulatory elements in mammalian gene group. BMC Genomics. 13(S3, https://doi.org/10.1186/1471-2164-13-S1-S3). 10.1186/1471-2164-13-S1-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla, E., Díaz Aquino V., Zambón D., Marin G.H., Mártire K., Roque G., Ichim T., Riordan N.H., Patel A., Sturla F., et al. 2011. Could metabolic syndrome, lipodystrophy, and aging be mesenchymal stem cell exhaustion syndromes? Stem Cells Int. 2011:943216. 10.4061/2011/943216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué, J., and Xi Q.. 2012. TGF-β control of stem cell differentiation genes. FEBS Lett. 586:1953–1958. 10.1016/j.febslet.2012.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué, J., Seoane J., and Wotton D.. 2005. Smad transcription factors. Genes Dev. 19:2783–2810. 10.1101/gad.1350705 [DOI] [PubMed] [Google Scholar]
- Mederacke, I., Hsu C.C., Troeger J.S., Huebener P., Mu X., Dapito D.H., Pradere J.P., and Schwabe R.F.. 2013. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun. 4:2823. 10.1038/ncomms3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederacke, I., Dapito D.H., Affò S., Uchinami H., and Schwabe R.F.. 2015. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat. Protoc. 10:305–315. 10.1038/nprot.2015.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X.M., Nikolic-Paterson D.J., and Lan H.Y.. 2016. TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 12:325–338. 10.1038/nrneph.2016.48 [DOI] [PubMed] [Google Scholar]
- Metzger, D., and Chambon P.. 2001. Site- and time-specific gene targeting in the mouse. Methods. 24:71–80. 10.1006/meth.2001.1159 [DOI] [PubMed] [Google Scholar]
- Netherton, S.J., and Bonni S.. 2010. Suppression of TGFβ-induced epithelial-mesenchymal transition like phenotype by a PIAS1 regulated sumoylation pathway in NMuMG epithelial cells. PLoS One. 5:e13971. 10.1371/journal.pone.0013971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden, D.A., and Mickliem H.S.. 1976. The fate of serially transplanted bone marrow cell populations from young and old donors. Transplantation. 22:287–293. 10.1097/00007890-197609000-00010 [DOI] [PubMed] [Google Scholar]
- Pardali, E., Sanchez-Duffhues G., Gomez-Puerto M.C., and Ten Dijke P.. 2017. TGF-β-Induced Endothelial-Mesenchymal Transition in Fibrotic Diseases. Int. J. Mol. Sci. 18:2157. 10.3390/ijms18102157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando, T.A. 2006. Stem cells, ageing and the quest for immortality. Nature. 441:1080–1086. 10.1038/nature04958 [DOI] [PubMed] [Google Scholar]
- Sato, Y., Murase K., Kato J., Kobune M., Sato T., Kawano Y., Takimoto R., Takada K., Miyanishi K., Matsunaga T., et al. 2008. Resolution of liver cirrhosis using vitamin A-coupled liposomes to deliver siRNA against a collagen-specific chaperone. Nat. Biotechnol. 26:431–442. 10.1038/nbt1396 [DOI] [PubMed] [Google Scholar]
- Shi, Z., Wakil A.E., and Rockey D.C.. 1997. Strain-specific differences in mouse hepatic wound healing are mediated by divergent T helper cytokine responses. Proc. Natl. Acad. Sci. USA. 94:10663–10668. 10.1073/pnas.94.20.10663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L.L., Liu F.P., and Wang D.W.. 2011. Transplantation of human umbilical cord blood mesenchymal stem cells improves survival rates in a rat model of acute hepatic necrosis. Am. J. Med. Sci. 342:212–217. 10.1097/MAJ.0b013e3182112b90 [DOI] [PubMed] [Google Scholar]
- Staahl, B.T., Benekareddy M., Coulon-Bainier C., Banfal A.A., Floor S.N., Sabo J.K., Urnes C., Munares G.A., Ghosh A., and Doudna J.A.. 2017. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 35:431–434. 10.1038/nbt.3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standish, R.A., Cholongitas E., Dhillon A., Burroughs A.K., and Dhillon A.P.. 2006. An appraisal of the histopathological assessment of liver fibrosis. Gut. 55:569–578. 10.1136/gut.2005.084475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai, S., Sakaida I., Nishina H., and Okita K.. 2005. Lesson from the GFP/CCl4 model--translational research project: the development of cell therapy using autologous bone marrow cells in patients with liver cirrhosis. J. Hepatobiliary Pancreat. Surg. 12:203–207. 10.1007/s00534-005-0977-0 [DOI] [PubMed] [Google Scholar]
- Vajda, N.A., Brimacombe K.R., LeMasters K.E., and Ladd A.N.. 2009. Muscleblind-like 1 is a negative regulator of TGF-beta-dependent epithelial-mesenchymal transition of atrioventricular canal endocardial cells. Dev. Dyn. 238:3266–3272. 10.1002/dvdy.22155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rossen, E., Vander Borght S., van Grunsven L.A., Reynaert H., Bruggeman V., Blomhoff R., Roskams T., and Geerts A.. 2009. Vinculin and cellular retinol-binding protein-1 are markers for quiescent and activated hepatic stellate cells in formalin-fixed paraffin embedded human liver. Histochem. Cell Biol. 131:313–325. 10.1007/s00418-008-0544-2 [DOI] [PubMed] [Google Scholar]
- Venturini, L., You J., Stadler M., Galien R., Lallemand V., Koken M.H., Mattei M.G., Ganser A., Chambon P., Losson R., and de Thé H.. 1999. TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 18:1209–1217. 10.1038/sj.onc.1202655 [DOI] [PubMed] [Google Scholar]
- Wang, P.P., Xie D.Y., Liang X.J., Peng L., Zhang G.L., Ye Y.N., Xie C., and Gao Z.L.. 2012. HGF and direct mesenchymal stem cells contact synergize to inhibit hepatic stellate cells activation through TLR4/NF-kB pathway. PLoS One. 7:e43408. 10.1371/journal.pone.0043408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, J., Chen Y., Wu Y., Wang Z., Zhou A., Zhang S., Lin K., Aldape K., Majumder S., Lu Z., and Huang S.. 2015. Tumour suppressor TRIM33 targets nuclear β-catenin degradation. Nat. Commun. 6:6156. 10.1038/ncomms7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Wang X., Osunkoya A.O., Iqbal S., Wang Y., Chen Z., Müller S., Chen Z., Josson S., Coleman I.M., et al. 2011. EPLIN downregulation promotes epithelial-mesenchymal transition in prostate cancer cells and correlates with clinical lymph node metastasis. Oncogene. 30:4941–4952. 10.1038/onc.2011.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R., Gong L., Li L., Guo L., Zhu D., Wu Z., and Zhou Q.. 2013. nm23-H1 is a negative regulator of TGF-β1-dependent induction of epithelial-mesenchymal transition. Exp. Cell Res. 319:740–749. 10.1016/j.yexcr.2012.10.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows six anti-fibrosis factor candidates.
shows primers for mRNA of hHGF or mHGF in mouse liver.