Abstract
Limb regeneration is the outcome of a complex sequence of events that are mediated by interactions between cells derived from the tissues of the amputated stump. Early in regeneration, these interactions are mediated by growth factor/morphogen signaling associated with nerves and the wound epithelium. One shared property of these proregenerative signaling molecules is that their activity is dependent on interactions with sulfated glycosaminoglycans (GAGs), heparan sulfate proteoglycan (HSPG) in particular, in the extracellular matrix (ECM). We hypothesized that there are cells in the axolotl that synthesize specific HSPGs that control growth factor signaling in time and space. In this study we have identified a subpopulation of cells within the ECM of axolotl skin that express high levels of sulfated GAGs on their cell surface. These cells are dispersed in a grid-like pattern throughout the dermis as well as the loose connective tissues that surround the tissues of the limb. These cells alter their morphology during regeneration, and are candidates for being a subpopulation of connective tissue cells that function as the cells required for pattern-formation during regeneration. Given their high level of HSPG expression, their stellate morphology, and their distribution throughout the loose connective tissues, we refer to these as the positional information GRID (Groups that are Regenerative, Interspersed and Dendritic) cells. In addition, we have identified cells that stain for high levels of expression of sulfated GAGs in mouse limb connective tissue that could have an equivalent function to GRID cells in the axolotl. The identification of GRID cells may have important implications for work in the area of Regenerative Engineering.
Keywords: axolotl, mouse, regeneration, heparan sulfate, morphogens, positional information
Lay summary:
The extracellular matrix (ECM) is important in controlling the spatial and temporal patterns of cell-cell signaling during regeneration. In this paper we identify a subpopulation of cells (GRID cells) within the ECM of axolotl skin that form a grid throughout the loose connective tissues of the limb. These cells are candidates for being the subpopulation of connective tissue cells that function to control pattern formation during axolotl limb regeneration. We also have identified a similar population of connective tissue cells in mammalian (mouse) tissues.
Future works:
Understanding the function of GRID cells will lead to the ability to induce and enhance regeneration in humans by the engineering of a biomimetic positional information grid to control the response of cells to endogenous growth factors.
Introduction
Limb regeneration is the outcome of a complex sequence of events that are mediated by interactions between cells derived from the tissues of the amputated stump. Among the signaling pathways involved, there is direct evidence for FGF/BMP signaling associated with nerves and the wound epithelium as the mediators of early blastema formation [1–4]. Subsequently, the cells within the blastema use signals to communicate their positional identity leading to blastema cell proliferation and pattern formation [2]. This signaling occurs between blastema cells derived from connective tissue fibroblasts of the stump tissues, and is mediated by sulfated glycosaminoglycans (GAGs) in the extracellular matrix (ECM) [2,4,5]. The challenge to discovering how to induce limb regeneration in mammals is to identify these signals, as well as the cells that are specialized to produce and receive the signals.
Within the blastema, there are two populations of cells that communicate with each other, and both are required for regeneration. One set of cells control the spatial arrangement of the regenerated tissues in order to insure that they are regenerated in the right position relative to each other. These cells are referred to as the “pattern forming” cells, and are derived from cells of the loose connective tissues, historically referred to as fibroblasts [2,6,7]. The cells that respond to the pattern-forming signals are the “pattern following” cells that remake the lost tissues such as muscle, bone, blood vessels and nerves. Much is known about the biology of the pattern following cells during regeneration. For example, satellite cells are lineage-committed, adult stem cells that are associated with muscle in both vertebrate and invertebrate animals [2,8]. In response to injury, these cells are activated and proliferate to give rise to progeny that are committed to the myogenic lineage.
In contrast to the pattern-following cells, relatively little is know about the biology of the pattern-forming cells. Experimentally, the presence of these cells is demonstrated by controlling the interactions between connective tissue fibroblasts from different positions within the limb, leading to the formation of supernumerary limb structures [2,7]. Since fibroblasts are associated with essentially all tissues, grafts of most tissues from one position to another results in the formation of supernumerary structures. However, when the fibroblasts can be removed from the graft prior to grafting (e.g. cartilage), the grafted tissue loses the ability to induce extra pattern [9]. Although the phenomenon of supernumerary limb formation is well documented, the underlying signaling mechanisms for pattern formation are less well understood, due in part to a lack of molecular markers to identify specific subpopulations of fibroblasts in the loose connective tissue.
One shared property of the signaling molecules that have been identified as being able to induce blastema formation and growth (e.g. FGF and BMP) [1,3,10], is that their activity is dependent on interactions with sulfated GAGs, heparan sulfate proteoglycan (HSPG) in particular, in the ECM [5,11,12]. The phenomenon of ECM mediated cell-cell signalling is best understood for FGF signaling which is dependent on the presence of HSPGs at the cell surface [11,12]. Given that cells in the dermis function as pattern-forming cells, and that cells in the dermis also synthesize HSPGs of the ECM, we hypothesized that the function of the pattern-forming cells in the axolotl is dependent on their ability to synthesize specific HSPGs that control growth factor signaling in time and space [5]. Consistent with this, we previously demonstrated that alteration of HS-mediated signaling leads to changes in growth and pattern formation during axolotl limb regeneration [5]. HSPG mediated signaling is spatially heterogeneous such that anterior and posterior ECM differ in their ability to mediate growth factor signaling. Using the axolotl limb as an assay for signaling activity, we also discovered that mouse limb dermis regulates pattern formation in the axolotl blastema by a HSPG-dependent and ontogenetic- dependent mechanism [5].
Our prior studies identifying HSPGs as mediators of pattern formation led us to look for and identify a subpopulation of cells within the ECM of axolotl skin that express high levels of sulfated GAGs on their cell surface. These cells are dispersed in a grid-like pattern throughout the dermis as well as the loose connective tissues that surround the tissues of the limb (e.g. myotubes, blood vessels, nerves, periosteum, and perichondrium). The cells alter their morphology during regeneration, and are candidates for being the subpopulation of connective tissue cells that function as the pattern-forming cells required for regeneration. Given their high level of HSPG expression, their stellate morphology, and their distribution throughout the loose connective tissues, we refer to these as positional information GRID (Groups that are Regenerative, Interspersed and Dendritic) cells. In addition to documenting the distribution of these cells in the connective tissues of axolotls, we have identified alcian blue-positive cells (staining for expression of sulfated GAGs) in mouse limb connective tissue that could have an equivalent function to GRID cells in the axolotl.
Methods and materials
Ethics statement
This study was carried out in accordance with the recommendations in the Guide for Care and Use of Laboratory Animals of the National Institutes of Health. The experimental work was conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee of the University of California Irvine (IACUC protocol #2007–2705).
Animals and surgical procedures
Experiments were performed on white axolotls (Ambystoma mexicanum) measuring 10–15 cm snout to tail tip that were spawned at the University of California Irvine or at the Ambystoma Genetic Stock Center at the University of Kentucky. The animals were maintained in 40% Holtfreter’s solution and were anesthetized prior to any procedure in a 0.1% solution of MS222 (ethyl 3-aminobenzoate methanesulfonate salt, Sigma), pH 7.4. Samples of mouse limb skin were obtained post-euthanasia from CD1 mice used for other studies.
Immunohistochemistry and Alcian blue staining
For staining tissue section samples, collected tissues were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) overnight, and treated with Decalcifier I (Surgipath) to decalcify the bones of the limb tissues in order to facilitate subsequent sectioning. Tissues were then dehydrated in graded ethanol, cleared in xylene, embedded in paraplast, and sectioned at 10 μm.
For staining whole-mount dermal skins, amputated limbs were rinsed in Ca+2/Mg+2 free Hanks solution at room temperature, and then incubated in 2.5 mM EDTA/ PBS for 15 min at 25°C to dissociate epidermal cells. The skin (epidermis-free dermis) was carefully peeled off from the limbs and fixed in 4% PFA/ PBS overnight.
For immunohistochemistry, samples were incubated with blocking solution (3% Bovine serum albumin in 1x PBS-Tween) at room temperature for 1 – 2 h, then incubated overnight at 4°C in primary antibody diluted with blocking solution. 10E4 (US Biological, 1:200), rabbit anti-collagen Type III antibody (ROCKLAND, 1:200), and rabbit anti-GFAP antibody (Sigma, 1:500 were used as a primary antibody. The bound antibodies were detected by Alexa Fluor 594 Goat anti-mouse IgG (H+L) and Alexa Fluor 488 Goat anti-rabbit IgG (H+L) diluted 1:400 with 1% axolotl powder (acetone extracted axolotl tissues) in blocking solution, and mounted in Vectashield with DAPI (Vector Laboratories).
For alcian blue staining of sulfated GAGs [13], samples were stained by 0.02% Alcian blue/ 0.3M MgCl2/ 0.1M sodium acetate/ 70% ethanol (pH 5.8) for 4 h at 37C, followed by optional standard eosin staining.
Degradation of heparan sulfate with nitrous acid
Heparan sulfate was degraded by deaminative cleavage by incubating whole-mount epidermis-free dermis in 0.24M nitrous acid in 1.8 M acetic acid (pH 1.5) at room temperature for 160 min. After 80 min, the nitrous acid was removed and fresh nitrous acid was re-applied for a further 80 min. The samples were washed with PBS and then stained with Alcian blue as described above [14].
Results
In previous studies, we discovered that the ECM influences pattern formation during axolotl limb regeneration [5]. In those experiments, decellularized ECM was grafted into wounds induced to form a blastema in order to determine if the grafted ECM could induce ectopic pattern formation. In the absence of grafted ECM, a blastema formed, but no skeletal structures were formed. In contrast, ECM from specific positions around the limb circumference induced ectopic limb structures in a position-dependent manner; i.e. posterior ECM induced limb pattern in anterior wounds, but anterior ECM did not [5]. Of importance for the present study, this positional-specific signaling was dependent on the presence of HSPGs, which function as critical co-factors for growth factor signaling [5,11,12]. Since the ECM in general, and HSPGs in particular are synthesized by cells within the ECM, we looked for and identified a subpopulation of cells within the ECM of axolotl skin that expressed high levels of sulfated GAGs on their cell surface.
Identification of cells within the ECM that have high levels of sulfated GAGs on their surface
We identified the presence and distribution of cells expressing high levels of sulfated GAGs (Figure 1) by staining with alcian blue under conditions that are specific for this class of macromolecules [13]. Alcian blue-positive cells were localized in the loose connective tissues of the limb. The morphology of these cells was complex, with multiple, branching cell processes that extended over distances that were many times the diameter of the cell nucleus (Figure 1a, arrows). When visualized at lower magnification of whole-mount preparations of dermal tissue, it was evident that these cells were arranged in a grid and their multiple cellular processes overlapped and appeared to make contact with processes from other alcian blue-positive cells (Figure 1b). The processes of these cells were diverse in their diameters (some regions thicker and others thinner), and it appeared that there were localized regions that were expanded, which might correspond to specialized points of contact and interaction between processes of different cells (arrowheads in Figure 1a; scattered throughout Figure 1b). An understanding of the specific structure and function of these putative sites of contact will require higher resolution imaging in conjunction with more specific cell-type specific markers in future studies. Given their high level of sulfated GAG expression that could regulate morphogen signaling during regeneration, their stellate morphology, and their distribution throughout the loose connective tissues, we refer to these cells as positional information GRID (Groups that are Regenerative, Interspersed and Dendritic) cells.
Figure 1.
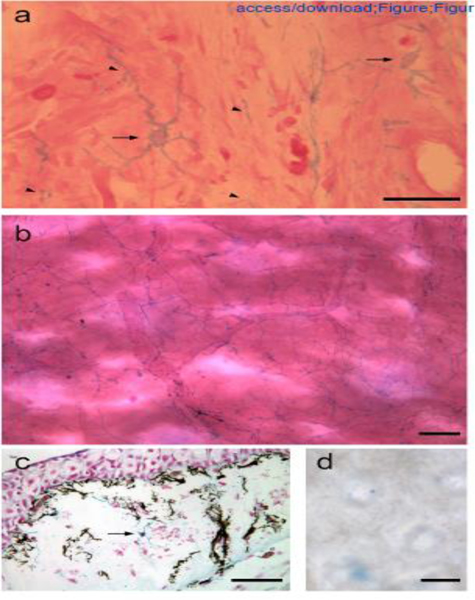
Alcian blue-positive GRID cells in the loose connective tissue of the dermis.
High magnification (a) and low magnification (b) images of whole-mount preparations of dermal tissue illustrating the dendritic morphology of GRID cells that form an interspersed cellular network. (c) GRID cells (arrow indicates an alcian blue-positive cell body) are distinct from melanocytes (black cell bodies and processes). (d) Nitrous acid treatment degraded HS, and alcian blue-positive GRID cells were no longer evident in dermal tissues. Examples of GRID cell bodies are indicated by arrows, and expanded regions of GRID cell processes are indicated by arrowheads (a). Scale bars: 50 μm (a, d), 100 μm (b, c).
As reported below (Figure 2), we have attempted to determine whether the alcian blue-positive GRID cells are an already identified subpopulation of cells within the connective tissues. Although the identity of these cells is unclear, it is evident that they are not pigment cells, which also have a dendritic morphology (Figure 1c). Dermal tissue from a wild-type axolotl with melanocytes, in contrast to white axolotls that were used for most of the experimental studies, contained alcian blue-positive GRID cells (Figure 1c, arrow) distinct from the melanin containing melanocytes. The specificity of alcian blue staining was confirmed by treatment with nitrous acid that cleaves HS side chains [14], and eliminated any staining of GRID cells in whole mount dermal tissues (compare Figures 1d and 1a,b).
Figure 2.
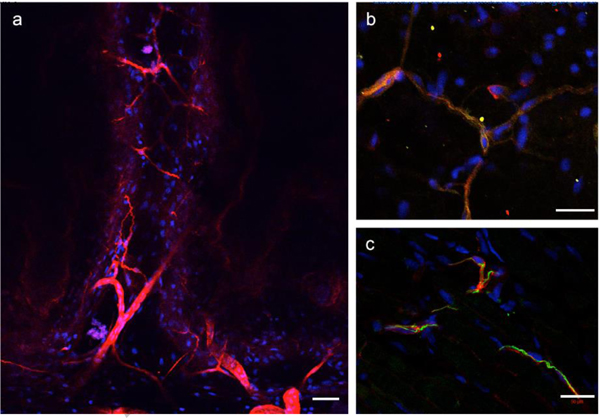
Heparan sulfate immunolocalization with 10E4 immunohistochemistry.
Whole-mount connective tissue from the dermis was imaged by confocal microscopy. (a) 10E4 positive cells formed an interspersed, dendritic network similar to what was observed with alcian blue staining. (b) Some 10E4-positive cells also expressed Collagen III (green, regions of co-expression were yellow/orange). (c) Some 10E4-positive cells were observed adjacent to cells expressing GFAP. Although cell process of both cell types were localized next to each other, cell process staining for both antigens (yellow/orange) were not observed. Nuclei of the cells were visualized by DAPI staining (blue). Scale bars: 100 μm (A), 50 μm (B, C).
In order to better characterize the GRID cells, we used a number of antibodies to screen for their affinity to cells within the ECM. Since the technique for alcian blue staining is not compatible with immunolocalization techniques, we could not double-stain for alcian blue-positive cells that also bound the antibodies that were tested. We identified three antibodies that localized to a subpopulation of GRID cells. In each case, the overall number of immuno-positive cells was less than expected based on the density of alcian blue-staining cells. We therefore presume that there is a diverse population of alcian blue-positive cells, and each of the three antibodies recognized a subpopulation of these cells. Further characterization of the cells within the loose connective tissues (fibroblasts) will allow for the identification and characterization of these subpopulations.
Given that alcian blue-staining under the conditions used in this study is specific for sulfated GAGs, we stained tissues with the antibody 10E4, which recognizes the 10E4 epitope that is present in many types of heparan sulfate, but does not react with hyaluronan, chondroitin sulfate, dermatan sulfate, or keratan sulfate. Cells throughout the loose connective tissues were positive for 10E4 staining (Figure 2a). As noted above, the density of 10E4-postivie cells was less compared to alcian-positive cells. Many of the loose connective tissue cells also were positive for expression of Collagen III, which is colocalized with expression of the epitope for the ERTR-7 antibody in mouse tissues [15], and was co-localized in some cells that were positive for 10E4 staining (yellow cell processes in Figure 2b). We did not detect immunolocalization in axolotl tissues with the ERTR-7 antibody. Finally, we detected a subpopulation of cells in loose connective tissues that were positive for GFAP (glial fibrillary acidic protein) expression. GFAP was originally reported as a marker for glial cells in the nervous system, but also is expressed by a number of other cells types. GFAP appeared to be expressed by cells that were associated with 10E4-positive cells, although the red (10E4) and green (GFAP) signals were not directly overlapping (no extensive regions of yellow staining) (Figure 2c).
GRID cells have a patterned distribution within the ECM
When viewed in whole-mount preparations, it was evident that GRID cells formed a network of interconnected cells dispersed through out the loose connective tissues (Figure 1). In sections, it was observed that these cells also were localized in the connective tissues associated with the differentiated tissues of the limb (Figure 3). These tissues included the dermis of the skin (Figure 3a), nerves (Figure 3b), muscle (Figure 3c), and blood vessels (Figure 3d). As in whole-mount preparations, it appeared that GRID cells made connections with other GRID cells; however, the specific nature of these contacts will require higher resolution imaging and utilization of more specific markers in future studies.
Figure 3.
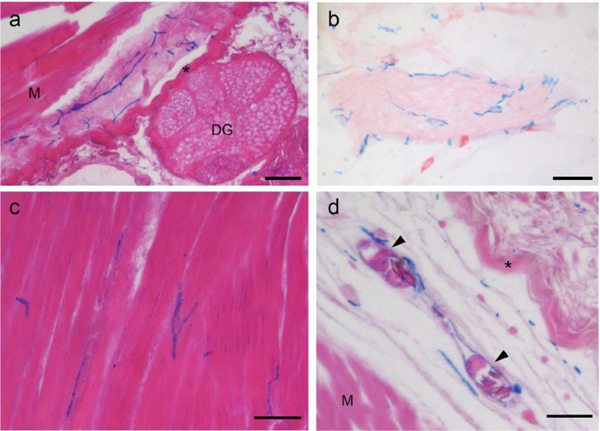
Alcian blue-positive GRID cells are localized within the connective tissues of differentiated tissues in the limb.
(a) GRID cells within the connective tissue of the dermis of the skin (M, skeletal muscle; DG, dermal skin gland; asterisk, dense collagen layer of the dermis). (b) Network of GRID cells surround a nerve. (c) Grid cells interspersed within skeletal muscles. (d) GRID cells within the loose connective tissue of the dermis surround blood vessels (M, skeletal muscle; asterisk, dense collagen layer of the dermis; arrowheads, nucleated erythrocytes within blood vessels). Scale bars: 50 μm.
GRID cells are not evident during early stages of blastema formation, and reappear at later stages
Although GRID cells were observed throughout the connective tissues of the uninjured limb, they were not evident at the early stages of blastema formation distal to the amputation plane (Figure 4a). It was not until later stages of regeneration (e.g. late bud blastema) that alcian blue-positive cells were observed again distal to the amputation plane (Figure 4b). At these later blastema stages, GRID cells (as visualized by 10E4 immunolocalization) were not uniformly distributed. At proximal regions of a late bud blastema where regenerated tissues were beginning to differentiate, there were many 10E4-positive cells (Figure 4c). In contrast, in the distal region, where blastema cells were still undifferentiated, only a few 10E4-positive cells were observed (Figure 4d). It may be the case that GRID cells, and their progeny were present in the early blastema but had lost their stellate morphology and were not expressing high levels of sulfated GAGs; therefore they were not visualized by our techniques. Alternatively, GRID cells may not contribute to the early blastema, and subsequently migrate from the stump into the proximal regions of the blastema as differentiation begins in a proximal to distal sequence [6,16,17]. Further experiments will be necessary to distinguish between these two hypotheses.
Figure 4.
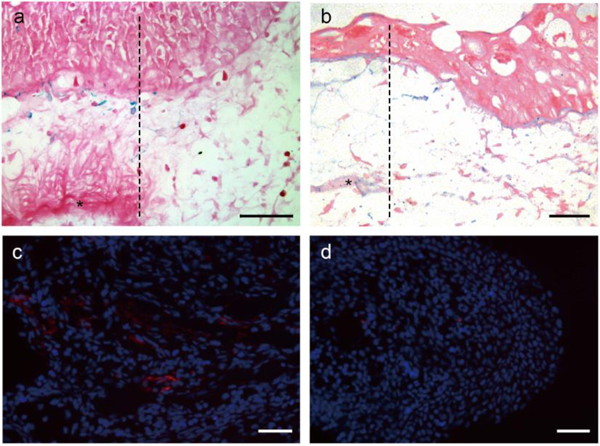
The distribution of alcian blue-positive GRID cells changes during regeneration.
(a) Alcian blue-positive GRID cells were distributed through the dermis of the uninjured stump, proximal to the plane of amputation; however, these cells were not observed distal to the amputation plane during the early stages of blastema formation (amputation plane is indicated by the dashed line, the stump with blue GRID cells is to the left, dense collagen layer of the dermis is indicated by an asterisk, and the basal region of an early bud blastema is to the right). (b) Alcian blue-positive cells are observed at later stages of regeneration (late bud blastema), but are more readily observed in proximal regions (c) than in distal regions (d). GRID cells are visualized in (c,d) by 10E4 immunostaining (red). Scale bars: 100 μm.
Alcian blue-positive cells are present in the connective tissues of neonatal mice
As in the axolotl, we observed the presence of alcian blue-positive cells in the loose connective tissues of neonatal mice (Figure 5). The abundance of these cells appeared to change ontogenetically such that many blue cells were observed at postnatal day 1 (PN1; Figure 5a), fewer cells were observed at PN6 (Figure 5b) and PN9 (Figure 5c), and even fewer in adult skin (Figure 5d). We attempted to determine if these cells in the mouse ECM had a stellate morphology that was comparable to GRID cells in the axolotl. Although there was variation in the morphology of the cell bodies, cell processes were not convincingly evident, even in sections that were not counterstained with eosin (Figure 5e, f). In most tissues, the cells exhibited a spherical morphology (Figure 5e; PN1); however, in some tissues the cells were more elongate (Figure 5f; PN9). Historically, it has been technically challenging to image cell processes (e.g. cytonemes and nanotubes) because of their very thin diameter (~200 nm), and because they are easily damaged by standard cell fixation [18,19]. The question of whether such processes are present in the alcian blue-positive cells we observed will require further studies utilizing advances in imaging technology and the availability of genetic tools, reporter genes and fluorescent-tagged components (see [20].
Figure 5.
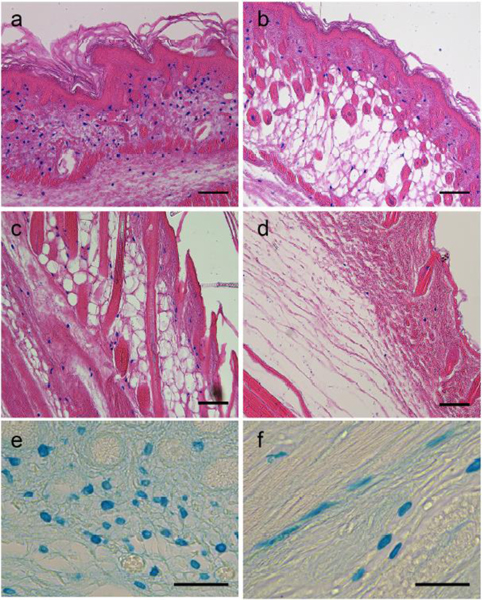
Alcian blue-positive cells in the connective tissues of mouse skin.
Alcian blue-positive cells were abundant throughout the dermal connective tissue of mouse skin at postnatal day 1 (a, PN1). These cells were observed at progressively decreasing abundance as the neonatal skin matured (b, PN6; c, PN9; and d, adult). The histology sections are oriented with the epidermis of the skin toward the upper-right of the images (a – d). At higher magnification without eosin counterstaining, the cell bodies were mostly round (e, PN1), although some appeared more elongate and stellate (f, PN9). Scale bars: 100 μm (A, B, C, D), 50 μm (E, F).
Genes involved in the synthesis and modification of sulfated GAGs are expressed in uninjured axolotl tissues, as well as during limb regeneration
Our earlier studies that assayed for the ability of HSPGs in the ECM to induce pattern formation led us to look for cells that expressed high levels of these molecules (the current study). These two studies have provided evidence that these ECM macromolecules are important for the regulation of pattern formation in axolotls as well as in mammals. To facilitate future investigations into the role of sulfated GAGs in axolotl regeneration, we searched the axolotl EST database for genes expected to be involved in both the synthesis of GAG chains and the modification of patters of sulfation (Supplemental Table 1). To date, we have identified a total of 29 candidate genes, and have designed primers for RT-PCR in order to characterize their patterns of transcription in limb skin, blastema tissues during stages of regeneration, and a diversity of non-limb tissues (Supplemental Figures 1 and 2; Supplemental Table 2). Expression of a subset of these genes was analyzed previously[5], and our recent reanalysis confirmed the patterns of expression for those genes. Collectively, the new data provide a set of reagents that can be utilized for further investigations of the function of sulfated GAGs in embryonic and regenerative development in the axolotl model.
Discussion
In this study we have identified a cell type that exhibits the physical and chemical properties predicted for cells that function in the control of positional information during regeneration. Numerous grafting studies have characterized cells with positional information as those with the ability to induce ectopic limb structures (e.g. supernumerary limbs), and demonstrated that these cells reside in the loose connective tissues of the limb (see [2]). In addition, positional information is a stable property of the ECM produced by these cells, such that decellularized matrix also can induce formation of ectopic limb pattern [5]. The ability of the ECM to do this is dependent on the presence of HSPGs, and this ability is spatially regulated around the limb circumference. This spatial pattern of HSPG modifications is hypothesized to arise as a consequence of the spatial and temporal regulation of expression of the enzymes responsible for modifying the patterns of HSPG sulfation [5]. Thus GRID cells are in the right place (the loose connective tissues) and express high levels of the right macromolecules (sulfated GAGs) required to function in the control of positional information.
It is instructive to think of there being two populations of cells involved in regeneration; the pattern-forming cells and the pattern-following cells [2,6,7]. By this view, the pattern-following cells have known functions; e.g. contractile myocytes for muscle contraction, and Schwann cells for ensheathing axons. However, in addition there must be cells that function to instruct the pattern-following cells as to when and where to migrate and differentiate in order to reform the correct limb pattern. The function of the pattern-forming cells is dependent on their ability to regulate growth factor (morphogen) signaling in time and space [2,4]. Given the essential role of sulfated GAGs, and HSPG in particular, in the regulation of morphogen signaling [11,12], as well as direct evidence for HSPG function in pattern formation during regeneration [5], an understand of the glycobiology of axolotl fibroblasts will be essential in understanding how pattern-forming cells function during regeneration.
Populations of fibroblasts within the loose connective tissues appear to be heterogeneous, as evidenced by immunohistochemistry data from this study (Collagen III, GFAP, and 10E4), as well as by tissue-specific and position-specific expression of HOX genes [21,22]. It may be that some fibroblasts function as pattern-forming cells, whereas others are pattern-following cells (differentiate to make the connective tissues, e.g. cartilage, ligaments, and tendons) during regeneration. Alternatively, pattern-forming fibroblasts may also give rise to progeny that can function as pattern-following cells. In the case of regeneration of the axolotl limb skeleton, the progenitors of both the pattern-forming and pattern- following cells reside in the connective tissue surround the uninjured skeletal elements (perichondrium), and the differentiated chondrocytes do not participate in the regenerative response [9]. The challenge moving forward is to validate markers for fibroblast subpopulations, and then utilize the axolotl regeneration models to determine which population is specialized for encoding positional information. Based on the available data, the GRID cells identified in this study are a good place to start looking.
Another predicted property of pattern-forming cells is that their behavior, and the expression of GAGs on their surface, must be dynamically regulated during regeneration. In order to make a blastema and regenerate a limb, cells in the stump must be able to proliferate and migrate [2,23]. In order to do either of these, the blastema progenitor cells cannot remain differentiated. For example GRID cells in the uninjured limb are dendritic, and therefore in order to proliferate and migrate distally, they must retract their cell processes, round up as they proliferate, and extend filopodia as they migrate. During this period of dedifferentiation, it is likely that they also change their patterns of expression of cell-surface GAGs. The observation that the expression of enzymes that synthesize and modify GAGs changes during the stages of regeneration is consistent with this presumption. Regardless, the patterns of GAGs expression prior to injury are reestablished at the end of regeneration.
The premise that positional information is reprogrammed during regeneration is based on a number of experimental studies. Early cell contribution studies demonstrated that about 25% of the cells of the blastema, and subsequently the regenerated limb, originated from progenitor cells from the opposite side of the limb [24]. The predicted plasticity of positional information was demonstrated experimentally by grafting of early bud blastemas that become reprogrammed to adopt the pattern of the site to which they are grafted [6,25]. In contrast, a grafted late bud blastema has stabilized the positional identity of the pattern-forming cells, and when grafted, an ectopic limb forms [6,25,26]. The acquisition and stabilization of positional identity is initiated proximally and progresses distally such that the basal region of a late bud blastema has cells with stabile information, at the same time that more distally cells are still positionally labile, like cells of the early bud blastema [6,17]. The lack of alcian blue-positive cells in the early bud blastema (Figure 4), as well as the higher density of 10E4-positive GRID cells in the proximal region as compared to the distal region of late bud blastemas is correlated with the temporal and spatial stabilization of positional information during regeneration.
The role of positional signaling during regeneration has been formalized by the Polar Coordinate Model (PCM), which is based on the hypothesis that there are cells with positional information that interact with each other through cell-cell contacts in order to reform the tissue patterns that are lost during injury [27,28]. The presence of long cell processes (see Figure 1) implicates GRID cells as being the cells that encode and communicate positional information as envisioned in the PCM. In recent years, it has become increasing evident that the spatial and temporal regulation of morphogen signaling by long cell processes is a widely utilized mechanism for long distance signaling between cells (see [20]). These signaling filopodia (e.g. cytonemes, airinemes) function as a conduit for the transport of morphogens between cells[19,20,29–31], and thus allow for direct physical contact at distance between signal-sending and signal receiving cells, as predicted by the PCM. In addition, the extension and retraction of these processes are regulated over time, as would be needed for regeneration. We presume that the processes of GRID cells are functional equivalent to the signaling filopodia of many other cells.
The regulation of morphogen signaling by HSPG can be both qualitative and quantitative. The ability of HSPGs to bind proteins is dependent on structural motifs, the type of sulfation, and the negative charge density of subdomains, which allows for HSPGs to modulate a variety of signaling pathways, as receptors, co-receptors, recruiters, and/or ligand sinks [32,33]. Many growth factors and morphogens that play fundamental roles in development and pattern formation, including Fgfs, Wnts, and Hedgehogs are regulated by HSPGs (see [32]). The complexity of HSPG chemistry and biology has led to the concept of there being an HSPG code [34] that can be regulated during the course of embryonic and regenerative development in a number of model systems. In addition, there is evidence for quantitative regulation, specifically in the context of axolotl limb regeneration [5]. Variations in concentration of HSPG in artificial ECM grafts (collagen plus HSPG) results in variations in the regeneration outcome, ranging from no blastema formation (inhibition of regeneration), to blastema formation with pattern formation, and finally to blastema formation but no induced pattern formation. In this same study, grafts of decellularized, mouse limb skin ECM from different stages of mouse development resulted in comparable variations in the regeneration outcome in the axolotl regeneration assay [5]. The regeneration response ranged from no effect (E11.5 graft), inhibition of regeneration (PN1 graft), induction of pattern formation (PN3/4 and PN7 grafts), and minimal effect on either blastema or pattern formation (PN9 grafts). Treatment of PN3/4 ECM grafts with heparin lyase, which degrades HSPG, abolished the pattern-inducing activity. At this point, it is unclear if there is positional information encoded by GRID cells in the dermis of mouse limbs that are functionally equivalent to axolotl GRID cells; however, we note that there is an ontogenetic decrease in the abundance of alcian blue-positive cells (see Figure 5, a–d) that is developmentally coincident with the period during which HSPG-dependent signaling occurs, as assayed previously [5].
Regenerative Engineering has been defined as the convergence of advanced materials science, stem cell science, developmental biology/morphogenesis, physics and clinical translation for the regeneration of complex tissues and organ systems [35]. Understanding the role of sulfated GAGs in controlling the spatial and temporal activity of growth factor/morphogen signaling may ultimately enable us to engineer a biomimetic GRID for enhanced regenerative therapies using a convergence of technologies. Rather than delivering growth factors to a wound at the right time, in the right place, and at the right dose, it may be possible to tune this engineered ECM to control the response of cells to endogenous growth factors. The challenge is to see the GRID in order to engineer the GRID, and the axolotl is an excellent model for regeneration that provides an opportunity to explore the nature of the GRID. By staining for sulfated GAGs in general, and specific modifications of HSPG in particular, we may be able to visualize the position information GRID cells can see. Further exploration of the GRID is presents as a new and exciting new dimension for the field of regenerative engineering.
Supplementary Material
Visualization of gels from RT-PCR analysis of expression of axolotl gene that are involved in the synthesis of GAG chains and in modification of their patters of sulfation in axolotl limb skin, as well as blastema tissue during limb regeneration.
Visualization of gels from RT-PCR analysis of expression of axolotl gene that are involved in the synthesis of GAG chains and in modification of their patters of sulfation in a diversity of tissues.
Results from a search of the axolotl EST database (ref) for genes expected to be involved in both the synthesis of GAG chains and in modification of their patters of sulfation. PCR primers were synthesized as indicated for RT-PCR analysis of expression (Supplemental Figure 1a, 1b, and Supplemental Table 2).
Summary of the relative levels of expression of axolotl genes (RT-PCR analysis illustrated in Supplemental Figures 1a, 1b) that are involved in the synthesis of GAG chains and in modification of their patters of sulfation in a diversity of tissues, as well as during limb regeneration.
Acknowledgements:
We wish to thank the members of the Bryant/Gardiner Lab for help with and encouragement of the research. Mouse limb skin tissue samples were kindly provided by Dr. David Rowe. Research was supported by an NIH Director’s Pioneer Award, and the National Science Foundation through its support of the Ambystoma Genetic Stock Center at the University of Kentucky, Lexington. Dr. Laurencin was the recipient of the Presidential Faculty Fellow Award from the National Science Foundation.
Footnotes
Competing Interests:
None of the authors have competing interests.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Makanae A, Mitogawa K, Satoh A. Co-operative Bmp- and Fgf-signaling inputs convert skin wound healing to limb formation in urodele amphibians. Developmental Biology. 2014;396:57–66. [DOI] [PubMed] [Google Scholar]
- 2.McCusker C, Bryant SV, Gardiner DM. The axolotl limb blastema: cellular and molecular mechanisms driving blastema formation and limb regeneration in tetrapods. Regeneration (Oxf). 2015;2:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vieira WA, Wells KM, Raymond MJ, De Souza L, Garcia E, McCusker CD. FGF, BMP, and RA signaling are sufficient for the induction of complete limb regeneration from non-regenerating wounds on Ambystoma mexicanum limbs. Dev Biol. 2019;451:146–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant SV, Gardiner DM. The relationship between growth and pattern formation. Regeneration (Oxf). 2016;3:103–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phan AQ, Lee J, Oei M, Flath C, Hwe C, Mariano R, et al. Positional information in axolotl and mouse limb extracellular matrix is mediated via heparan sulfate and fibroblast growth factor during limb regeneration in the axolotl (Ambystoma mexicanum). Regeneration (Oxf). 2015;2:182–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCusker CD, Gardiner DM. Positional information is reprogrammed in blastema cells of the regenerating limb of the axolotl (Ambystoma mexicanum). Tsonis PA, editor. PLoS ONE. Public Library of Science; 2013;8:e77064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCusker CD, Gardiner DM. Understanding positional cues in salamander limb regeneration: implications for optimizing cell-based regenerative therapies. Dis Model Mech. Company of Biologists; 2014;7:593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–5. [DOI] [PubMed] [Google Scholar]
- 9.McCusker CD, Diaz-Castillo C, Sosnik J, Q Phan A, Gardiner DM. Cartilage and bone cells do not participate in skeletal regeneration in Ambystoma mexicanum limbs. Dev Biol. 2016;416:26–33. [DOI] [PubMed] [Google Scholar]
- 10.Satoh A, Mitogawa K, Makanae A. Regeneration inducers in limb regeneration. Dev Growth Differ. 2015;57:421–9. [DOI] [PubMed] [Google Scholar]
- 11.Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–8. [DOI] [PubMed] [Google Scholar]
- 12.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. Cold Spring Harbor Lab; 2011;3:a004952–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott JE, Dis JDAR, 1965. Differential staining of acid glycosaminoglycans (mucopolysaccharides) by alcian blue in salt solutions. His-tochemie. 1965; 5: 221–233. Fletcher CDM .... [DOI] [PubMed] [Google Scholar]
- 14.Conrad GW, Hamilton C, Haynes E. Differences in glycosaminoglycans synthesized by fibroblast-like cells from chick cornea, heart, and skin. J Biol Chem. 1977;252:6861–70. [PubMed] [Google Scholar]
- 15.Marrero L, Simkin J, Sammarco M, Muneoka K. Fibroblast reticular cells engineer a blastema extracellular network during digit tip regeneration in mice. Regeneration (Oxf). 2017;4:69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neufeld DA, Day FA. Perspective: a suggested role for basement membrane structures during newt limb regeneration. Anat. Rec. 1996;246:155–61. [DOI] [PubMed] [Google Scholar]
- 17.McCusker CD, Athippozhy A, Diaz-Castillo C, Fowlkes C, Gardiner DM, Voss SR. Positional plasticity in regenerating Amybstoma mexicanum limbs is associated with cell proliferation and pathways of cellular differentiation. BMC Dev. Biol. BioMed Central; 2015;15:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodeen WJ, Marada S, Truong A, Ogden SK. A fixation method to preserve cultured cell cytonemes facilitates mechanistic interrogation of morphogen transport. Development. 2017;144:3612–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramírez-Weber FA, Kornberg TB. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 1999;97:599–607. [DOI] [PubMed] [Google Scholar]
- 20.González-Méndez L, Gradilla A-C, Guerrero I. The cytoneme connection: direct long-distance signal transfer during development. Development. 2019;146:dev174607. [DOI] [PubMed] [Google Scholar]
- 21.Rinn JL, Wang JK, Allen N, Brugmann SA, Mikels AJ, Liu H, et al. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22:303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardiner DM, Muneoka K, Bryant SV. The migration of dermal cells during blastema formation in axolotls. Developmental Biology. 1986;118:488–93. [DOI] [PubMed] [Google Scholar]
- 24.Tank PW, Connelly TG, Bookstein FL. Cellular behavior in the anteroposterior axis of the regenerating forelimb of the axolotl,Ambystoma mexicanum. Developmental Biology. 1985; 109:215–23. [DOI] [PubMed] [Google Scholar]
- 25.Iten LE, Bryant SV. The interaction between the blastema and stump in the establishment of the anterior--posterior and proximal--distal organization of the limb regenerate. Dev Biol. 1975;44:119–47. [DOI] [PubMed] [Google Scholar]
- 26.Stocum DL. The urodele limb regeneration b blastema: a self-organizing system. i. Differentiation in vitro. Dev Biol. 1968;18:441–56. [DOI] [PubMed] [Google Scholar]
- 27.French V, Bryant PJ, Bryant SV. Pattern regulation in epimorphic fields. Science. 1976;193:969–81. [DOI] [PubMed] [Google Scholar]
- 28.Bryant SV, French V, Bryant PJ. Distal regeneration and symmetry. Science. 1981;212:993–1002. [DOI] [PubMed] [Google Scholar]
- 29.Kornberg TB, Roy S. Cytonemes as specialized signaling filopodia. Development. Oxford University Press for The Company of Biologists Limited; 2014;141:729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eom DS, Parichy DM. A macrophage relay for long-distance signaling during postembryonic tissue remodeling. Science. American Association for the Advancement of Science; 2017;355:1317–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eom DS, Bain EJ, Patterson LB, Grout ME, Parichy DM. Long-distance communication by specialized cellular projections during pigment pattern development and evolution. Elife. eLife Sciences Publications Limited; 2015;4:2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin X Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–21. [DOI] [PubMed] [Google Scholar]
- 33.Poulain FE, Yost HJ. Heparan sulfate proteoglycans: a sugar code for vertebrate development? Development. 2015;142:3456–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bülow HE, Berry KL, Topper LH, Peles E, Hobert O. Heparan sulfate proteoglycan-dependent induction of axon branching and axon misrouting by the Kallmann syndrome gene kal-1. Proc Natl Acad Sci U S A. 2002;99:6346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurencin CT, Khan Y Regenerative Engineering. CRC Press, Taylor & Frances Group. Boca Raton. FL. 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Visualization of gels from RT-PCR analysis of expression of axolotl gene that are involved in the synthesis of GAG chains and in modification of their patters of sulfation in axolotl limb skin, as well as blastema tissue during limb regeneration.
Visualization of gels from RT-PCR analysis of expression of axolotl gene that are involved in the synthesis of GAG chains and in modification of their patters of sulfation in a diversity of tissues.
Results from a search of the axolotl EST database (ref) for genes expected to be involved in both the synthesis of GAG chains and in modification of their patters of sulfation. PCR primers were synthesized as indicated for RT-PCR analysis of expression (Supplemental Figure 1a, 1b, and Supplemental Table 2).
Summary of the relative levels of expression of axolotl genes (RT-PCR analysis illustrated in Supplemental Figures 1a, 1b) that are involved in the synthesis of GAG chains and in modification of their patters of sulfation in a diversity of tissues, as well as during limb regeneration.


