Abstract
We engineered a machine learning approach, MSHub, to enable auto-deconvolution of gas chromatography-mass spectrometry (GC-MS) data. We then designed workflows to enable the community to store, process, share, annotate, compare, and perform molecular networking of GC-MS data within the Global Natural Product Social (GNPS) Molecular Networking analysis platform. MSHub/GNPS performs auto-deconvolution of compound fragmentation patterns via unsupervised non-negative matrix factorization and quantifies the reproducibility of fragmentation patterns across samples.
Given its ease of use and low operational cost, gas chromatography-mass spectrometry (GC-MS) has applications with broad societal impact, such as detection of metabolic disease in newborns, toxicology, doping, forensics, food science, and clinical testing. The predominant ionization technique in GC-MS is electron ionization (EI), in which all compounds are ionized by high energy (70eV) electrons. Because fragmentation occurs with ionization, EI GC-MS data are subjected to spectral deconvolution, a process that separates fragmentation ion patterns for each eluting molecule into a composite mass spectrum.
The 70eV for ionizing electrons in GC-MS has been the standard, making it possible to use decades-old EI reference spectra for annotation 1. There are ~1.2 million reference spectra that have been accumulated and curated over a period of >50 years 2. Many tools and repositories for GC-MS data have been introduced 3–15; however, much of GC-MS data processing is restricted to vendor-specific formats and software8. Currently, deconvolution requires setting multiple parameters manually 3–5, or computational skills to run the software 7. Also, the lack of data sharing in a uniform format precludes data comparison between laboratories and prevents taking advantage of repository-scale information and community knowledge, resulting in infrequent reuse of GC-MS data 8,11–15.
Although batch modes exist, deconvolution quality is currently not enhanced by utilizing information from all other files. To leverage across-file information, improve scalability of spectral deconvolution, and eliminate the need for manually setting the deconvolution parameters (m/z error correction of the ions, peak shape - slopes of raising and trailing edges, peak RT shifts, and noise/intensity thresholds), we developed an algorithmic learning strategy for auto-deconvolution (Figure 1a–f). We deployed this functionality within GNPS/MassIVE (https://gnps.ucsd.edu)16 (Figure 1f–i). To promote analysis reproducibility, all GNPS jobs performed are retained in the “My User” space and can be shared as hyperlinks.
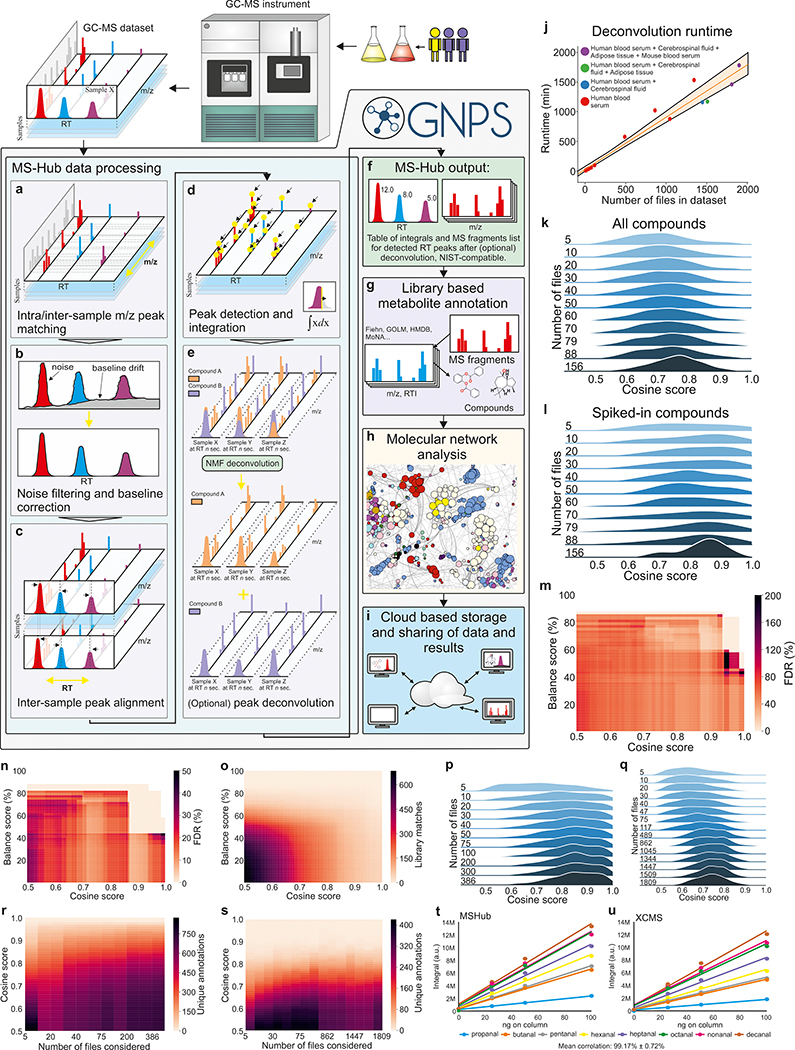
Figure 1. The processing pipeline and performance.
a) Spectra are aligned and binned, noise is filtered and b) Baseline corrected c) Common profile across the dataset and peaks in RT dimension are aligned using Fast Fourier Transform (FFT)-accelerated correlation. d) Generation of both peak integrals for all samples and their common fragmentation patterns. e) Separation of overlapping peaks with patterns across samples using NMF. f) Peak integrals for all samples and canonical fragmentation patterns. g) Annotation with public or private libraries. h) Molecular networks. i) Data and results are shared between users. j) Linear dependence of the MSHub processing time. k) Distributions of library matching scores with an increased volume of data (datasets with known spiked compounds Test1-Test11, Table S1) for all matches and l) for the spiked compounds only. m) False discovery rate for annotations (Test11) of the top match and (n) top ten matches. o) Number of library matches for spiked compounds. p), q) Cosine improves as higher volume of the data enhances deconvolution quality for the top match of biological samples: breath (non-derivatized, ICL1-ICL11, Table S1) (p) and human and mouse blood serum, adipose tissue and cerebrospinal fluid (silylated, datasets UCD1-UCD16, Table S1) (q). r) The unique annotations across datasets ICL1-ICL11 and s) datasets UCD1-UCD16; no balance score filtering applied. t, u) Quantitative comparison of XCMS (t) and MSHub (u). NMF - Non-negative matrix factorization, FFT - Fast Fourier Transform, RT - retention time.
This user-independent ‘automatic’ parameter optimization is accomplished via fast Fourier transformation, multiplication, and inverse Fourier transformation for each ion across entire data sets, followed by an unsupervised non-negative matrix factorization (one layer neural network). Then, the compositional consistency of spectral patterns for each spectral feature deconvoluted across the entire data set can be summarized as a “balance score”. The balance score (mathematical definition in the Methods) quantifies reproducibility of the deconvoluted fragmentation patterns across the data, which, in turn, gives insight into how well the spectral feature is explained by the available data. Thus the balance score provides an orthogonal metric of deconvoluted spectral quality. We refer to the dataset spectral deconvolution tool within the GNPS environment as “MSHub”.
All MSHub algorithms use efficient HDF5 technologies. The Fourier transform with multiplication improves MSHub’s efficiency, resulting in deconvolution times that scale linearly with the number of files (Figure 1j, Supplementary Fig. 1a, Supplementary Fig. 2, Supplementary Fig. 4). We achieved this performance using out-of-core processing, a technique used to process data that are too large to fit in a computer’s main memory (RAM): MSHub uploads files one at a time into the RAM module, data are then processed and deleted from memory, iteratively. Because only one sample is stored in the memory, the load is constant (Supplementary Fig. 2a–f). As machine learning approaches gain improved performance with increased volumes of information, including more data into analysis leads to better scores of spectral matches (Figure 1k,l and Supplementary Fig. 1b). The spectral library match scores increase and their distributions become narrower indicating better quality of results (Figure 1p,q). More files deconvoluted in MSHub leads to fewer chimeric spectra, resulting in higher quality spectral features, and an increase in the number of annotations with improved scores (Figure 1r,s). MSHub performs as well or better as other deconvolution tools (Figure 1t,u and Supplementary Fig. 3, Supplementary Fig. 4, Supplementary Fig. 5). Linear scaling for MSHub makes it the only tool amenable to repository-scale operation in its present form (Supplementary Table 2). GNPS saves deconvoluted data as a summary file, so the deconvolution step does not need to be re-performed for any future analyses.
Once the summary file is generated by GNPS-MSHub or imported from another deconvolution tool, the spectra can be searched against public, private or commercial libraries. Matches are narrowed down based on user-defined filtering criteria such as number of matched ions, Kovats index, balance score, cosine score, and abundance. We provide freely available reference data of 19,808 spectra for 19,708 standards, a ~29% increase of free public libraries. All annotations should be considered level 3 (a molecular family) annotation17. When multiple annotations can be assigned, GNPS provides all candidate matches within the user’s filtering criteria.
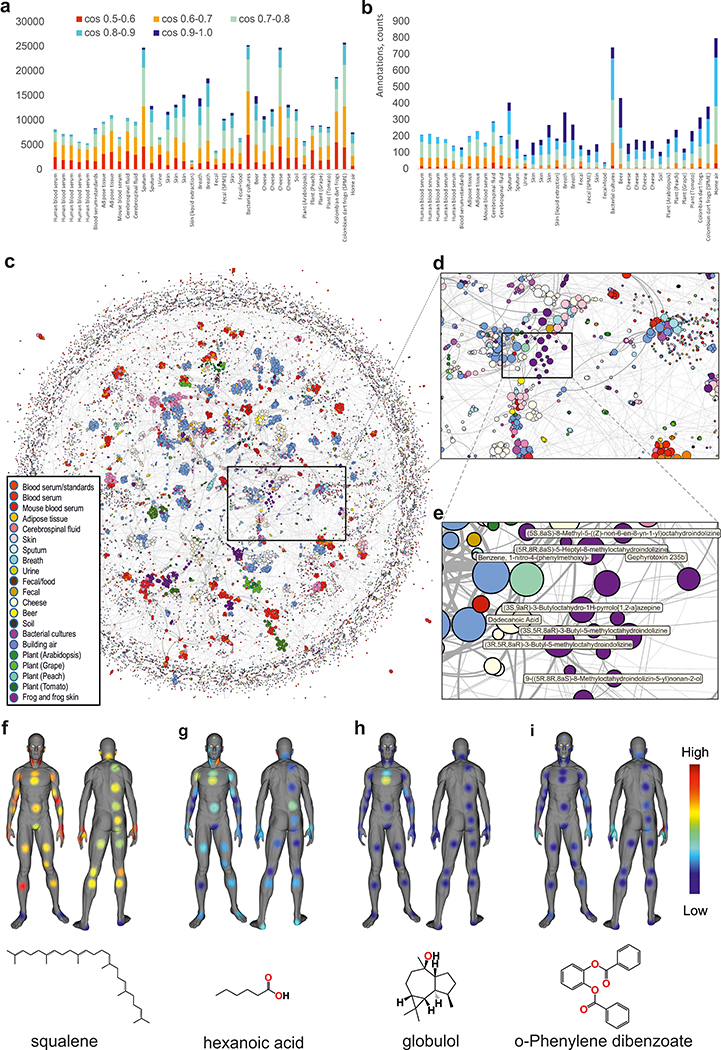
One of the developments that enabled finding structural relationships within mass spectrometry data is spectral alignment, which forms the basis for molecular networking 18. GNPS has now expanded to include GC-MS-specific molecular networking16. GNPS-based GC-MS analysis enables data co- and re-analysis, as the processing is agnostic to the data origin. To showcase this ability, we built a global network of various public GC-MS datasets and applied a balance score of 65% (Figure 2 a,b, Supplementary Fig. 6) to ensure that only good quality deconvoluted spectra are matched against the reference library (Figure 2c–e, Supplementary Fig. 9, Supplementary Fig. 10). Molecular networking can further guide the annotation at the family level by utilizing information from connected nodes rather than focusing on individual annotations (Supplementary Fig. 7, Supplementary Fig. 8). One can visualize aspects such as derivatized vs. non-derivatized, candidate compound class or subclass, instrument type or other metadata and inspect individual clusters of nodes (Supplementary Fig. 9). For example, we observed a cluster that belonged to dart frogs from the Dendrobatoidea superfamily, while the long-chain ketones are found in cheese and beer (Figure 2e, Supplementary Fig. 10a). The output from GNPS can be exported for use in statistical analysis environments and for data visualization (e.g., Supplementary Figs. 7–10), including molecular cartography 19 (Figure 2f–i).
Figure 2: Analysis and molecular networking of GC-MS data.
Annotated spectra a) without filtering and b) with a 65% balance score filtering. c) Global network containing 35,544 nodes from 8,489 files in 38 GNPS datasets. The size of the node is proportional to the number of nodes that connect, the edge thickness is proportional to the cosine score (Figure S6). The annotation is the top match with cosine above 0.65. d) Zoomed-in region e) Cluster of compounds from dart frog skin samples - all nodes are alkaloids. f) human surface volatilome visualized with ‘ili19. Molecular distributions for squalene g) hexanoic acid, a malodour molecule h) globulol, common uses in perfume i) phenylenedibenzoate, common in skin care products.
GNPS/MassIVE lowers the expertise threshold required for analysis and encourages FAIR practices20 by promoting re-use of GC-MS data. To highlight the broader utility of GNPS GC-MS based analysis, videos were created (Supplemental Videos 1–6). This work aims to democratize scientific analyses. GC-MS is often the only mass spectrometry method in non-metabolomics laboratories or laboratories with fewer resources, including those from developing countries. GNPS-based GC-MS allows free access to data and reference data, and to powerful computing infrastructures.
Online Methods:
Tutorials and general note
The tools are accessible through gnps.ucsd.edu. The documentation to use the GC-MS interface can be found here: https://ccms-ucsd.github.io/GNPSDocumentation/gcanalysis/.
The tutorials for the deconvolution with can be accessed here: https://ccms-ucsd.github.io/GNPSDocumentation/gc-ms-deconvolution/ while the library search and molecular networking instructions can be found here: https://ccms-ucsd.github.io/GNPSDocumentation/gc-ms-library-molecular-network/.
The tutorial for spectral libraries upload can be found here: https://ccms-ucsd.github.io/GNPSDocumentation/batchupload/
The GNPS workflows can be launched with recommended default settings or adjusted according to user’s needs. The ranges and impact of settings are described in the tutorial.
The results can be inspected and quality filters applied according to the user’s criteria.
The tutorial also describes how user can utilize various other aspects of GNPS functionality that include:
Data upload and storage
Data sharing
Sharing analysis by sharing workflows
Reproducing analyses
Saving and sharing reference spectra
Using GNPS analysis links for publishing
Using GNPS/MassIVE repository for providing access to data along with the publication when required by journal
The video tutorials for GNPS use for GC-MS data and examples of networking application videos can be accessed at:
Tutorial for the use of GNPS for analysis of GC-MS data.
https://www.youtube.com/watch?v=KIOim2h8i64
GNPS for GC-MS in breathomics: using molecular networking to combine different datasets.
https://www.youtube.com/watch?v=bDZj7NI-ZGw
GNPS for GC-MS in petroleomics: using molecular networks to find incorrect annotations.
https://www.youtube.com/watch?v=r7DSsL03Hbk
GNPS for GC-MS in biology: using molecular networks for compound discovery in dart frogs.
https://www.youtube.com/watch?v=eNLPrAjuX6w
GNPS for GC-MS in microbiology: using networks to explore chemistry of cheese.
https://www.youtube.com/watch?v=fWus3zhKbOA
GNPS for GC-MS in biochemistry: use of networking to discover antifungals produced by B. Subtilis.
Use of the GNPS GC-MS workflows
GNPS GC-MS environment
The GNPS leverages the repository infrastructure now has expanded to include GC-MS-specific deconvolution, reference spectra matching and molecular networking tools. The new analysis workflows not only solved the scaling of analysis, but are also configured to promote data analysis reproducibility, as an analysis performed in GNPS is retained in the account-specific job tab and can be shared as a hyperlink. The user’s own or someone else’s shared analysis can be precisely reproduced by clicking the “clone” button. In addition, we have enabled the community to upload and share reference spectra which then continuously accumulate leading to continuous improvements of annotations. GNPS also gives the ability to explore all public data sets together with studies in one’s private space for a particular research problem (e.g. drug discovery). There are no other GC-MS deconvolution and annotation infrastructures that also work with the data in a repository. The scalability, reproducibility, capture of knowledge and the ability to efficiently reuse data in the public domain make the GC-MS infrastructure in GNPS unique compared to other existing open or commercial resources. GNPS promotes Findable, Accessible, Interoperable, and Reusable (FAIR) use practices for mass spectrometry data20.
The community infrastructure can be accessed at https://gnps.ucsd.edu under the header “GC-MS EI Data Analysis”.
Deconvolution
Currently, 1D EI GC-MS data are amenable. We recommend to use a minimum of 10 files in the dataset for deconvolution with MSHub. If the user only has fewer than 10 files, spectral deconvolution and alignment should be performed using alternative methods (e.g. MZmine, OpenChrom, AMDIS, MZmine/ADAP, MS-DIAL, BinBase, XCMS/XCMS Online, MetAlign, SpecAlign, SpectConnect, PARAFAC2, MeltDB, eRah). After using one of those tools, molecular networking can be performed in the same fashion as for MSHub (detailed description is given in the Supplementary Notes), as the library search GNPS workflow accepts input from other tools into the GNPS/MassIVE environment. GNPS directly supports deconvolution output from MZmine/ADAP and MS-DIAL. The quantitative table of the deconvolution output can be used for statistical analysis with external tools.
Library search
Once the .mgf file is generated by GNPS-MSHub or imported from another deconvolution tool, the spectral features can be searched against public libraries (currently GNPS has Fiehn, HMDB, MoNA, VocBinBase) or the user’s own private or commercial libraries (such as NIST 2017 and Wiley) and the freely available reference data of 19,808 spectra for 19,708 standards released with this manuscript. Users can also upload their own libraries to GNPS as well to share them with the community. Although the possible candidate annotations can be further narrowed by retention index (RI), they should still be considered level 3, a molecular family, annotation according to the 2007 metabolomics standards initiative (MSI)17. Calculation of RIs is enabled and encouraged but not enforced. When multiple annotations can be assigned, GNPS provides all candidate matches within the user’s filtering criteria.
Filtering the results
The balance score is a new metric which will be available when MSHub deconvolution is used. A fragmentation pattern of a compound found to be the same in different measurements would result in a high balance score. Missing or chimeric peaks would change randomly across files and would result in a low balance score. Even when a compound is present in a few samples, as long as the spectral patterns, irrespective of compound abundances, are conserved across samples it would result in a high balance score.
Cosine and balance score should be jointly used as spectral matching filters for processing of the final results. The effect of filtering can be seen on the Figure 1m–o and S3d,e. For the test dataset shown on Figure 1m,n, the lowest FDR of the top match is achieved with the combined threshold values of cosine >0.9 and balance score >60% (Figure 1m). A more conservative balance score value of >80% essentially ensures the lowest observed FDR, even for poor cosine scores (here referred to as match scores). Conversely, even the high match score by itself may still result in unacceptably high FDR if the balance score is poor (Figure 1m,n). The high match score reflects that a library spectrum exists that is similar to the query spectrum, while a high balance score is reflective of the high confidence in deconvolution of the spectral pattern. A well-deconvoluted pattern as defined by the balance score is more likely to give better matches against the spectral library. Selecting higher values of both metrics ensures the best spectra are used and are matched to most likely annotations. The “optimal” thresholds, i.e. the values that minimize mis-annotations without being excessively restrictive, are data-specific, but we recommend to use the above values as a good starting point.
Molecular networks
No matter how the spectral library is searched in GC-MS, due to the absence of a parent mass, a list of spectral matches is more likely to contain mis-annotations, both related (isomers, isobars) or less frequent, entirely unrelated compounds1. However, to spot mis-assignments at the molecular family level, we propose to explore deconvoluted GC-MS data via molecular networking, a strategy that has been effective for LC-MS/MS data16. In the case of EI, unlike in LC-MS/MS where the precursor ion mass is known, the molecular ion is often absent. For this reason, the molecular networks are created through spectral similarity of the deconvoluted fragmentation spectrum without considering the molecular ion. We explored molecular networking patterns for the EI data (Figure S7) and observed that the EI-based cosine similarity networks are predominantly driven by structural similarity based on chemical class annotations (Figure S7a). These EI networks can be used to visualize chemical distributions and guide annotations (Figure S8). Some examples of molecular networking applications are discussed in the Supplemental Videos.
3D mapping of volatilome
The sample collection and GC-MS analysis are described above in the “Skin volatilome analysis” section of Supplementary Notes. Feature tables from the deconvolution jobs for headspace and liquid injection were downloaded from GNPS and combined into a single table. The coordinates for 3D model were picked for all of the sampled spots and added into the feature table as described in the tutorial (https://ccms-ucsd.github.io/GNPSDocumentation/gcanalysis/). The chemical distributors were then visualized using ‘ili19. The chemical annotations of features have been cross-referenced from the library search jobs as described in the tutorial. Using balance filters at 50% and >0.9 cosine, we arrived at annotations that, once visualized, revealed the distributions of skin volatiles (Figure 2f–i). For example, squalene was found on all locations, but less on the feet. Hexanoic acid was most abundant on the chest and armpits. Globulol, a perfume ingredient this individual used on the chest, was most intense on the chest, while phenylene dibenzoate, a skincare ingredient, was found on the face and hands.
The 3D model, feature table used for mapping and snapshots shown on the Figure 2f–i are available at: https://github.com/aaksenov1/Human-volatilome-3D-mapping-
Generation of molecular networks
The data were collected across multiple studies as described in the Supplementary Notes. All of the datasets (Table S1) were processed on GNPS MSHub deconvolution workflow as described in the tutorial. The figures were generated as described in the Supplementary Notes.
Testing and validation
All modules have been tested and validated individually to determine possible fail points and the results validated by manually reviewing the annotations that are obtained. The full pipeline was also tested for a variety of datasets, including those collected for this study (“GC-MS analysis for validation studies” section of the Supplementary Notes) and data from several previously published studies and unpublished public data. A variety of GC-MS data are represented, including different types of mass analyzers (both high and low resolution instruments), different modes of sample introduction, and analysis of both derivatized and non-derivatized samples. The goal was to ensure that both feature finding and library matching workflows are operational for all of these scenarios and that the results are consistent with those expected. We have manually verified that the molecules that are known to be present in the dataset are indeed identified and reported by the workflow. The testing information is summarized in Table S1.
Comparison of deconvolution tools
We have compared the deconvolution performance of MSHub alongside MZmine2/ADAP3 and MS-DIAL4. These tools were chosen because they satisfy the following criteria: are open, specifically designed for GC-MS data, can perform multi-file processing, are being routinely used by the metabolomics community, and are actively being developed and maintained. The detailed description of the procedure and parameters are given in the Supplementary Notes.
Generating input files with the alternative workflows
The Mzmine/ADAP and MS-DIAL workflows are the alternative options to perform spectral deconvolution on GC-MS data explicitly supported to be compatible with GNPS library search workflow. For better integration, we have added a new module to MZmine (version 2.52 and later) to export the quantification table (.csv) and the spectra summary file (.mgf) for the GNPS GC-MS workflow. Furthermore, a new MZmine module was also developed to enable the creation of the Kovats RI marker file compatible with the GNPS workflow. The detailed directions are given in the GNPS documentation: https://ccms-ucsd.github.io/GNPSDocumentation/gc-ms-deconvolution/
Generation of plots
All plots were generated in Python 3.7.3, using NumPy 1.16.4, Pandas 0.25.0, RDKit 2019.03.4, and lxml 4.3.4 for data analysis purposes; and Matplotlib 3.1.0 and Seaborn 0.9.0 for visualization purposes The detailed description is given in the Supplementary Notes.
Data and code availability
All of the data used in preparation of this manuscript are publicly available at the MassIVE repository at the UCSD Center for Computational Mass Spectrometry website (https://massive.ucsd.edu). The dataset accession numbers are: #1 (MSV000084033), #2 (MSV000084033), #3 (MSV000084034), #4 (MSV000084036), #5 (MSV000084032), #6 (MSV000084038), #7 (MSV000084042), #8 (MSV000084039), #9 (MSV000084040), #10 (MSV000084037), #11 (MSV000084211), #12 (MSV000083598), #13 (MSV000080892), #14 (MSV000080892), #15 (MSV000080892), #16 (MSV000084337), #17 (MSV000083658), #18 (MSV000083743), #19 (MSV000084226), #20 (MSV000083859), #21 (MSV000083294), #22 (MSV000084349), #23 (MSV000081340), #24 (MSV000084348), #25 (MSV000084378), #26 (MSV000084338), #27 (MSV000084339), #28 (MSV000081161), #29 (MSV000084350), #30 (MSV000084377), #31 (MSV000084145), #32 (MSV000084144), #33 (MSV000084146), #34 (MSV000084379), #35 (MSV000084380), #36 (MSV000084276), #37 (MSV000084277), #38 (MSV000084212).
All of the GNPS analysis jobs for all of the studies are summarized in Supplementary Table 1.
The source code of the MSHub software, including low- and high resolution data processing versions is available online at Github (version used in GNPS) (https://github.com/CCMS-UCSD/GNPS_Workflows/tree/master/mshub-gc/tools/mshub-gc/proc) and at BitBucket (standalone version in MSHub developers’ repository, both high and low resolution: https://bitbucket.org/iAnalytica/mshub_process/src/master/). Scripts used to parse, filter, organize data and generate the plots in the manuscript are available online at Github (https://github.com/bittremieux/GNPS_GC_fig). Script for merging individual .mgf files into a single file for creating global network is available at Github: https://github.com/bittremieux/GNPS_GC/blob/master/src/merge_mgf.py)
The 3D model, feature table with coordinates used for the mapping and snapshots shown on the Figure 4a–d are available at: https://github.com/aaksenov1/Human-volatilome-3D-mapping-. The GC-MS adapted MolNetEnhancer code with an example Jupyter notebook can be found here: https://github.com/madeleineernst/pyMolNetEnhancer.
Supplementary Material
Acknowledgments:
The conversion of the data from different repositories was supported by R03 CA211211 on reuse of metabolomics data, to build enabling chemical analysis tools for the ocean symbiosis program, the development of a user-friendly interface for GC-MS analysis was supported by the Gordon and Betty Moore Foundation through Grant GBMF7622. The UC San Diego Center for Microbiome Innovation supported the campus wide SEED grant awards for data collection that enabled the development of some of this infrastructure. PCD was supported by National Sciences Foundation (NSF) (grant IOS-1656475), and the U.S. National Institutes of Health (NIH) (grants U19 AG063744 01, P41 GM103484, R03 CA211211, R01 GM107550). KV and IL are very grateful for the support of Vodafone Foundation as part of the project DRUGS/DreamLab. ME was supported by the University of Corsica. LFN was supported by the NIH (R01 GM107550), and the European Union’s Horizon 2020 program (MSCA-GF, 704786). AB was supported by the National Institute of Justice Award 2015-DN-BX-K047. Additional support for data acquisition and data storage was provided by P41 GM103484 Center for Computational Mass Spectrometry, the collection of data from the HomeChem project was supported by the Sloan Foundation. GBH, SD, IL, KV and IB are grateful for the support of the OG cancer breath analysis study by the NIHR London Invitro Diagnostic Co-operative and Imperial Biomedical Research Centre, Rosetrees and Stonegate Trusts and Imperial College Charity. DV acknowledges support by ERC-Consolidator Grant No. 724228 (LEMAN). IB acknowledges the contribution of Qing Wen and Dr Michelangelo Colavita for the production of the training video. CC was supported by the Research Foundation Flanders (FWO), with support from the industrial research fund of Ghent University. WB was supported by the Research Foundation Flanders (FWO). AAO acknowledges the support of Fulbright Commission and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET-Argentina). The work of RL and PLB on the dataset 30 was supported by the Metaboscope, part of the “Platform 3A” funded by the European Regional Development Fund, the French Ministry of Research, Higher Education and Innovation, the region Provence-Alpes-Côte d’Azur, the Departmental Council of Vaucluse and the Urban Community of Avignon. SA and ARF acknowledge the PlantaSYST project by the European Unions Horizon 2020 research and innovation programme (SGA-CSA No 664621 and No 739582 under FPA No. 664620). VV acknowledges the support by the National Institute On Alcohol Abuse and Alcoholism award R24AA022057. MG and RC acknowledge the support of the Krupp Endowed Fund grant. A portion of mass spectra in the public reference library was produced within the framework of the State Task for the Topchiev Institute of Petrochemical Synthesis RAS and with the support of the RUDN University Program 5–100. RSB acknowledges support of the State Task for the Topchiev Institute of Petrochemical Synthesis RAS. LNK acknowledges support of the RUDN University Program 5–100. IM acknowledges support of the Israel Science Foundation project number 1947/19 and European Research Council under the European Union’s Horizon 2020 research and innovation program (project number 640384). JS has been supported by NIH/NIAMS R03AR072182, The Colton Center for Autoimmunity, Rheumatology Research Foundation, The Riley Family Foundation and The Snyder Family Foundation. JM acknowledges support from 2017 Group for Research and Assessment of Psoriasis and Psoriatic Arthritis (GRAPPA) Pilot Research Grant and NIH/NIAMS T32AR069515. RG is grateful to the Azrieli Foundation for the award of an Azrieli Fellowship. JJJvdH acknowledges support from an ASDI eScience grant, ASDI.2017.030, from the Netherlands eScience Center-NLeSC. BA was supported by the NSF through the Graduate Research Fellowship Program. GC-MS analyses for collection of the dataset MSV000083743 were supported by the Pacific Northwest National Laboratory, Laboratory Directed Research and Development Program, and were contributed by the Microbiomes in Transition Initiative; data were collected in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy (DOE) Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle Memorial Institute for the DOE under contract DEAC05–76RLO1830. Authors are grateful to Dr. Ricardo da Silva for his contribution to developing the first prototype of the EI data network and his continuous assistance with further development and testing of the infrastructure. Authors are also grateful to Drs. Marina Vance and Delphine Farmer who have organized the sampling for HomeChem indoor chemistry project (https://indoorchem.org/projects/homechem/) that allowed to collect samples for the dataset MSV000083598. Brandon Ross has assisted with collecting data for the dataset MSV000084348. GC-MS analyses for collection of the datasets MSV000084211 and MSV000084212 were supported by the announcement N757 Doctorados Nacionales and project EXT-2016–69-1713 from Departamento Administrativo de Ciencia, Tecnología e Innovación (COLCIENCIAS), the seed project INV-2019–67-1747 and FAPA project of Chiara Carazzone from the Faculty of Science at Universidad de los Andes, and the grant No. FP80740–064-2016 of COLCIENCIAS. Authors are grateful to Lida M. Garzón, Pablo Palacios, Marco Gonzalez and Jack Hernandez for their contributions collecting the samples, and to Jhony Oswaldo Turizo for designing and manufacturing the amphibian electrical stimulator. AS and XD acknowledge the support by the National Cancer Institute award U01CA235507. Authors are grateful to Dr. Steffen Neuman for the feedback regarding the XCMS deconvolution tool.
Footnotes
Competing interests
Pieter C. Dorrestein is a scientific advisor for Sirenas LLC. Mingxun Wang is a consultant for Sirenas LLC and the founder of Ometa labs LLC. Alexander A. Aksenov is a consultant for Ometa labs LLC.
References
- 1.Stein S Analytical Chemistry vol. 84 7274–7282 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Aksenov AA, da Silva R, Knight R, Lopes NP & Dorrestein PC Nature Reviews Chemistry vol. 1 (2017). [Google Scholar]
- 3.Smirnov A et al. Analytical Chemistry vol. 91 9069–9077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsugawa H et al. Nat. Methods 12, 523–526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amigo JM, Skov T, Bro R, Coello J & Maspoch S TrAC Trends in Analytical Chemistry vol. 27 714–725 (2008). [Google Scholar]
- 6.Kessler N et al. Bioinformatics 29, 2452–2459 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domingo-Almenara X et al. Analytical Chemistry vol. 88 9821–9829 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Skogerson K, Wohlgemuth G, Barupal DK & Fiehn O BMC Bioinformatics 12, 321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akiyama K et al. In Silico Biol. 8, 339–345 (2008). [PubMed] [Google Scholar]
- 10.Tautenhahn R, Patti GJ, Rinehart D & Siuzdak G Analytical Chemistry vol. 84 5035–5039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horai H et al. J. Mass Spectrom 45, 703–714 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Nucleic Acids Res. 44, D463–70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll AJ, Badger MR & Harvey Millar A BMC Bioinformatics 11, 376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haug K et al. Nucleic Acids Research vol. 41 D781–D786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hummel J et al. The Handbook of Plant Metabolomics 321–343 (2013) doi: 10.1002/9783527669882.ch18. [DOI] [Google Scholar]
- 16.Wang M et al. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sumner LW et al. Metabolomics vol. 3 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Gupta N, Bandeira N & Pevzner PA Mol. Cell. Proteomics 8, 53–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Protsyuk I et al. Nat. Protoc 13, 134–154 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson MD et al. Sci Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data used in preparation of this manuscript are publicly available at the MassIVE repository at the UCSD Center for Computational Mass Spectrometry website (https://massive.ucsd.edu). The dataset accession numbers are: #1 (MSV000084033), #2 (MSV000084033), #3 (MSV000084034), #4 (MSV000084036), #5 (MSV000084032), #6 (MSV000084038), #7 (MSV000084042), #8 (MSV000084039), #9 (MSV000084040), #10 (MSV000084037), #11 (MSV000084211), #12 (MSV000083598), #13 (MSV000080892), #14 (MSV000080892), #15 (MSV000080892), #16 (MSV000084337), #17 (MSV000083658), #18 (MSV000083743), #19 (MSV000084226), #20 (MSV000083859), #21 (MSV000083294), #22 (MSV000084349), #23 (MSV000081340), #24 (MSV000084348), #25 (MSV000084378), #26 (MSV000084338), #27 (MSV000084339), #28 (MSV000081161), #29 (MSV000084350), #30 (MSV000084377), #31 (MSV000084145), #32 (MSV000084144), #33 (MSV000084146), #34 (MSV000084379), #35 (MSV000084380), #36 (MSV000084276), #37 (MSV000084277), #38 (MSV000084212).
All of the GNPS analysis jobs for all of the studies are summarized in Supplementary Table 1.
The source code of the MSHub software, including low- and high resolution data processing versions is available online at Github (version used in GNPS) (https://github.com/CCMS-UCSD/GNPS_Workflows/tree/master/mshub-gc/tools/mshub-gc/proc) and at BitBucket (standalone version in MSHub developers’ repository, both high and low resolution: https://bitbucket.org/iAnalytica/mshub_process/src/master/). Scripts used to parse, filter, organize data and generate the plots in the manuscript are available online at Github (https://github.com/bittremieux/GNPS_GC_fig). Script for merging individual .mgf files into a single file for creating global network is available at Github: https://github.com/bittremieux/GNPS_GC/blob/master/src/merge_mgf.py)
The 3D model, feature table with coordinates used for the mapping and snapshots shown on the Figure 4a–d are available at: https://github.com/aaksenov1/Human-volatilome-3D-mapping-. The GC-MS adapted MolNetEnhancer code with an example Jupyter notebook can be found here: https://github.com/madeleineernst/pyMolNetEnhancer.