Abstract
Rubus phoenicolasius Maxim. is a traditional Tibetan medicine and widely used in the clinical pharmacology. In current study, the complete chloroplast genome of R. phoenicolasius was reported. The total length of the genome was 155,144 bp with the GC content of 37.9%. We predicted 130 genes in the genome including 84 protein-coding genes, 37 tRNA genes, 8 rRNA genes and 1 pseudogene. 17 genes were duplicated in the IR regions including 7 tRNA, 4 rRNA and 6 protein-coding genes. Phylogenomic analysis revealed that R. phoenicolasius forms a strong supported branch with R. amabilis and R. coreanus under the Rosaceae clade.
Keywords: Rubus phoenicolasius, chloroplast genome, phylogenetic analysis
Rubus phoenicolasius Maxim. (urn:lsid:ipni.org:names:895791-1), belonging to Rosaceae, is mainly distributed in China, Japan and Korea, and is naturalized in Europe and North America (Wu et al. 2003). As a traditional Tibetan medicine, R. phoenicolasius has long been used for the treatment of rheumatism, irregular menstruation and kidney ailments (Yang 1991). In recent years, most research concerning R. phoenicolasius mainly focused on its chemical composition (Luo et al. 2014), pharmacological activity (Liu et al. 2014), genetic diversity (Innis et al. 2011), and little is known about the chloroplast genome of R. phoenicolasius. In the current study, we report the complete chloroplast genome of R. phoenicolasius based on the next-generation sequencing method.
A wild individual of R. phoenicolasius was collected from Qinghai, China (N 32°39′33″, E 100°57′35″). The specimen was deposited at Qinghai-Tibetan Plateau Museum of Biology (HNWP, Xiaofeng Chi, xfchi@nwipb.cas.cn) under the voucher number Chi202002. The total genomic DNA was extracted from the fresh leaves with the modified CTAB method (Doyle 1987). The DNA sample was randomly fragmented to construct paired-end libraries according to the Illumina standard protocol and the VAHTS Universal DNA Library Prep Kit (Vazyme Biotech, Nanjing, China) was used for preparing the library. Sequencing was conducted on the Illumina Novaseq platform (San Diego, CA, USA). The Q20 and Q30 of the clean data were 97.65% and 92.92%, respectively, which showed that the quality of sequencing was fine. The complete chloroplast genome was assembled by SPAdes v3.10.1 (Bankevich et al. 2012) with the kmer setting at 55, 87 and 121. Annotation was performed on CPGAVAS2 (Shi et al. 2019) coupled with manual adjustment of start/stop codons and intron/exon borders after BLAST searches.
The total length of the R. phoenicolasius chloroplast genome was 155,144 bp including two inverted repeats (IR, 25,973 bp), a large single-copy region (LSC, 84,618 bp) and a small single-copy region (SSC, 18,580 bp). The GC content of the genome is 37.9% with the LSC of 35.6%, the IR of 42.9% and SSC of 31.9%. 130 genes was predicted, including 84 protein-coding genes, 37 tRNA genes, 8 rRNA genes and 1 pseudogene (ycf1). Seven tRNA, four rRNA and six protein-coding genes were duplicated in the IR regions.
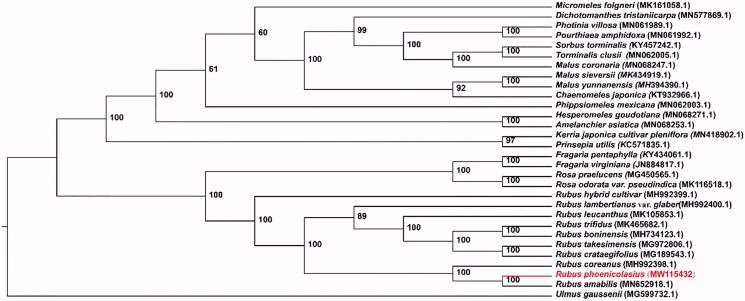
Phylogenetic analysis was performed on the PhyloSuite platform (Zhang et al. 2020). The whole plastome sequences of R. phoenicolasius and 28 species in Rosaceae was used for the analysis. Meanwhile Ulmus gaussenii (Ulmaceae) was used as the outgroup. Sequences were aligned using MAFFT (Kazutaka and Standley 2013), and ModelFinder was employed to select the best model for phylogenetic analysis. A maximum likelihood (ML) analysis was conducted on IQ-TREE based on the TVM + F+R3 nucleotide substitution model with 1000 replications (Figure 1). The phylogenetic tree showed that R. phoenicolasius forms a strong supported branch with R. amabilis and R. coreanus under the Rosaceae clade. We expect that the chloroplast genome of R. phoenicolasius will be a valuable resource for future studies on conservation genetics, taxonomy, and phylogeny involving this particular species.
Figure 1.
IQ-TREE based on 30 complete chloroplast genome sequences. Numbers associated with the branches are bootstrap values.
Funding Statement
This work was supported by Applied Basic Research of Qinghai Province [2020-ZJ-749].
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The chloroplast genome and raw sequencing data in this study are available in GenBank (https://www.ncbi.nlm.nih.gov/) under the accession numbers of MW115432 and SRR13021958.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Innis AF, Forseth IN, Whigham DF, Mccormick MK.. 2011. Genetic diversity in the invasive rubus phoenicolasius as compared to the native rubus argutus using inter-simple sequence repeat (ISSR) markers. Biol Invasions. 13(8):1735–1738. [Google Scholar]
- Kazutaka K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liao ZX, Liu SJ, Sun JY, Yao GY, Wang HS.. 2014. A new pregnane glycoside from Rubus phoenicolasius and its antiproliferative activity. Nat Prod Res. 28(21):1843–1846. [DOI] [PubMed] [Google Scholar]
- Luo GF, Liu C, Ji LJ.. 2014. Study on the flavonoid constituents of Rubus phoenicolasius maxim. Northwest Pharm J. 29(1):5–8. [Google Scholar]
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C.. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZY, Raven PH, Hong DY.. 2003. Flora of China, Vol. 9. Beijing: (China): Science Press, St. Louis: Missouri Botanical Garden Press. [Google Scholar]
- Yang YC. 1991. Flora Tebitan medicine. Xininng (China): Qinghai People's Publishing House. [Google Scholar]
- Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT.. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The chloroplast genome and raw sequencing data in this study are available in GenBank (https://www.ncbi.nlm.nih.gov/) under the accession numbers of MW115432 and SRR13021958.