Abstract
The complete chloroplast genome of Quercus virginiana was sequenced with Illumina HiSeq 2000 platform. It was a typical quadruple structure as other plants of Quercus with 161,221 bp in length, including a large single-copy (LSC: 90,553 bp) region and a small single-copy (SSC: 19,016 bp) which were separated by a pair of inverted repeats (IRa, b: 25,826 bp) region. The overall GC content is 36.9%. A total of 131 genes was annotated which contained 86 protein-coding genes including the Trans splicing gene of rps12, 37 tRNA genes, and 8 rRNA genes. ML phylogenetic analysis compared with 17 expressed chloroplast genomes revealed that Q. virginiana was a sister to other species of Quercus, which were grouped together with five species of Section Quercus and another 12 species of Quercus were divided into another group.
Keywords: Quercus virginiana, chloroplast genome, phylogenetic analysis
Quercus virginiana Mill. is a multipurpose tree species of Quercus in Fagaceae, one of the commonest and best known species in the coastal region of the southeastern United States. Which was the strategic resource tree of the United States, because of it’s the hardest, strongest and toughest wood of all oaks (Elias 1971). It is also an excellent evergreen broad-leaved tree species in beach wetland, saline alkali land greening and coastal windbreak forest construction (Harms 1990). Quercus virginiana has been widely and long-term introduced in the Yangtze River Delta of China (Chen et al, 2007). And the characteristics of adaptability of introduced species, such as cold resistance (Zhang et al. 2018), light adaptability (Wang et al. 2019) and mycorrhizal types (Jin et al. 2020) have been revealed at present. However, the genomic information of Q. virginiana lacked that would be helpful to the study of scientific introduction and efficient utilization.
The fresh leaves of Q. virginiana were collected from the living individual permanently conserved in the Quercus gene bank (36.77°N, 117.471°E) which was introduced and bred in 2014. The specimens were preserved in the herbarium of Shandong Forest Germplasm Resources Center (barcode SDF2019L0012). Total genomic DNA (saved in DNA library of Shandong Forest Germplasm Resources Center with the code of fjnyl2020cp03) was extracted by the Plant DNA extraction Kit (TIANGEN, Beijing, China) according to the requirements of the reagent company.
Paired-end reads were constructed according to the Illumina library preparation protocol and sequenced on an Illumina HiSeq 2000 platform. The whole chloroplast genome of Q. virginiana was assembled by MITObim v1.8 (Hahn et al. 2013) and was annotated in DOGMA (http://dogma.ccbb.utexas.edu/). The whole chloroplast genome of Q. virginiana and other 17 published plastomes of Quercus were conducted by using MAFFT v7.429 (Katoh and Standley 2013), with Juglans cathayensis as outgroup. Maximum-likelihood (ML) phylogenetic tree with 1000 bootstrap replicates was inferred using IQ-TREE v1.6.12 (Lam-Tung et al. 2015) and TVM + F+R2 model.
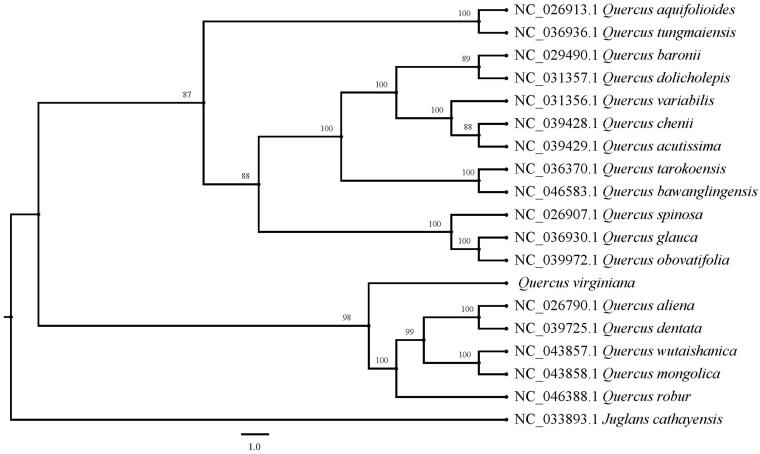
The chloroplast genome of Q. virginiana (GenBank accession number MT916773) was also a typical quadruple structure with 161,221 bp in length that contains a large single-copy (LSC: 90,553 bp) region and a small single-copy (SSC: 19,016 bp), which were separated by a pair of inverted repeats (IRa, b: 25,826 bp) region. The overall GC content was 36.9%. A total of 131 genes were annotated, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among these, 15 genes had a single intron respectively, clpP and ycf3 had two introns respectively. While rps12 had Trans splicing function. The chloroplast genome of Q. virginiana was similar to that of Q. aliena, Q. dentate, Q. wutaishanica, Q. mongolica and Q. robur, which were grouped together as the member of the Section Quercus. While the other 12 species of Quercus were divided into another group. ML phylogenetic analysis compared with 17 expressed chloroplast genomes of Quercus revealed that Q. virginiana was a sister to other Quercus species (Figure 1).
Figure 1.
A maximum-likelihood (ML) tree of Q. virginiana and other 17 related species of Quercus based on the complete chloroplast genome sequences with J. cathayensis as outgroup. The accession numbers are showed in the figure, and the numbers behind each node are bootstrap support values.
Funding Statement
The study was financially supported by “Collection, Conservation, and Accurate Identification of Forest Tree Germplasm Resources” of Shandong Provincial Agricultural Elite Varieties Project (2019LZGC018).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT 916773. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA694782, SRR13528289, and SAMN17574357 respectively.
References
- Chen Y, Chen Y, Huang Y, Sun H, Chen D.. 2007. Preliminary study on Quercus virginiana introduction in Eastern China. Forest Research. 20(4):542–546. [Google Scholar]
- Elias TS. 1971. The genera of Fagaceae in the southeastern United States. J Arnold Arbor. 52:159–195. [Google Scholar]
- Hahn C, Bachmann L, Chevreux B.. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms WR. 1990. Quercus virginiana Mill. live oak. In: Burns RM, Hokala BH, editors.. Silvics of North America Vol. 2: hardwoods. Agriculture handbook No. 654. Washington: USDA Forest Service, p. 751–754. [Google Scholar]
- Jin W, Yang Y, Sun H, Chen L, Yuan Z.. 2020. Diversity of ectomycorrhizal fungi a seed collecting forest of Quercus virginiana. Scientia Silvae Sinicae. 56(1):120–132. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam-Tung N, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32 (1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Shi X, Chen Y, Sun H, Wang T, Chen Y.. 2019. Effects of shading on growth, photosynthesis and chlorophyll fluorescence parameters in leaves of Quercus virginiana seedlings. Forest Research. 32(5):99–106. [Google Scholar]
- Zhang J, Lu F, Shen C, Wang G, Meng G, Tian J, Luo F, Chen Y, Qian L, Liu M.. 2018. Study on cold resistance of introduced evergreen broad-leaved tree species in Xuzhou region. Forest Resour Manage. 6(6):84–89. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT 916773. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA694782, SRR13528289, and SAMN17574357 respectively.