Abstract
Achimenes Pers. is well known for its floral diversity and widely used in horticulture, but its phylogenetic position is still poorly understood. And most research about the complete chloroplast genome sequence focused on the Old World species; therefore, we think it is necessary to examine taxa of the New World in more detail. This study determined the complete chloroplast genome of Achimenes cettoana H.E. Moore. The cp genome was 153,011 bp in a total length containing two inverted repeats (IRs) of 25,162 bp separated by a large single-copy (LSC) region of 84,669 bp and a small single-copy (SSC) region of 18,018 bp. The whole cp genome of A. cettoana contains 112 genes, including 79 protein-coding genes, 29 tRNA genes, and four rRNA genes. This plastid genome is firstly reported in the New World Gesneriaceae, which will be a valuable resource for future studies on breeding, conservation genetics, and phylogeny of Gesneriaceae.
Keywords: Achimenes, chloroplast genome, phylogenetics
Based on molecular phylogenetic studies, the first detailed and overall classification of Gesneriaceae was made by Weber et al. (2013), three subfamilies were recognized: Sanangooideae (monospecific with Sanango racemosum G.S. Bunting & J.A. Duke), Gesnerioideae, and Didymocarpoideae. Both Subfam. Sanangooideae and Subfam. Gesnerioideae are distributed in the New World (Titanotrichum Soler. excluded), and Subfam. Didymocarpoideae is distributed in the Old World. Achimenes Pers. (belonging to the Subfam. Gesnerioideae) is an herbaceous genus of about 24 species (IPNI 2020; Tropicos 2020), distributed from southern Mexico through Central America (Roa et al. 2002; Roalson and Roberts 2016).
The DNA sequence of the chloroplast genome can be used as a valuable resource for future studies on breeding, conservation genetics and phylogeny. Within Gesneriaceae, all chloroplast genomes reported belong to Subfam. Didymocarpoideae and no complete chloroplast sequence for the New World Gesneriaceae have been published to date.
The members of the Gesneriad Society collected seeds of Achimenes cettoana, which grows on rocks in the stream of a shaded ravine, from Chiapas, Mexico (GPS: 16.74 N, 92.69 W). Then seeds were shared to GCCC, and were being cultivated in the nursery of GCCC since 2015. Achimenes cettoana is a compact plant and adapts well to cultivation (Moore 1961), especially for a windowsill or under artificial light. The voucher specimen (HZJ001) was deposited at the Herbarium of Guangxi Institute of Botany, CAS (IBK). The chloroplast DNA of A. cettoana was extracted by the CTAB method (Doyle and Doyle 1987) and sequenced using the Illumina Hiseq 4000 sequencing platform (Majorbio Company (http://www.majorbio.com/, China). We used Map to Reference function in Geneious R11 (Kearse et al. 2012) to exclude nuclear and mitochondrial reads using the plastid genome of Petrocodon jingxiensis (GenBank-MK887172) as reference. Putative chloroplast reads were used for de novo assembling construction using Geneious R11. Generated contigs were concatenated into larger ones using the Repeat Finder function in Geneious R11. The original data were again mapped to the larger contigs to extend their boundaries until all contigs were able to concatenate to one contig. The inverted repeat (IR) region was determined using the Repeat Finder function in Geneious R11 and was inverted and copied to obtain the complete chloroplast sequence. DOGMA program was used for annotation of the complete chloroplast genome (Wyman et al. 2004) and the annotation was corrected with Geneious R11 (Kearse et al. 2012). The complete chloroplast genome of A. cettoana was 153,011 bp in length (MT627324), and the GC content was 38.2%. Large single-copy (LSC) and small single-copy (SSC) contained 84,669 bp and 18,018 bp, while IRs were 25,162 bp in length. The plastid genome included 79 protein-coding genes, 29 tRNA genes, and four rRNA genes.
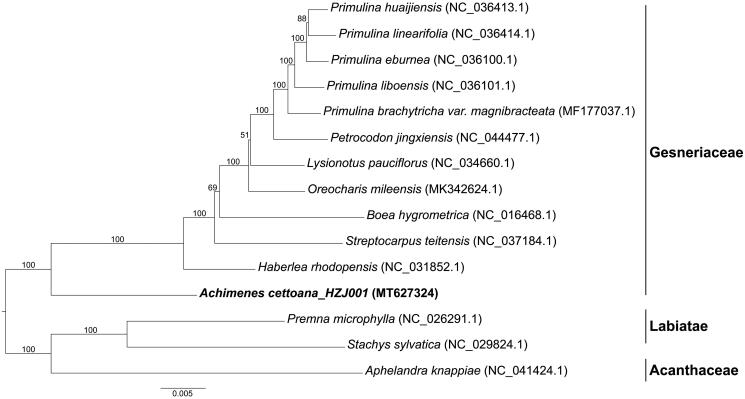
To analyze the phylogenetic position of A. cettoana, the concatenated sequences of protein-coding genes of A. cettoana and 11 other species from Gesneriaceae were aligned using MAFFT version 7.307 (Katoh and Standley 2013) and 3 species from Labiatae and Acanthaceae were selected as outgroup. A phylogenetic tree (Figure 1) was constructed with the maximum likelihood (ML) method and the TVM + F + R2 evolutionary model using RAxML (Stamatakis 2014). The result was congruent with previous studies that established that Achimenes is a member of the New World Gesneriaceae (Clark et al. 2012; Weber et al. 2013). The topology of this molecular tree supports the division of this subfamily and shows that A. cettoana is a representative of Subtr. Gloxiniinae and is a sister group of Gesneriinae (Clark et al. 2012). The newly reported chloroplast genome of A. cettoana allows developing markers for further studies on resolving the relationship within the Subfam. Gesnerioideae and particularly, the genus Achimenes.
Figure 1.
Phylogenetic tree reconstructed by maximum likelihood (ML) analysis based on concatenated sequences of protein-coding genes from 15 taxa of Lamiales, numbers upon branches are assessed by ML bootstrap.
Acknowledgments
We would like to thank Michael LoFurno (Adjunct Professor, Temple University, Philadelphia, PA, USA) for his editorial assistance.
Funding Statement
This study was supported by Guilin Science and Technology Foundation. [20170107-2/20180107-6], Guangxi Science and Technology Project [Guike AD20159091], Basal Research Fund of GXIB [Guizhiye20009], 21st Talent project of ‘Ten-Hundred-Thousand’ in Guangxi, and the Foundation of Guangxi Key Laboratory of Plant Conservation and Restoration Ecology in Karst Terrain [19-050-6].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT627324.
References
- Clark JL, Funke MM, Duffy AM, Smith JF.. 2012. Phylogeny of a neotropical clade in the Gesneriaceae: more tales of convergent evolution. Int J Plant Sci. 173(8):894–916. [Google Scholar]
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull. 19:11–15. [Google Scholar]
- IPNI. 2020. The international plant names index [accessed 3 July 2020]. http://www.ipni.org.
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore HE. 1961. Achimenes cettoana (Gesneriaceae). Baileya. 8:144. [Google Scholar]
- Roa AR, Skog LE, Ramirez A.. 2002. Novae Gesneriaceae neotropicarum X: a new species of Achimenes from Mexico. Novon. 12(3):382–384. [Google Scholar]
- Roalson EH, Roberts WR.. 2016. Distinct processes drive diversification in different clades of Gesneriaceae. Syst Biol. 65(4):662–684. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropicos. 2020. Tropicos.org. Missouri botanical garden. [accessed 3 July 2020]. http://www.tropicos.org.
- Weber A, Clark JL, Möller M.. 2013. A new formal classification of Gesneriaceae. Selbyana. 31(2):68–94. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]
- Xin ZB, Fu LF, Fu ZX, Li S, Wei YG, Wen F.. 2019. Complete chloroplast genome sequence of Petrocodon jingxiensis (Gesneriaceae). Mitochondrial DNA Part B. 4(2):2771–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT627324.