ABSTRACT
Single treatment of plants with pathogens like Pseudomonas syringae can trigger systemic acquired resistance (SAR) that lasts several days to several weeks in Arabidopsis thaliana. Similar primed resistances were described for abiotic stresses like drought and heat stress. Most studies about plant resistance to ultraviolet (UV)-radiation used low UV-B radiations over a long period. These experimental designs make it difficult to distinguish acclimation effects from real cellular memory which facilitate transcriptional and other responses to a second UV-radiation after a latent phase. Here we present a novel UV-B priming system. We demonstrate that a single UV-B treatment, which causes neither visible damage nor accumulation of pigments, can stimulate resistance against UV-B stress. After a second damaging UV-B treatment, UV-primed plants showed significantly reduced damage in comparison to non-primed plants. Furthermore, the acquirement of the induced UV-B resistance was impaired in uvr8-6 mutants suggesting that the UV-B receptor is essential for UV-B stress memory in Arabidopsis. We discuss advantages and limits of our UV-B priming system which will be a powerful tool to investigate UV-B memory in future studies.
KEYWORDS: Abiotic plant resistance, acquired plant resistance, UVR8, NRP1, primed genes, WRKY30, CHS
Introduction
Plants use sunlight as source of energy and as environmental signal to regulate growth and development.1,2 UV-B, which is an integral part of natural sunlight, is involved in photomorphogenesis but can also cause damage and even necrosis.3 In Arabidopsis thaliana, UV RESISTANCE LOCUS8 (UVR8) encodes the well-known UV-B photoreceptor.4,5 uvr8 mutants are highly sensitive to UV-B that generates more cellular damage than in wild-type.6,7 The expression of many UV-B response genes are dependent on UVR8 including genes, which encode enzymes like CHALCONE SYNTHASE (CHS) involved in flavonoid and anthocyanin biosynthesis.6 Nevertheless, other pathways can induce expression of UVR8-independent UV-B response genes like WRKY30.3
In plant research, priming systems are composed of minimum three steps: (I) Priming treatment, which usually does not cause visible changes to the plant, (II) second treatment that may or may not cause damage, and (III) measurement of the readout that can be at mRNA or protein levels, or changes to the plant phenotype including damages. For example, priming by treatment of distal Arabidopsis leaves with acibenzolar S-methyl (BTH), a synthetic analog of the stress phytohormone salicylic acid (SA), sets the histone modifications H3K4me3 at the promoters of WRKY transcription factors.8 This activation-related histone modifications do not per se lead to higher expression of the WRKY genes but when the primed plants are stressed by water injection, the transcription rates of the WRKYs significantly increase in comparison to non-primed plants.8 Comparable to the WRKYs, some response genes of drought and heat stress are also highly ‘trainable’ genes that are associated with high H3K4me3 levels after a priming stress treatment, which contributes to transcriptional memory in plants.1,9–11
Recent studies about acquired resistance and photomorphogenic responses to UV-B used low UV-B radiations over a long period.12,13 The described phenomena therefore are rather classical acclimation effects. Hence, we present here a novel UV-B priming system that allows to investigate the memory of a single UV-B radiation event which facilitates transcriptional and other responses to a second UV-B radiation after a latent phase.
Results and discussion
We designed our UV-B priming system in analogy to the BTH/water stress system of Jaskiewicz et al., 2011.8 The UV-B priming system included three steps (Figure 1a): (I) a short non-damaging UV-B treatment (priming, +/) of 14 days old seedlings, (II) 3 days later, a second treatment with damaging UV-B stress (/+), and (III) 3 h after the second treatment, measurement of expression changes and of the damage, few days later. UV-B trainable genes should be significantly higher expressed in primed plants (+/+), whereas primed plants should have significant reduced damage (Figure 1a). We chose to apply high UV-B radiation (35 µW/cm2) which can damage the leaves of the seedlings. In order to determine the maximal time period in this setting, which induces no visible damage, we performed a time course with Col-0 seedlings (Figure 1b). Three days after UV-B treatment, plants radiated for 15 min or less did not show any visible damage, while 20 min UV-B treatment caused damage of at least one primary leaf by 22% of the Col-0 plants. In the same time-course, all plants showed damage if they were UV-B treated for 50 min or longer (Figure 1b). Taking a safety reserve into account, we specified the duration of UV-B priming to 10 min, and the second damaging treatment to 60 min. Since UV-B responses are widely dependent on UVR8 activity,14,15 we used uvr8-6 mutants as control (Figure 1c-Figure 1f). The damage by UV-B were significantly reduced in primed (+/+) in comparison to non-primed Col-0 plants (-/+), whereas primed (+/+) uvr8-6 plants were equally damaged as non-primed uvr8-6 plants (-/+). These findings suggest that Arabidopsis plants can acquire a certain resistance to UV-B stress after a single treatment with UV-B, and that this type of UV-B memory seems to depend on UVR8 signal transduction.
Figure 1.
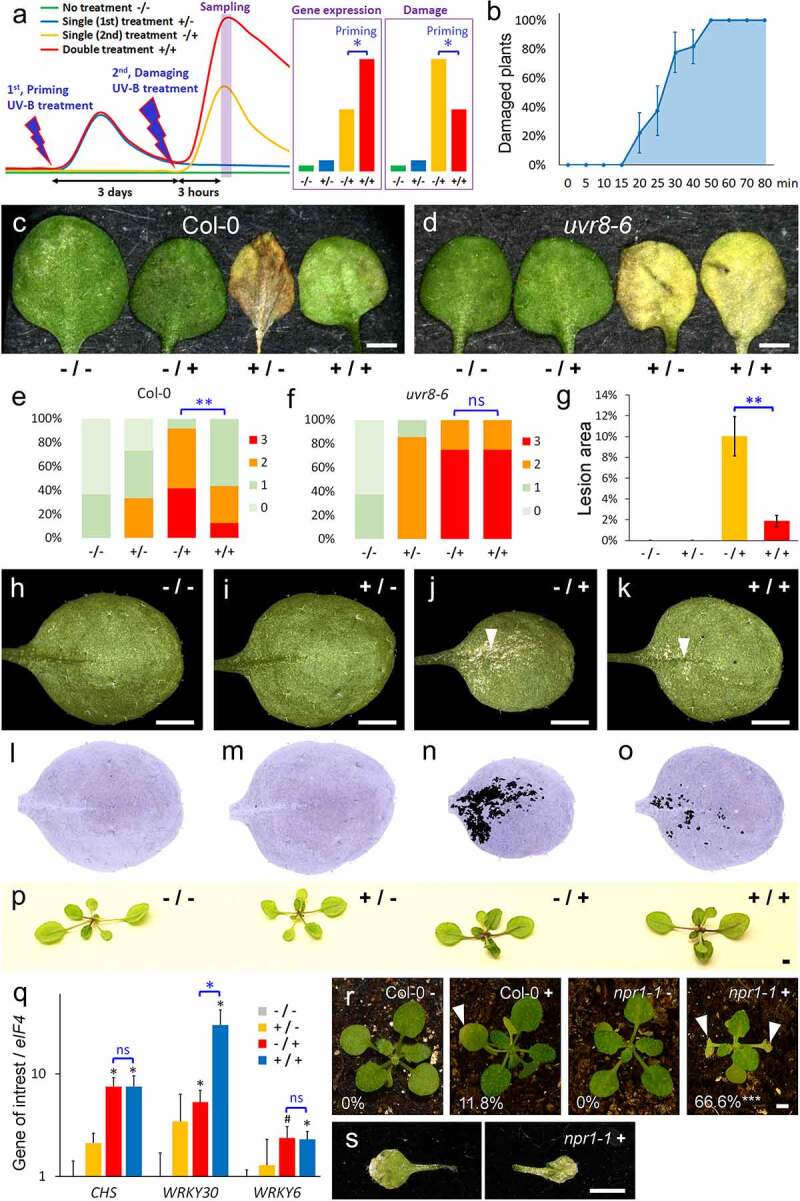
UV-B priming system. (a) Conceptional model of the UV-B priming system. The plant material of all treatments is harvested to one time point (Sampling). Priming: A gene is primed if its expression is significantly higher after double treatment (+/+) than without priming (1st) treatment (-/+), whereas primed plants show reduced damage. (b) Percentage of plants with at least one damaged primary leaf dependent of the duration (min, minutes) of a single high dose UV-B treatment; N = 9. (c-f) Damage of Col-0 (c,e) and uvr8-6 primary leaves (d,f) exposed to the four treatments of the UV-B priming system four days after UV-B second treatment (high dose). The plants were classified into four damage categories (e,f); N ≥ 12 (Col-0) or 4 (uvr8-6). (g-o) Damage of Col-0 primary leaves exposed to the four treatments, five days after the second UV-B treatment (low dose). Arrow heads mark lesions in leaf surface. (g) N = 9. (l-o) False color images of (h-k). (p) Pigment accumulation in Col-0 plants exposed to the four treatments, seven days after second UV-B treatment (low dose). Equal treated plants displayed similar colored leaves, whereas -/+ and +/+ plants were not distinguishable (N = 10 plants, each treatment). (q) Quantitative RT-PCR analyses of mRNA expression in seedlings (17 DAG, 3 hours after 2nd treatment) normalized by eIF4. The expression levels are indicated as the mean of relative fold changes of three biological replicates; values are scaled to -/- = 1; the black bars represent the standard errors. (r-s) Damage of primary leaves of Col-0 (N = 17) and npr1-1 plants (N = 15) exposed to UV-B priming treatment three days after UV-B second treatment (+, high dose). Note the limited damage of one primary leaf in Col-0 + in comparison to the necrotic leaves in npr1-1 + (arrow heads). (s) Detached primary leaves of the plant in (R, npr1-1 +). (a-s) Asterisks indicate significant changes in comparison to -/-, whereas the blue asterisks indicate significant changes between -/+ and +/+ (Student’s t test: #, P = .06; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). All scale bars = 2 mm
Previously, we used low UV-B radiation in an almost identical experimental setting (Figure 1g-Figure 1q). Priming with 20 minutes low UV-B radiation (+/-) did not cause any visible damage or accumulation of pigments like flavonoids and anthocyanins (Figure 1i,Figure 1m,Figure 1p). The second treatment (-/+) with low UV-B radiation caused rather lesions on the leaf surface (Figure j,n) than programmed cell death of whole primary leaves, which we observed after high UV-B radiation (Figure 1c). Nevertheless, primed leaves (+/+) had significantly less lesions than non-primed leaves (-/+; Figure 1g). In order to identify trainable genes, we test the expression pattern of the known UV-B response genes CHS3,6 and WRKY303 by qRT-PCR (Figure 1q). Surprisingly, the expression levels of CHS were identical in primed (+/+) and non-primed plants after the second stress (-/+). Also long-term observation did not reveal any differences in the darker leaf color between primed (+/+) and non-primed plants (-/+), and therefore the accumulation of pigments depended only on the second UV-B stress. These findings suggest that differential pigmentation is not involved in the resistance to UV-B damage (Figure 1p). On the contrary, WRKY30 expression was significantly increased in primed (+/+) in comparison to non-primed Col-0 plants (-/+; Figure 1q). Nonetheless, previous studies showed that the induction of WRKY30 by UV-B is independent of UVR8 activity3 making it very unlikely that the trainable WRKY30 expression contributes to the observed UVR8-dependent UV-B resistance.
In our review Müller-Xing et al. (2014), we proposed that SA could have the function of a short-term memory for UV-B but also other stresses.1 WRKY30 and other WRKYs like WRKY6 are known targets of SA signaling.16 Hence, the expression pattern of SA inducible WRKYs could correspond to similar pattern of SA levels. To test this hypothesis, we tested also the expression of WRKY6, but whose mRNA levels were identical in primed (+/+) and non-primed Col-0 plants (-/+; Figure 1q). SALICYLIC ACID INSENSITIVE 1/NONEXPRESSER OF PR GENES 1 (NPR1) encodes the main receptor of SA.17 Therefore, we used npr1-1 mutants18 in our UV-B priming system for a more direct test of the role of SA signaling in UV-B memory (Figure 1r). Unexpectedly, the priming stress of 10 min triggered strong damage to primary leaves of two-third of the npr1-1 plants but not in the wild-type control (Figure 1r-Figure 1s), which made it impossible to test UV-B memory of npr1-1 in our priming system. These findings suggesting that NPR1-dependent SA signaling is rather essential for a general UV-B resistance than for the UV-B memory, which we observed in our UV-B priming system.
Conclusion and outlook
With our UV-B priming system, we provide a powerful tool to investigate UV-B memory in future studies. Nevertheless, it may be useful to optimize the system first, for example, by using filters, which can cut off unintended wave lengths. Notably, we had difficulties to achieve robust results with narrow band UV-B lamps. Our data suggest that UVR8 is a key component of this type of UV-B memory, whereas SA signaling did not appear to be important for acquiring but essential for general UV-B resistance. Since loss of UVR8 impaired the acquired UV-B resistance, testing other components of the UVR8 signal pathway seems a worthwhile approach for the UV-B priming system. Furthermore, the system can be used as platform for the identification of other genes involved in UV-B stress memory either by whole genome expression analysis of all four treatments (Figure 1a) or a mutagenesis screen for mutants impaired in the UV-B stress memory.
Methods
Arabidopsis wild-type (Col-0), uwr8-6 (SALK_033468) and npr1-118 plants were grown at 21°C under long-day (16 h light/8 h dark). For UV-B treatment in the priming system, we used two UV-B broadband lamps (290–315 nm) from Koninklijke Philips Electronics N.V. or from Shanghai Chenchen Lighting Electric Appliances Co., Ltd: high UV-B doses (35 µW/cm2 measured by UV-B illuminometer MC01080376 Beijing Shida Photoelectric Technology Co., Ltd) with lamp-to-plant distance of 32 cm; low UV-B doses with lamp-to-plant distance of 120 cm (about 7% of high UV-B dose). 14 days after germination, half of the plants was UV-B treated for 10 min with high UV-B dose or 20 min with low UV-B dose (1st, priming treatment; Figure 1a). After 3 days, a subset of the plants (-/+ and +/+) was radiated for 1 h with high or low UV-B doses, respectively (2nd, damaging treatment; Figure 1a. For expression analysis, plant material of all four treatments was harvested 3 h after the 2nd treatment. The phenotype including damage analysis took place four to 7 days later. RNA isolation, cDNA synthesis and qRT-PCR including eIF4 primers were descript before.19 We used primers for CHS (5ʹ-CACTGCTAACCCTGAGAACC-3ʹ and 5ʹ-ACTTGTCGCACATGCGC-3ʹ), WRKY6 (5ʹ-GAAGCTCCGATGATAAGCGA-3ʹ and 5ʹ-AACGTTGAACTTGTTTGCGA-3ʹ) and WRKY30 (5ʹ-CAAGTTTCTCAGGGTGGAGG-3ʹ and 5ʹ-TGACTTCTTCGAACTCTTGATGAC-3ʹ) in the gene expression analysis. In this study, all values represent the mean ± standard error. One-tailed Student’s t tests, resulting in P values, were employed to assess statistical significance between pairs of values.
Acknowledgments
We thank for the support by Prof Dr Daniel Schubert since parts of the work by RMX were performed in his former Lab at the Heinrich-Heine-University, Düsseldorf.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Project No. 31640054 and 31771602), the Fundamental Research Funds for the Central Universities, China (Grant No. 2572016DA03) and Natural Science Foundation of Heilongjiang Province of China, General Program (Grant No. C2016007) to QX and RMX. We are also grateful for financial support from the Northeast Forestry University Starting Grant for Distinguished Young Scholar to RMX. Older parts of this study was supported by a FP7 Marie Curie-IEF fellowship (Plant-Memory) of the EU and by a return fellowship of the German Academic Exchange Service (DAAD) to RMX.
Authors contribution
RMX conceived and initiated the work, YX and RMX performed the experiments, QX and RMX wrote the manuscript.
Disclosure of potential conflicts of interest
Contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the funding agency.
References
- 1.Müller-Xing R, Xing Q, Goodrich J.. Footprints of the sun. Memory of UV and light stress in plants. Front Plant Sci. 2014;5:1. doi: 10.3389/fpls.2014.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourbousse C, Barneche F, Laloi C.. Plant chromatin catches the sun. Front Plant Sci. 2019;10:1728. doi: 10.3389/fpls.2019.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown BA, Jenkins GI.. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 2008;146:576–4. doi: 10.1104/pp.107.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang T, Yang Y, Liu H. Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytol. 2019;221:1247–1252. doi: 10.1111/nph.15469. [DOI] [PubMed] [Google Scholar]
- 5.Rizzini L, Favory -J-J, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schafer E, Nagy F, Jenkins GI, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science (New York, N Y). 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 6.Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA. 2005;102:18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130:234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaskiewicz M, Conrath U, Peterhänsel C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 2011;12:50–55. doi: 10.1038/embor.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Y, Fromm M, Avramova Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat Commun. 2012;3:740. doi: 10.1038/ncomms1732. [DOI] [PubMed] [Google Scholar]
- 10.Song Z-T, Zhang -L-L, Han -J-J, Zhou M, Liu J-X. Histone H3K4 methyltransferases SDG25 and ATX1 maintain heat stress gene expression during recovery in Arabidopsis. Plant J. 2020. doi: 10.1111/tpj.15114. [DOI] [PubMed] [Google Scholar]
- 11.Lämke J, Brzezinka K, Altmann S, Bäurle I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. Embo J. 2016;35:162–175. doi: 10.15252/embj.201592593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao X, Liu W, Yang H-Q, Jenkins GI. A dynamic model of UVR8 photoreceptor signalling in UV-B-acclimated Arabidopsis. New Phytol. 2020;227:857–866. doi: 10.1111/nph.16581. [DOI] [PubMed] [Google Scholar]
- 13.Findlay KM, Jenkins GI. Regulation of UVR8 photoreceptor dimer/monomer photo-equilibrium in Arabidopsis plants grown under photoperiodic conditions. Plant Cell Environ. 2016;39:1706–1714. doi: 10.1111/pce.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin R, Ulm R. How plants cope with UV-B. From perception to response. Curr Opin Plant Biol. 2017;37:42–48. doi: 10.1016/j.pbi.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins GI. Photomorphogenic responses to ultraviolet-B light. Plant Cell Environ. 2017;40:2544–2557. doi: 10.1111/pce.12934. [DOI] [PubMed] [Google Scholar]
- 16.Besseau S, Li J, Palva ET. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot. 2012;63:2667–2679. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Zhang D, Chu J, Boyle P, Wang Y, Brindle I, De Luca V, Després C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012;1:639–647. doi: 10.1016/j.celrep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/S0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Zheng Y, Xing Q, Ardiansyah R, Zhou H, Ali S, Jing T, Tian J, Song XS, Li Y, et al. Ectopic expression of the transcription factor CUC2 restricts growth by cell cycle inhibition in Arabidopsis leaves. Plant Signal Behav. 2020;15:1706024. doi: 10.1080/15592324.2019.1706024. [DOI] [PMC free article] [PubMed] [Google Scholar]


