ABSTRACT
Drought is one of the main abiotic factors that affect agricultural productivity, jeopardizing food security. Modern biotechnology is a useful tool for the generation of stress-tolerant crops, but its release and field-testing involves complex regulatory frameworks. However, gene editing technology mediated by the CRISPR/Cas9 system is a suitable strategy for plant breeding, which can lead to precise and specific modifications in the plant genome. The aim of the present work is to produce drought-tolerant plant varieties by modifying the trehalase gene. Furthermore, a new vector platform was developed to edit monocot and dicot genomes, by modifying vectors adding a streptomycin resistance marker for use with the hypervirulent Agrobacterium tumefaciens AGL1 strain. The gRNA design was based on the trehalase sequence in several species of the genus Selaginella that show drought tolerance. Arabidopsis thaliana carrying editions in the trehalase substrate-binding domain showed a higher tolerance to drought stress. In addition, a transient transformation system for gene editing in maize leaves was characterized.
KEYWORDS: Gene editing, drought tolerance, editing vectors, Trehalose
Introduction
Drought is one of the main abiotic factors affecting crop productivity and, together with increasing scarcity of water for irrigation and consumption, puts food security at risk around the world. Drought tolerance is a quantitative trait, and several biotechnological strategies have been used to obtain new varieties, including genetically modified plants that express genes conferring adaptation to water deficit. These examples include the modification of the anabolic pathway of the disaccharide trehalose, given its role in tolerance to abiotic stress.1,,2 Trehalose (α-D-glucopyranosyl-1,1-α-D-glucopyranoside) is a non-reducing disaccharide consisting of two D-glucose units linked by an α, α-1,1 bond. Plants, like many other eukaryotes, possess only one pathway for trehalose biosynthesis, compared to the multiple pathways found in prokaryotes and archaea.3 The plant trehalose biosynthetic pathway begins with the condensation of UDP-glucose and glucose 6-phosphate, where trehalose 6-phosphate synthase (TPS) catalyzes the formation of trehalose 6-phosphate. Trehalose 6-phosphate is converted to trehalose by trehalose 6-phosphate phosphatase (TPP). In Arabidopsis, there are 11 ORFs for TPS and 10 for TPP.4 In plants, the catabolic pathway consists of a single reaction, where the enzyme trehalase (TRE1, EC 3.2.1.28) hydrolyzes trehalose into two molecules of D-glucose.2,5
Trehalose plays an important role in microorganisms, as a carbon source as well as a means of carbohydrate storage and transport, and as osmoprotector against different types of stress including water limitation, osmotic shock, freezing, salinity, oxidation, and radiation. In general, low concentrations of trehalose are found in plants. However, drought-tolerant plants such as Selaginella lepidophylla, S. tamariscina, Myrothamnus flabellifolia, Sporobolus stapfianus, Fagus sylvatica, and Botrychium lunaria accumulate remarkably high levels of trehalose (up to 20% of its carbohydrate content). Trehalose helps maintain cell viability through different mechanisms, such as the substitution of water molecules that interact with proteins and membranes, acting as a vitrifying agent that preserves the structure and function of biomolecules, and reducing the radius of the solvation layer of proteins.6–10 Phosphorylated trehalose acts as a signal molecule, and carbon and energy source.11 Overexpression of trehalose anabolic enzymes can lead to low productivity, possibly due to the signaling activity of trehalose-6 phosphate (T6P).12 A promising strategy for obtaining drought resistant varieties is to decrease the activity of the trehalase, either by decreasing the amount of enzyme or its activity, thus, allowing the accumulation of trehalose. In A. thaliana, high levels of trehalase transcripts are found in seeds, siliques, and developing floral organs.13 We used the CRISPR/Cas9 system14,15 for the modification of the trehalase-binding site employing modified binary vectors compatible with an A. tumefaciens hypervirulent strain. The edited plants showed a drought tolerant phenotype. Parallel experiments in maize allowed the transient expression of binary vectors to test their functionality in monocots.
Materials and methods
Plant growth conditions
Wild-type seeds of Arabidopsis thaliana ecotype Columbia-0 were stratified for two days at 4°C in 0.05% agarose, sowed in sterilized soil and kept in growth chamber (Conviron A1000, Conviron, Winnipeg, Canada) at 22°C with long day conditions (16 h light/8 h dark, long day photoperiod conditions), 70% of relative humidity and watered with Miracle-Gro Water Soluble Universal Fertilizer, following the manufacturer’s recommendations. Soil employed was Sunshine Mix (Sungro, Santa Maria, CA). Hybrid maize plants VT-401 (obtained from Instituto de Investigación y Capacitación Agropecuaria, Acuícola y Forestal, México) and B73 were grown at the greenhouse and employed for transient expression studies.
Bioinformatic analysis and phylogeny
To build a plant trehalase phylogeny, sequences employed were retrieved from the Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html), (https://www.uniprot.org/) or GenBank (https://www.ncbi.nlm.nih.gov/) databases.
The sequence alignment was performed using MUSCLE and the phylogenetic trees were made with the MEGA 7.0 software16 using the maximum likelihood method (by pairs op: 35, Ext: 0.7, Multiple op: 15, Ext: 0,3) with 1000 bootstraps for statistical validation.17 Structural bioinformatic analysis of plant trehalases was performed using Prosite (http://prosite.expasy.org), Interpro (https://www.ebi.ac.uk/interpro/) and Pfam (http://pfam.xfam.org/) databases, in order to identify catalytic and substrate-binding domains. The representation of the consensus sequence of the substrate-binding domain was carried out with WebLogo software (http://weblogo.berkeley.edu/).18
Structural modeling and docking studies
Structural models were predicted by homology using several plant trehalases, including Selaginella moellendorffii (SmTRE), Arabidopsis thaliana (AtTRE), Cucumis melo (CmTRE), and Zea mays (ZmTRE) using the SWISS-MODEL platform (http://swissmodel.expasy.org/)19 based on the crystal structure of the Enterobacter cloacae trehalase (PDB: 5Z6H). Accession numbers of the plant trehalases employed are shown in Table S3. When necessary, the models were refined using the Galaxy Refine Server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE)20 and validated using PROCHEK v.3.5 (https://servicesn.mbi.ucla.edu/PROCHECK/)21 and ProSA (https://prosa.services.came.sbg.ac.at/prosa.php).22 The homology models were visualized using UCSF Chimera23 and docking simulations were carried out with AutoDock Vina24 using trehalase as receptor and trehalose as ligand, generating 10 different poses for each molecular docking. The best pose was selected and analyzed based on the energy score and the RMSD parameter of each model.
Plasmid constructs
Arabidopsis U6-26 RNA polymerase III expression cassette, used for the gRNA transcription, was excised from vector pBUN501 by digestion with HindIII, dephosphorylation with rSAP (New England Biolabs, Beverly MA) and then inserted into pBUN411 and pBUN6I11 vectors, generating the pBUN4U6 and pBUN6U6 vectors, respectively. Similarly, the Oryza sativa snRNA U3 expression cassette was cloned from pBUN411 to pBU501, generating the pBUN5U3 vector. Then, an additional cassette for spectinomycin resistance derived from the PCR®8/GW/TOPO® plasmid (Invitrogen, Carlsbad CA) was inserted into pBUN411, pBUN5U3, pBUN6I11, pBUN4U6, pBUN501, and pBUN6U6, through PCR amplification with Phusion DNA Polymerase (New England Biolabs, Beverly MA) employing primers ClaSmR-F and ClaISmR-R (Table S1) which contain ClaI restriction sites. The PCR conditions were 94°C for 3 min, 30 cycles at 94°C for 30 s, 65°C for 30 s, 72°C for 1 min, and a final extension of 72°C for 5 min. The PCR product was digested with ClaI and the vector digested with AclI, dephosphorylated with rSAP and subsequently ligated. pBUN411, pBUN501 pBUN6I11 were kindly provided by Prof. Qi-Jun Chen (Addgene plasmid 50581, 50582, and 50580).
The design of gRNAs was carried out with the Deskgen software (https://www.deskgen.com/landing/) identifying the best candidates in the trehalase genes of Z. mays (GRMZM2G162690) and A. thaliana (AT4G24040), considering the proximity to amino acids involved in catalytic or substrate-binding activity with the highest on-target and the lowest off-target value, excluding those with potential off-target in coding regions. These gRNAs were subsequently cloned by digestion with BsaI-HF®, generating the pBUN4U6SM-AtTRE1 and pBUN4U3SM-ZmTRE1 vectors, according to previous reports.25
Additionally, a 1973 bp fragment corresponding to the maize polyubiquitin gene promoter (ZmUbi1 promoter) was amplified by PCR from pBUN4U3 using the ZmUbip1-F and ZmUbip1-R primers (Table S1) and then cloned into the T/A PCR®8/GW/TOPO® plasmid. The PCR conditions were 94°C for 3 min, 30 cycles at 94°C for 30 s, 62°C for 30 s, 72°C for 2 min and a final extension of 72°C for 10 min. LR Cloning into pBGWFS7.0 was carried out with LR Clonase (Invitrogen), generating vector pBGWFS7-ZmUbip1.
All plasmid constructs were verified by PCR and Sanger sequencing and subsequently introduced into A. tumefaciens AGL1 strain by electroporation with an Equibio electroporator and 1 mm cuvettes (EquiBio EasyJect Optima, Ashford, UK), which was then grown at 28°C for 48 h in LB medium containing 50 mg/L carbenicillin, 25 mg/L kanamycin, and 50 mg/L spectinomycin.
Transient expression assays
Seeds of Z. mays genotypes B73 and VT401 were surface sterilized with 70% ethanol and incubated in a growth chamber for 3 days at 30°C with moistened paper towels. The seeds that showed elongation were selected, dissected at the base of the coleoptile and incubated with a suspension of A. tumefaciens AGL1, which harbored pBGWFS7-ZMUbi1p or pBUN4U3SM-ZmTRE1 for 48 h in the growth chamber. Plants that showed emerging leaves were transferred to potted soil for further development and characterization as previously described.26 The quantification of copy number of the transformed lines was carried out by digital droplet PCR (ddPCR) as described.27 In addition, 6-week-old N. benthamiana and 4-week-old Arabidopsis leaves were vacuum infiltrated with A. tumefaciens AGL1 harboring pBGWFS7-ZMUbi1p to assess the functionality of the ZmUbi1 promoter in dicots 7 days after infiltration by confocal microscopy and Western Blot using anti-GFP polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz CA), essentially as described in other reports.28–30
Generation of trehalase edited plants
Five to six-week-old A. thaliana Col-0 plants were transformed with A. tumefaciens AGL1 carrying the plasmid pBUN4U6SM-AtTRE1 using the floral dip method.31 T1 seeds were selected by weekly spraying of glufosinate ammonium (300 µM) for 3 weeks. Genomic DNA was extracted from glufosinate-tolerant plants using the CTAB method32 and PCR was carried out with Cas9-F and Cas9-R primers, which amplify a 424 bp product and corresponds to a fragment of the gene-editing cassette (Table S1). Determination of copy number in F1 plants was performed by ddPCR as previously described.33 Subsequently, single copy T-DNA lines were selected. F2 plants were subjected to heat shock to improve editing efficiency.34 PCR was carried out in heat-recovered plants with the AtTRE1-F and AtTRE1-R primers, which amplify a 346 bp fragment of the TRE1 gene. The PCR conditions were 94°C for 3 min, 30 cycles at 94°C for 30 s, 65°C for 30 s, 72°C for 30 s, and a final extension of 72°C for 3 min. Subsequently, genome editing was determined using the T7 endonuclease assay, following the manufacturer’s instructions (New England Biolabs) and PCR products were cloned in pCR®2.1/GW/TOPO® (Invitrogen) for Sanger sequencing. Stable and transgene-free edited homozygous plants were selected, and the progeny of these plants were used for genotypic and physiological characterization.
Molecular and physiological characterization of edited lines
Homozygous T-DNA mutant lines genotyped for the 5ʹ-UTR of the trehalase gene (AT4G24040), SALK_147073, and SALK_151791 from the Arabidopsis Biological Resource Center (ABRC) were used to compare them with the edited and A. thaliana wild type lines. Edited, T-DNA insertional mutants, and wild type genotypes were analyzed 3 weeks after germination and the transcription levels of trehalase mRNA were determined through RT-qPCR using RTqPCR-AtTRE1-F and RTqPCR-AtTRE1-R primers, and as an endogenous control, specific primers for ubiquitin 10 (UBQ10-F and UBQ10-R)35 (Table S1). Drought stress was applied by suspending irrigation for 14 days as described.27,36
Results
In silico analysis to determine the editing site for the TRE1 gene.
An analysis of the amino acid sequences of trehalase was performed in order to identify potential editing targets, through structural characterization, including substrate binding, active and conserved structural sites (Table S2). We constructed a plant trehalase phylogeny tree derived from amino acid sequence accessions (Table S3), including the 25 amino acid conserved signature corresponding to the substrate-binding domain. This analysis allowed to group the sequences according to their phylogeny in dicots, monocots, other embryophytes and chlorophytes (Figure 1). The consensus sequence of this domain revealed the presence of serine near the carboxy end of the domain in almost all accessions (Figure 2a). However, this serine is replaced by a threonine in the genus Selaginella, which includes several drought-tolerant species (Figure 2b). Thus, this region was selected as a target for editing (Figure 2c,d)
Figure 1.
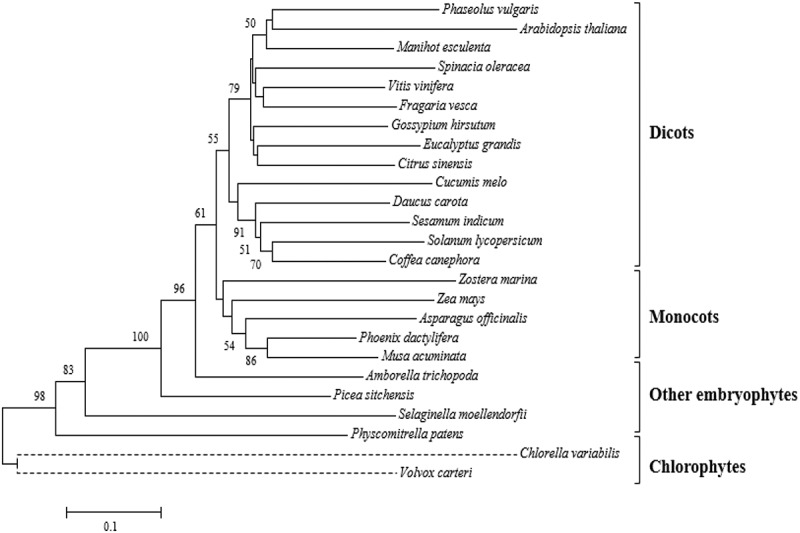
Phylogeny of the substrate-binding domain of plant trehalases
Phylogenetic analysis was performed by the neighbor joining method. The optimal tree whose sum of branch length = 5.46274126 is shown. The percentage of replicas of that tree, in which the associated taxa are grouped in more than 50% of the cases (1000 replicas) are shown in the tree nodes. Evolutionary distances were calculated using the Poisson corrected method. The analysis was performed based on a sequence of 25 amino acids corresponding to the substrate binding domain.
Figure 2.
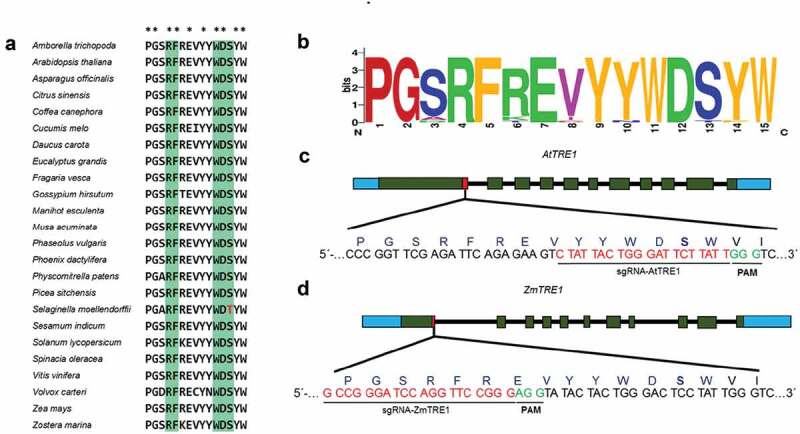
Design of sgRNA for TRE1 edition, located in the substrate binding domain of the trehalase enzyme
(A) Conserved amino acids in blue are the conserved signatures of the substrate binding site. Asterisks are in conserved motifs in this domain. The S→T change present in Selaginella is shown in red. (B) WebLogo of the consensus sequence of extant Viridiplantae accessions; position 13 indicates the overrepresentation of the amino acid Serine in the substrate-binding site, whereas in drought-tolerant plants such as S. moellendoffii, it is replaced by threonine. (http://weblogo.berkeley.edu/). (C) and (D) Position of the sgRNA in Arabidopsis and maize TRE1 genes, respectively. Green boxes correspond to exons, solid black lines, to introns. Red box shows the position of the sgRNA, and PAM is shown in green.
Design and cloning of gRNA
The substrate-binding domain was selected as a target for the design of the gRNA, and several high score gRNAs were identified in A. thaliana and Z. mays according to the algorithms described previously.37,38 For A. thaliana, the best gRNA was located on chromosome 4 (12,488,941 bp), with a GC% of 30 and without off-targets (Figure 2c). For maize, the gRNA were located on chromosome 1 (80,007,736 bp) with a GC% of 80 and without off-targets.
Structural modeling and docking studies
For the analysis of conservation structure of plant trehalases and to verify whether the substitution of threonine for serine in S. moellendorffii is involved in a decrease in the binding capacity to the substrate, we generated structural homology models for A. thaliana, C. melo, S. moellendoffii, and Z. mays. The models were compared with the crystallized structure of Enterobacter cloacae trehalase (PDB: 5Z6H) and sequence identities of 32.45% to 36.81% were obtained. The structural characterization of plant trehalases revealed a classic (α/α)6-barrel fold structure where Asp and Glu residues are responsible for the catalytic activity (Figure 3a). The conserved substrate binding signature and other sequences conforms a binding pocket for the trehalase-trehalose interaction as previously reported in glycosidase structures.39,40 Subsequently, we carried out molecular docking interactions between four plant trehalases using trehalose as ligand (Figure 3b-e).
Figure 3.
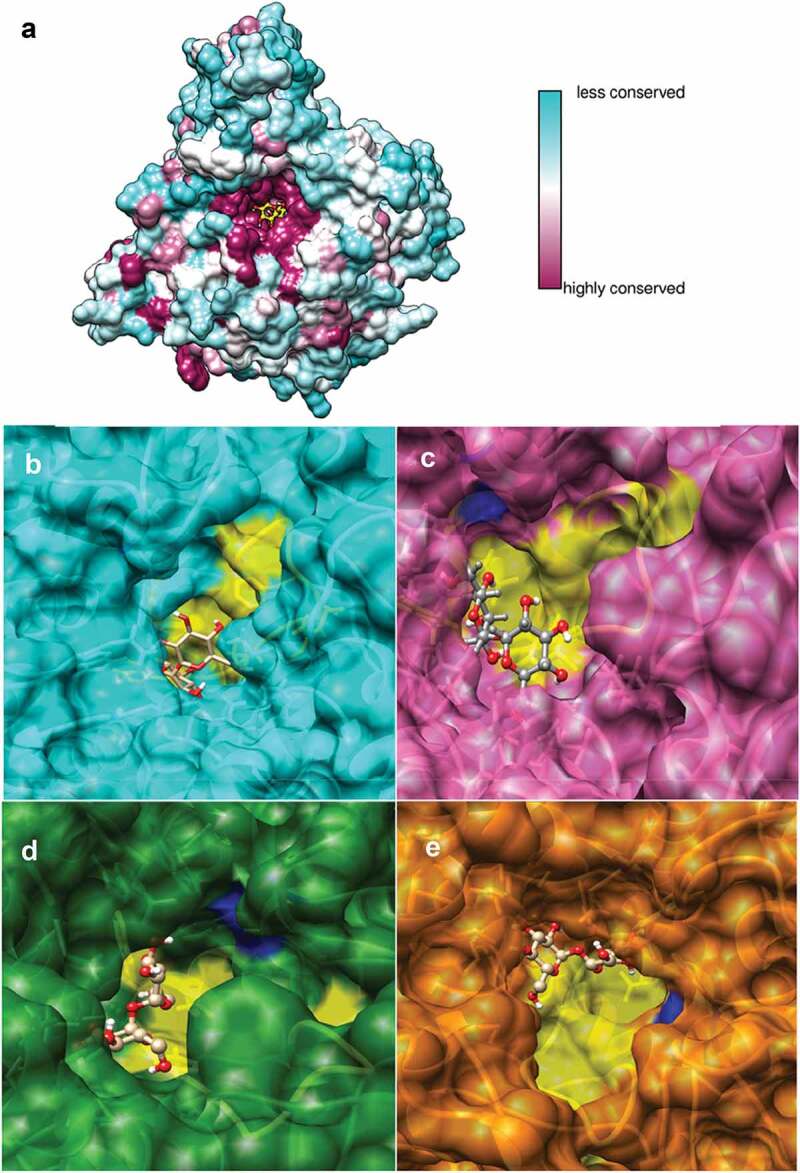
Structural modeling and molecular docking of different plant trehalases interacting with their substrate
Sequence conservation of plant trehalases mapped onto AtTRE1 homology model where highly conserved residues are shown in purple and variable residues in cyan. The trehalose molecule is shown in yellow (A). Docking interactions between different plant trehalases and trehalose for different plant species: A. thaliana (B), S. moellendorfii (B), C. melo (C) and Z. mays (D). Yellow surfaces were highlighted to show conserved substrate binding signatures. Blue regions show catalytic residues for trehalase enzyme activity.
The structure of plant trehalases in the tested species is quite conserved but its binding affinity was considerably lower in S. moellendoffii, supporting the notion that an increased accumulation of trehalose in this species is due to a lower affinity for its substrate (Table 1).
Table 1.
Parameters of molecular docking for plant trehalases interacting with trehalose
| Docking interaction | Energy (Kcal/mol) | RMSD (Å) |
|---|---|---|
| SmTRE-trehalose | −6.6 | 1.660 |
| AtTRE-trehalose | −7.9 | 0.008 |
| CmTRE-trehalose | −8.0 | 0.062 |
| ZmTRE-trehalose | −8.0 | 0.017 |
Construction of vectors for genome editing
The pBUN411, pBUN501, and pBUN6I11 vectors25 harbor resistance to kanamycin and spectinomycin, but since the cloning site of the gRNA is within the spectinomycin cassette, this marker is inactivated. We modified these vectors by adding a second spectinomycin resistance cassette outside the T-DNA. The addition of this cassette allowed the positive selection of the recombinant hypervirulent strain of A. tumefaciens AGL1, which already harbors resistance to kanamycin. The native Cas9 is harbored by pBUN4U3SM (monocots) and pBUN4U6SM (dicots), the Cas9 nickase version (nCas9)41,42 by the pBUN5U3SM vector (monocots) and pBUN5U6SM vector (dicots), and dCas9-KRAB version sequence (Cas9 lacking nuclease activity, fused to the transcriptional repressor Krüppel Associated Box) is contained in the pBUN6U3SM (monocots) and pBUN6U6SM (dicots) vectors (Table S4). The plasmids were modified in order to harbor more versatile features, such as selective markers for Agrobacterium selection, and functional promoters for gRNA expression in both monocots and dicots.
Vector functionality of the monocot promoter ZmUbi1p in dicots
To assess the functionality of the monocot promoter in the novel binary vectors for gene editing in dicots, pBGWFS7-ZMUbi1p was transformed into A. tumefaciens AGL1 and used to infiltrate Arabidopsis and N. benthamiana leaves. The transient expression of GFP encoded in this construct was monitored by confocal microscopy. Fluorescence images were obtained 7 days after infiltration and are presented in Figure 4a-f. Control Arabidopsis and N. benthamiana plants are shown in (Figure 4a,d), where autofluorescent stomata are indicated with arrows. GFP-associated fluorescence was observed in mesophyll cells (Figure 4b,c,f), epidermis (Figure 4e) and vascular bundles (figure 4f). Transient expression of GFP in leaves was confirmed by Western blot, using total protein extracts (Figure 4g). The recombinant vector expresses the GFP-GUS fusion, with a molecular weight of 95.3 kDa, present in both Arabidopsis and N. benthamiana extracts. Positive control is GFP, with a molecular weight of 27 kDa.
Figure 4.
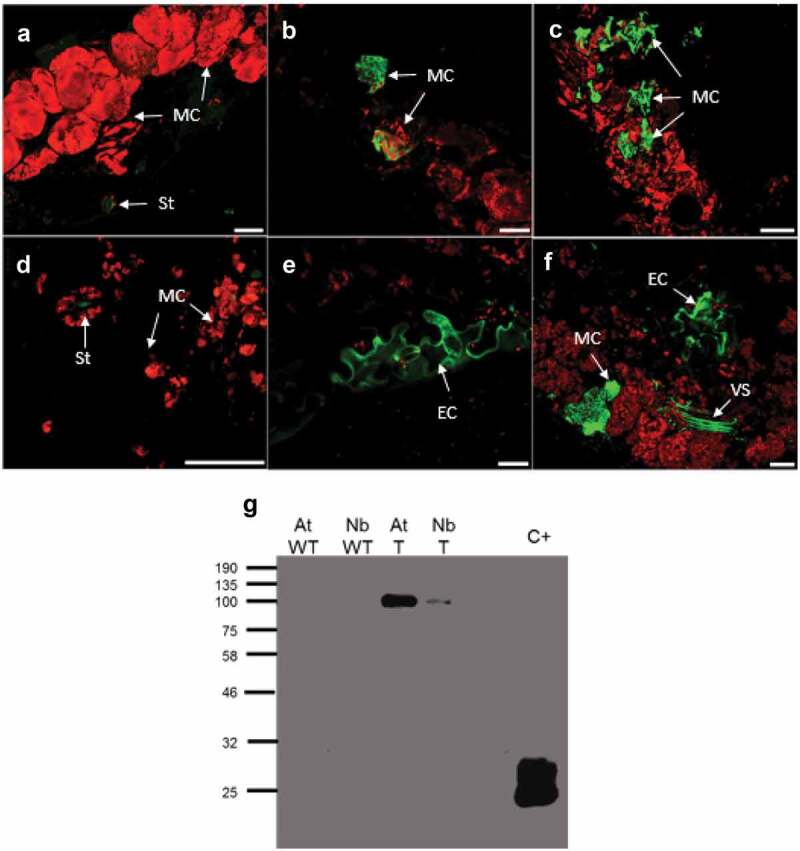
Expression analysis of pBGWFS7-ZmUbi1p in Arabidopsis and N. benthamiana leaves
(A-F): Confocal microscopy images. (A) Arabidopsis control, B-C: Arabidopsis expression GFP driven by ZmUbi1p. D) N. benthamiana negative control. E-F) N. benthamiana expressing GFP pBGWFS7-ZmUbi1p driven. White bars indicate 20 µm. MC, mesophyll cells; EP, epidermal cells; VS, vascular strand; St, auto fluorescent stomata. (G) Detection of GFP-GUS fusion protein by Western Blot, expressed under the ZmUbip1 promoter. AT: Arabidopsis, NB: N. benthamiana, WT: Wild type, T: Transformed, C+: Positive control of GFP expressed in N. benthamiana employing the ZYMV vector.28
Maize transient genetic transformation and selection of transformants
Transient transformation was carried out using coleoptiles, which were incubated with A. tumefaciens AGL1, as previously described. Germinated seeds of B73 (Figure 5a) and VT401 (Figure 5b) are shown, in which leaf primordia were dissected to allow Agrobacterium access the apical meristem. One month after transformation, plants expressing herbicide resistance were identified by topically applying glufosinate ammonium (300 µM) (Figure 5c,d). The leaves of the transformed plants did not show yellowing or necrosis in the tested area, in contrast to control plants. Once herbicide-resistant plants were selected, the transgenes were identified by end-point PCR (data not shown). Twelve independent lines with resistance to herbicide were obtained in which the transgene was also detected, from a total of five independent lots and from a total of 200 analyzed plants. This transient genetic transformation technique showed an efficiency of 6%. In a parallel test, GUS expression was observed after transient transformation with the pBGWFS7-ZMUbi1p vector in analyzed plants (figure 5f,g). Histological analysis in longitudinal sections of coleoptiles shows expression in all tissues, being more intense in meristems, in contrast to control plants (Figure 5e). Transiently transformed maize plants were tested to determine the number of inserts, using digital PCR (ddPCR). The values obtained suggest that the transformed plants are mostly chimeras, since less than one copy was consistently found per haploid genome (Fig. S1).
Figure 5.

Genetic transformation of maize coleoptiles
Coleptiles of var. B73 (A) and var. VT401 (B) were used as explants for transformation with A. tumefaciens AGL1 carrying pBUN4U3SM vector. Topical application of ammonium glufosinate in control maize leaf (C) and transformed plant with A. tumefaciens AGL1 carrying pBUN4U3SM (D). Histochemical GUS analysis of control (E) and transformed coleoptiles (F, G). Bars in (A, B) = 1 cm. (E, F, G) are 1 mm.
Selection and characterization of the AtTRE1 edited gene
Arabidopsis plants transformed with the construct expressing Cas9 and AtTRE1 gRNA were selected initially based on their herbicide resistance. Eight resistant plants were analyzed by end-point PCR and the number of inserted copies in each independent line was determined by ddPCR. The values indicate the presence of one copy of the transgene per haploid genome, which suggests that there is one insertion per cell (Fig. S2). The plants were self-crossed and homozygous lines harboring the edition and lacking the CRISPR/Cas9 constructs were selected for further characterization.
Identification of potential edited sites in the genomes of analyzed lines
A 346 bp fragment was amplified in F2 plants harboring the region corresponding to the gRNA; T7 endonuclease assays performed on PCR fragments from independent transformant lines showed the presence of putative edited sequences, which were selected for further analysis. The amplicons were then cloned and sequenced to identify possible changes. Plants with indels were identified. Three of these had insertion of one to two bases: lines LE04 (+02, AA), LE02 (+01, A) and LH01 (+01, C). Four lines showed deletions of one to twelve bases: LB01 (−01, G), LF08 (−02, TA), LG20 (−06, ATTCTT) and LA02 (−12, GGGATTCTTTATT), while line LA01 had a substitution of TCT for TTT, which caused the change from serine to phenylalanine (Figure 6).
Figure 6.
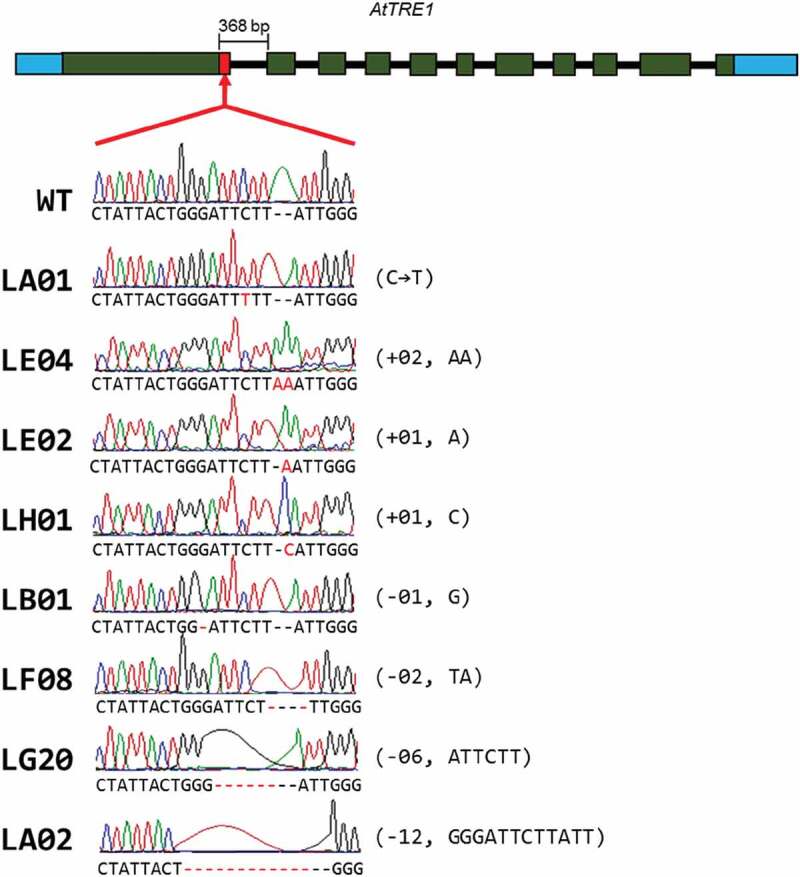
Sequence analysis of Arabidopsis edited lines in the Arabidopsis TRE1 gene (AT4G24040)
Upper panel represents organization of the TRE1 gene, in which the edited region is highlighted. Red box represents the substrate-binding domain, the target region for editing. Independent lines were termed LA01, LE04, LE02, LH01, LB01, LF08, LG20, and LA02. The aligned chromatograms are displayed, in which red dashed lines indicate deletions, while insertions are indicated in red capital letters. Sequence modifications are indicated to the right of the chromatogram.
Phenotypic comparison of Arabidopsis mutant T-DNAs with the edited line
The phenotype of edited lines was compared to T-DNA insertion mutants in the TRE1 gene. These lines are SALK_147073 (TRE1 silenced line) and SALK_151791 (overexpressing line); the insertion in both cases is in the 5ʹ UTR; however, while SALK_147073 shows wild-type levels of TRE1 expression, the latter behaves as an overexpressing line.43 These lines were genotyped by PCR to confirm their insertion site and their homozygous nature. The rosette diameter of these lines was compared to wild type Arabidopsis (Figure 7a), in which the overexpressing line (tre1OE) displayed a reduced diameter, while silenced (tre1-) and edited LA01 (C→T) displayed a larger rosette diameter. Also, accumulation of TRE1 mRNA was determined by RT-qPCR in rosette leaves (Figure 7b); overexpressing lines accumulated higher TRE1 mRNA levels, relative to control plants, while lower levels were observed in silenced lines. The edited line showed the same transcript levels as wild type plants, in agreement with a previous report.43 These results show that editing of the TRE1 gene did not affect the accumulation of its transcript.
Figure 7.
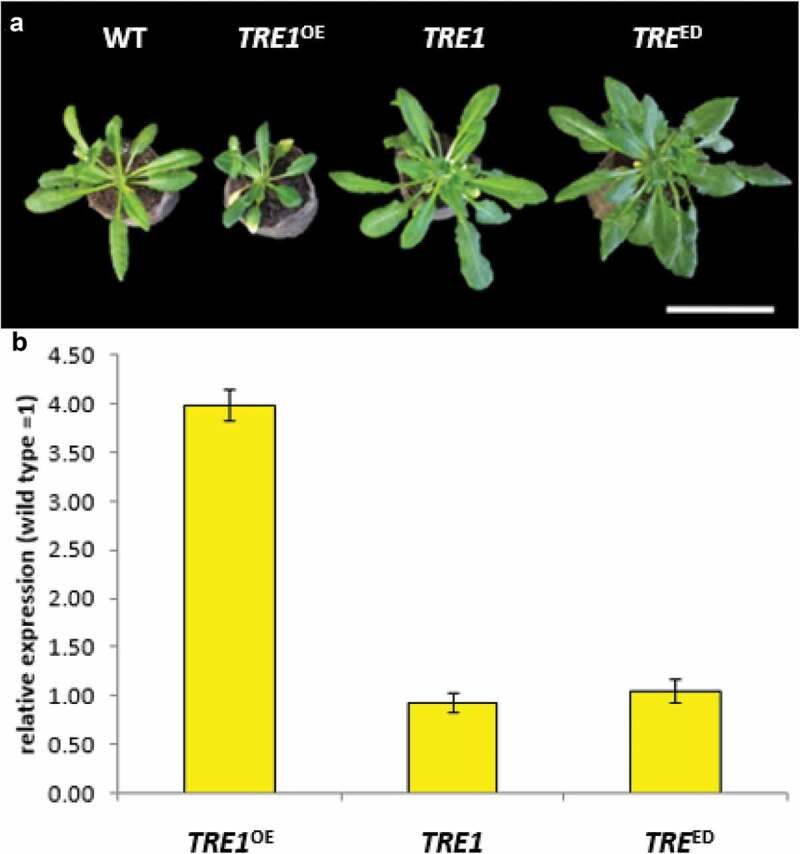
Comparison of TRE1 edited Arabidopsis with TRE1 mutant plants
(A) Vegetative growth of Wild Type (WT), Overexpressing line SALK_151791 (TRE1OE), silenced line SALK_147073 (TRE1), Edited line LA01, C→T (ED). Bar in (A), 5 cm. (B). TRE1 mRNA relative expression normalized with wild type Arabidopsis. Abbreviations as in (A).
TRE1-edited plants show increased drought tolerance relative to control and silenced plants
The growth of Arabidopsis edited in the TRE1 gene was similar to that of wild-type plants under normal irrigation conditions, and no delay in vegetative growth was observed. However, when irrigation was suspended, early flowering was observed in wild-type and overexpressed accessions (Figure 8). In addition, control and overexpressing lines showed loss of turgor and wilting two weeks after the onset of drought stress, while the edited and silenced lines maintained their turgor. Despite being irrigated after two weeks, the control plants were unable to recover from the applied stress, as they had reached the permanent wilting point, caused by drought stress (data not shown). In contrast, the edited and silenced plants grew normally and completed their developmental cycle to produce viable flowers and seeds.
Figure 8.

Phenotype of WT, SALK T-DNA mutant and TRE1-edited Arabidopsis subjected to a drought regime to for two weeks
(A) Control, wild type plants. (B) Arabidopsis TRE1 line LA01 (C→T). Bar is 5 cm.
Discussion
Trehalose accumulation is a hallmark of conserved drought responses in a wide range of taxa, from bacteria to animals, where it protects proteins and cellular structures from osmotic stress. Therefore, it has been suggested that decreasing trehalase activity would result in higher levels of trehalose and thus, in drought tolerance. This hypothesis has been tested experimentally in plants grown in vitro.11 Furthermore, this strategy has been used to confer drought tolerance in maize through genetic transformation with a construct that expresses an antisense RNA for TRE1, in open field trial.27 These plants displayed remarkable drought tolerance compared to control plants, supporting the idea that reduce trehalase levels or activity leads to tolerance to water deficit. However, this promising strategy for conferring drought tolerance to relevant crops faces tremendous regulatory hurdles in most countries. Also, a careful strategy is required, since hyperaccumulation of trehalose may lead to aberrant phenotypes (given the signaling role of a T6P, which is derived from trehalose).44,45
In order to search for editable sequences that could lead to a decrease in trehalase activity, a phylogenetic-structural approach was carried out. The phylogenetic analysis of the trehalase protein family included species that show natural tolerance to drought, such as the resurrection plants from the genus Selaginella. The signature of the substrate-binding domain PGXRFXEXYXWDSXW, showed a remarkable degree of conservation between different plant taxa. Interestingly, a threonine residue in S. moellendorffii replaces the almost conserved serine residue in plants at position 13 within this motif, decreasing its binding affinity, as was validated by molecular docking studies. Based on this evidence, the CRISPR/Cas9 gene-editing system was proposed to carry out the serine to threonine modification, in which a gRNA was designed to target the substrate-binding domain, close to the relevant Ser/Thr residue described. The gRNAs were designed, selecting these sequences based on their proximity to the substrate-binding domain, and the designed gRNAs with no theoretical off-targets were selected.
Arabidopsis genetic transformation is reproducible and highly efficient; in contrast, maize transformation is time-consuming and much less efficient. To improve the latter, a hypervirulent strain of A. tumefaciens, AGL1, was employed, which constitutively overexpresses virD and E genes. The modified vectors were shown to be functional in transient expression assays in maize coleoptiles, as assessed by reporter gene expression and herbicide tolerance. The plants obtained through coleoptile explants were shown to be a genetic mosaic, since a lower proportion of the transgene with respect to the endogenous gene was observed in ddPCR assays. However, this assay confirmed the functionality of the selection markers and promoters directing the expression of the reporter genes. Thus, these vectors will be helpful for gene editing in monocots.
Comparison between T-DNA insertion mutants, wild type and edited Arabidopsis plants indicates that the accumulation levels of TRE1 mRNA in edited plants are like wild-type plants. Given the role of T6P, in different signaling pathways, such as flowering and general growth regulation, it is not surprising that the T-DNA insertion line, which over-accumulates trehalase mRNA (and, conceivably, protein that can be phosphorylated), shows an abnormal phenotype consistent with the resulting levels of this signaling molecule.43
Along with other lines of evidence, this suggests that the effect of altering trehalase activity is not as straightforward. It is possible that the observed effects are also due to accumulation of trehalose metabolites, besides T6P and trehalose itself. To compare plant phenotypes with changes in the physiology of the metabolic pathway of trehalose synthesis, the phenotypes of these materials were characterized under vegetative growth conditions. The silenced plants displayed a larger diameter than the wild type plants. In contrast, the trehalase overexpressing lines displayed a low growth phenotype. The edited genotype showed a vegetative development like the silenced plant, which tests the hypothesis that changes in the substrate-binding domain could influence the enzyme activity. The identification of eight genetically independent lines, with edition in the substrate binding domain, showed the generation of biallelic genotypes, which were self-crossed to obtain plants without the CRISPR/Cas9 tool and, to have homozygous plants for the edition, which were then characterized. In the observed lines, DNA sequenced showed a substitution, insertions, and deletions. Frameshift mutations were observed in five independent lines, while deletions that conserved the frameshift were obtained in two lines (−6 and 12 nucleotides). Of interest was the change of the conserved serine by the amino acid phenylalanine. Physiological evaluation of the obtained lines showed differences in growth, although TRE1 mRNA levels remained comparable with wild plants. This finding is expected, because of the precision of the editing technique, as no changes were introduced within the promoter region that could affect transcript accumulation. Water stress tolerance was evident in edited and silenced Arabidopsis plants. Interestingly, when irrigation was stopped, the edited and silenced lines showed drought symptoms later than wild type and overexpressing plants. Indeed, plants with symptoms of wilting and yellowing were observed first in the controls, two weeks after applying water stress. Likewise, early flowering was observed in the control plants, which is consistent with a flowering stress-induced phenotype described previously.46,47
Several types of stress, including drought, are capable of inducing flowering. Indeed, drought stress under long day conditions (LD) can induce flowering in A. thaliana Col-0, 48 due to a photoperiod-induced accumulation of Flowering Locus T (FT) transcripts in buds. Likewise, early flowering induced by drought has been also observed in Brassica rapa and Mimulus guttatus.49,50 Interestingly, plants harboring the edited TRE1 gene do not display an early-flowering phenotype in contrast to wild type plants, suggesting a less severe stress impact due to trehalose accumulation. On a speculative note, it is possible that since trehalose content is increased in the edited plants, this could be phosphorylated, thus also functioning as a signal molecule. T6P may confer resistance to pathogens and provides abiotic stress tolerance, 51 supporting the previous notion. Gene editing is a very versatile tool to accurately generate greater genetic diversity. In particular, the editing of discrete changes in the trehalase gene can generate plants with tolerance to abiotic stress, a highly desired phenotype, due to both increasing water demand and also because its dwindling availability worldwide for agricultural purposes.
Supplementary Material
Acknowledgments
The authors thank the reviewers of this manuscript for their many helpful recommendations.
Funding Statement
This work was supported by the Consejo Nacional de Ciencia y Tecnología (CONACyT) under grant [FC-01234 to BX-C. LN-M] was supported by a CONACyT fellowship [426541].
Disclosure of Potential Conflicts of Interest
BX-C and RR-M have filed a patent application for silencing of trehalase in maize through RNAi.
Author contributions
BX-C, RR-M and JJH-M designed the experiments. LN-M conducted the experiments. BV-H conducted the protein modeling and molecular docking simulations. BX-C, RR-M, and LN-M analyzed the data and wrote the manuscript. BX-C supervised the experiments.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Almeida AM, Cardoso LA, Santos DM, Torné JM, Fevereiro PS.. Trehalose and its applications in plant biotechnology. Vitro Cell Dev Biol Plant. 2007;43:1–12. doi: 10.1007/s11627-006-9024-3. [DOI] [Google Scholar]
- 2.Barraza A, Sánchez F.. Trehalases: a neglected carbon metabolism regulator? Plant Signal Behav. 2013;8:e24778. doi: 10.4161/psb.24778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul MJ, Primavesi LF, Jhurreea D, Zhang Y. Trehalose metabolism and signaling. Annu Rev Plant Biol. 2008;59:417–441. doi: 10.1146/annurev.arplant.59.032607.092945. [DOI] [PubMed] [Google Scholar]
- 4.Leyman B, Van Dijck P, Thevelein JM. An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci. 2001;6:510–513. doi: 10.1016/S1360-1385(01)02125-2. [DOI] [PubMed] [Google Scholar]
- 5.Figueroa-Soto CG, Terán-Acuña E, Valenzuela-Soto EM. Changes in trehalase activity are associated with the hydric status of Selaginella lepidophylla plants. Biotecnia. 2014;16:15–19. doi: 10.18633/bt.v16i1.27. [DOI] [Google Scholar]
- 6.Bianchi G, Gamba A, Limiroli R, Pozzi N, Elster R, Salamini F, Bartels D. The unusual sugar composition in leaves of the resurrection plant Myrothamnus flabellifolia. Physiol Plant. 1993;87:223–226. doi: 10.1111/j.1399-3054.1993.tb00146.x. [DOI] [Google Scholar]
- 7.Müller J, Boller T, Wiemken A. Trehalose and trehalase in plants: recent developments. Plant Sci. 1995;112:1–9. doi: 10.1016/0168-9452(95)04218-J. [DOI] [Google Scholar]
- 8.Gu W, Zhang A, Sun H, Gu Y, Chao J, Tian R, Duan JA. Identifying resurrection genes through the differentially expressed genes between Selaginella tamariscina (Beauv.) spring and Selaginella moellendorffii Hieron under drought stress. PLoS ONE. 2019;14:e0224765. doi: 10.1371/journal.pone.0224765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pampurova S, Van Dijck P. The desiccation tolerant secrets of Selaginella lepidophylla: what we have learned so far? Plant Physiol Biochem. 2014;80:285–290. doi: 10.1016/j.plaphy.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Yobi A, Wone BWM, Xu W, Alexander DC, Guo L, Ryals JA, Oliver MJ, Cushman JC. Comparative metabolic profiling between desiccation-sensitive and desiccation-tolerant species of Selaginella reveals insights into the resurrection trait: comparative metabolomics of desiccation tolerance. Plant J. 2012;72:983–999. doi: 10.1111/tpj.12008. [DOI] [PubMed] [Google Scholar]
- 11.Van Dijck P, Mascorro-Gallardo JO, De Bus M, Royackers K, Iturriaga G, Thevelein JM. Truncation of Arabidopsis thaliana and Selaginella lepidophylla trehalose-6-phosphate synthase unlocks high catalytic activity and supports high trehalose levels on expression in yeast. Biochem J. 2002;366:63–71. doi: 10.1042/bj20020517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schluepmann H, Van Dijken A, Aghdasi M, Wobbes B, Paul M, Smeekens S. Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol. 2004;135:879–890. doi: 10.1104/pp.104.039503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller J, Aeschbacher RA, Wingler A, Boller T, Wiemken A. Trehalose and trehalase in Arabidopsis. Plant Physiol. 2001;125:1086–1093. doi: 10.1104/pp.125.2.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amitai G, Sorek R. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 2016;14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Li J, Xia L. Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement. Front Plant Sci. 2016;7:1928. doi: 10.3389/fpls.2016.01928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. doi: 10.1086/286013. [DOI] [Google Scholar]
- 18.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo-Cassarino T, Bertoni M, Bordoli L, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heo L, Park H, Seok C. GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41(W1):W384–W388. doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 22.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(suppl_2):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Chen QJ. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abhishek A, Kumari R, Karjagi CG, Kumar P, Kumar B, Dass S, Sai Kumar R, Ramteke PW. Tissue culture independent Agrobacterium tumefaciens mediated in planta transformation method for tropical maize (Zea mays.L). Proc Natl Acad Sci India Sect B Biol Sci. 2016;86:375–384. doi: 10.1007/s40011-014-0454-0. [DOI] [Google Scholar]
- 27.Agreda-Laguna KA, Cabrera-Ponce JL, Medrano RR, Garzon-Tiznado JA, Xoconostle-Cazares B. Trehalose accumulation provides drought tolerance to genetically modified maize in open field trials. Pak J Agric Sci. 2018;55:1009–1020. doi: 10.21162/PAKJAS/18.6901. [DOI] [Google Scholar]
- 28.Lin MK, Belanger H, Lee YJ, Varkonyi-Gasic E, Taoka KI, Miura E, Xoconostle-Cázares B, Gendler K, Jorgensen RA, Phinney B, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen LJ, Zou WS, Wu G, Lin HH, Xi DH. Tobacco alpha-expansin EXPA4 plays a role in Nicotiana benthamiana defence against Tobacco mosaic virus. Planta. 2018;247:355–368. [DOI] [PubMed] [Google Scholar]
- 30.Del Toro F, Tenllado F, Chung BN, Canto T. A procedure for the transient expression of genes by agroinfiltration above the permissive threshold to study temperature‐sensitive processes in plant–pathogen interactions. Mol Plant Pathol. 2014;15:848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 32.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:39–40. [Google Scholar]
- 33.Ruiz-Salas JL, Ruiz-Medrano R, Montes-Horcasitas MC, Agreda-Laguna KA, Hinojosa-Moya J, Xoconostle-Cázares B. Vascular expression of trehalose phosphate synthase1 (TPS1) induces flowering in Arabidopsis. Plant Omics. 2016;9:344–351. doi: 10.21475/poj.09.05.16.pne188. [DOI] [Google Scholar]
- 34.LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, Jacob Y. Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 2018;93:377–386. doi: 10.1111/tpj.13782. [DOI] [PubMed] [Google Scholar]
- 35.Soppe WJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJ. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6:791–802. doi: 10.1016/S1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- 36.Ramírez-Ortega FA, Herrera-Pola PS, Toscano-Morales R, Xoconostle-Cázares B, Ruiz-Medrano R. Overexpression of the pumpkin (Cucurbita maxima) 16 kDa phloem protein CmPP16 increases tolerance to water deficit. Plant Signal Behav. 2014;9:973823. doi: 10.4161/15592324.2014.973823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kötzler MP, Hancock SM, Withers SG. Glycosidases: functions, families and folds. In: eLS. Chichester, NJ/USA: John Wiley & Sons, Ltd; 2014. p. eLS.14. doi: 10.1002/9780470015902.a0020548.pub2. [DOI] [Google Scholar]
- 40.Cheng Q, Gao H, Hu N. A trehalase from Zunongwangia sp.: characterization and improving catalytic efficiency by directed evolution. BMC Biotechnol. 2016;16(1):9. doi: 10.1186/s12896-016-0239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 42.Shen H, Strunks GD, Klemann BJ, Hooykaas PJ, de Pater S. CRISPR/Cas9-induced double-strand break repair in Arabidopsis non homologous end-joining mutants. G3. 2017;7:193–202. doi: 10.1534/g3.116.035204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Houtte H, Vandesteene L, Lopez-Galvis L, Lemmens L, Kissel E, Carpentier S, Feil R, Avonce N, Beeckman T, Lunn JE, et al. Overexpression of the trehalase gene AtTRE1 leads to increased drought stress tolerance in Arabidopsis and is involved in abscisic acid-induced stomatal closure. Plant Physiol. 2013;161:1158–1171. doi: 10.1104/pp.112.211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iturriaga G, Suárez R, Nova-Franco B. Trehalose metabolism: from osmoprotection to signaling. Int J Mol Sci. 2009;10:3793–3810. doi: 10.3390/ijms10093793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M. Trehalose metabolism in plants. Plant J. 2014;79:544–567. doi: 10.1111/tpj.12509. [DOI] [PubMed] [Google Scholar]
- 46.Hwang K, Susila H, Nasim Z, Jung JY, Ahn JH. Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol Plant. 2019;12:489–505. doi: 10.1016/j.molp.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Kooyers NJ. The evolution of drought escape and avoidance in natural herbaceous populations. Plant Sci. 2015;234:155–162. doi: 10.1016/j.plantsci.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 48.Riboni M, Robustelli Test A, Galbiati M, Tonelli C, Conti L. Environmental stress and flowering time: the photoperiodic connection. Plant Signal Behav. 2014;9(7):e29036. doi: 10.4161/psb.29036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franks SJ, Sim S, Weis AE. Rapid evolution of flowering time by an annual plant in response to climate fluctuation. Proc Natl Acad Sci USA. 2007;104:1278–1282. doi: 10.1073/pnas.0608379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jordan CY, Ally D, Hodgins KA. When can stress facilitate divergence by altering time to flowering? Ecol Evol. 2015;5(24):5962–5973. doi: 10.1002/ece3.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brodmann D, Schuller A, Ludwig-Müller J, Aeschbacher RA, Wiemken A, Boller T, Wingler A. Induction of trehalase in Arabidopsis plants infected with the trehalose-producing pathogen Plasmodiophora brassicae. Mol Plant Microbe Interact. 2002;15:693–700. doi: 10.1094/MPMI.2002.15.7.693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


