Abstract
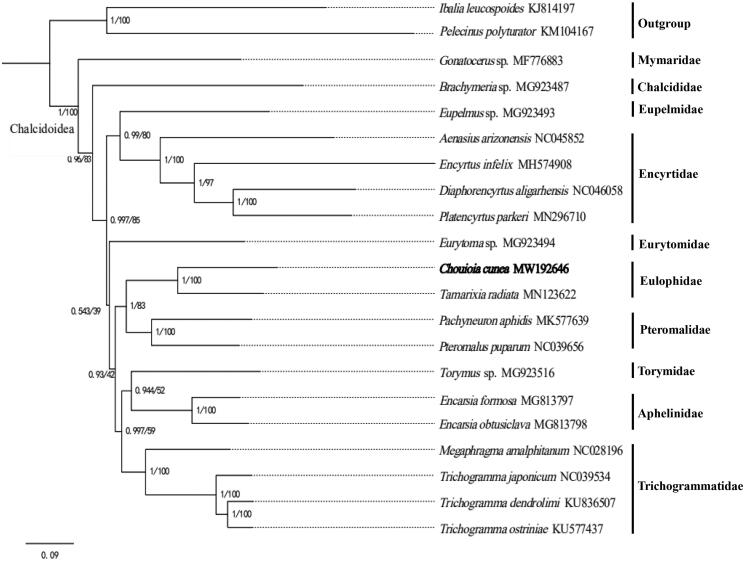
Chouioia cunea Yang 1989 is a parasitic wasp and natural enemy of several lepidopteran pests during their pupal stage. In this study, we sequenced and analyzed the mitochondrial genome of C. cunea, and obtained a complete DNA molecule that is 14,930 bp in size with 13 protein-coding genes (PCGs), two ribosomal RNA genes (rRNAs), and 22 transfer RNA genes (tRNAs) (GenBank accession number MW192646). All the 13 PCGs started with typical ATN (ATA, ATG, and ATT) and terminated with the stop codon TAA or TAG. Phylogenetic analysis showed that C. cunea formed the sister group with Tamarixia radiata, which belongs to the same family.
Keywords: Chalcidoidea, gene rearrangement, mitochondrial genome, Chouioia cunea Yang, phylogenetic relationship
Chouioia cunea Yang (Eulophidae: Chalcidoidea) was first collected from pupal of the fall webworm (Hyphantria cunea Drury), which can also be used for biological control of a variety of Lepidopteran pests such as c Fabricius, Clania variegata Snellen, Stilpnotia candida Staudinger, S. salicis (L.), Micromilalopha troglodyta (Graeser), and Ivela ochropoda Fabricius (Zhao et al. 2016; Xin et al. 2017). Insect mitochondrial genome has many characteristics which can make it play an important role in molecular evolution, phylogenetics, and population genetics. However, only T. radiata has been sequenced in Eulophidae (Du et al. 2019).
In this study, individuals of C. cunea were collected from a field in Hainan province and reared in the Environment and Plant Protection Institute, China Academy of Tropical Agriculture Sciences, Hainan, China (110°20′9″N, 19°59′21″E). The samples were preserved in 95% ethanol at −20 °C in herbarium of Post-Entry Quarantine Station for Tropical Plant, Haikou Customs District, PR China with an accession number IN07040201-0001-0020. Single sample were used for genomic DNA extraction. The mitogenome sequence of C. cunea was generated using Illumina HiSeq X TEN Sequencing System and assembled by MitoZ software without parameters (Meng et al. 2019).
The mitochondrial genome of C. cunea is 14,930 bp long, including 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and one partial non-coding AT-rich region with a length of 220 bp. All the genes were distributed on two coding chains, of which 27 genes were encoded on majority strand (J-chains) and the rest were transcribed on minority chains (N-chains). The overall base composition of the mitogenome sequence is 44.8% A, 40.3% T, 8.2% C, 6.7% G, with a high AT bias of 85.1%. Compared with ancestral insect mitochondrial genome, the mitogenome of C. cunea exhibits a dramatic mitochondrial gene rearrangement, of which is consistent with previous studies (Wu et al. 2020). A total of 27 genes have been rearranged in the C. cunea, including seven PCGs (trnC, trnK, trnD, ATP8, ATP6, NADH3, trnR), 20 tRNAs. The gene block (NADH3-trnG-COX3-ATP6-ATP8-trnD-trnQ-trnK-COX2-trnL2-COX1) was inverted in the mitochondrial genome of C. cunea.
All the 13 PCGs of C. cunea started with the conventional ATN codons, including three ATAs (ND1, ND3, and ND4L), five ATTs (ND2, ND5, ND6, COX2, and ATP8), five ATGs (COX1, ND4, CYTB, COX3, and ATP6). Twelve PCGs terminate with the stop codon TAA, whereas ND1 end with TAG. The length of 22 tRNA genes is between 58 and 69 bp, and all them have the typical clover-leaf structure, except for trnS1 and trnR which lack dihydrouridine (DHU) arm. It is a common phenomenon that trnS1 lacks DHU arm in the mitochondrial genome of many insects (Yuan et al. 2015; Xiong et al. 2019). Two rRNA genes (s-rRNA and l-rRNA) located at trnV/trnA and trnA/trnL1 regions, with the lengths of 754 and 1331 bp, respectively.
To validate the phylogenetic status of C. cunea, we selected the mitochondrial DNA sequences of 18 closely related taxa of Chalcidoidea in NCBI, and extracted the sequences by Phylosuite software (Zhang et al. 2020). Pelecinus polyturator and Ibalia leucospoides were used as outgroups. The analyses were performed with Bayesian inference and maximum likelihood in Phylosuite (Nguyen et al. 2015; Zhang et al. 2020). The result showed that C. cunea was clustered with T. radiata (Figure 1), which belongs to the same family. To sum up, the results of this study can provide essential and important DNA molecular data for further phylogenetic and evolutionary analysis of Chalcidoidea.
Figure 1.
Phylogenetic relationships of 13 mitochondrial protein-coding genes sequences within Chalcidoidea were performed using Bayesian/ML methods.
Funding Statement
This study was supported by the Hainan Natural Science Foundation Innovation Research Team Project [2019CXTD409]; National Key Research and Development Project [2019YFD03001]; Ministry of Agriculture and Rural Affairs's International Exchange and Cooperation Project ‘Joint Survey and Development Evaluation of Agricultural Resources in Tropical Countries Along the Belt and Road’ [BARTP-08-LBQ-TJH]; Special Project for Basic Scientific Research Business Expenses of Chinese Academy of Tropical Agricultural Sciences [1630042017011].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW192646. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA691215, SRP301245, and SRS8004285, respectively.
References
- Du YM, Song X, Liu XJ, Ouyang ZG, Lu ZJ.. 2019. Mitochondrial genome of Tamarixia radiata (Hymenoptera: Chalcidoidea: Eulophidae) and phylogenetic analysis. Mitochondrial DNA Part B. 2(4):2839–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng GL, Li YY, Yang CT, Liu SL.. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Haeseler VA, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YF, Yang HL, Feng ZB, Li BY, Zhou WB, Song F, Li H, Zhang LM, Cai WZ.. 2020. Novel gene rearrangement in the mitochondrial genome of Pachyneuron aphidis (Hymenoptera: Pteromalidae). Int J Biol Macromol. 149:1207–1212. [DOI] [PubMed] [Google Scholar]
- Xin B, Liu PX, Zhang S, Yang ZQ, Daane KM, Zheng YA.. 2017. Research and application of Chouioia cunea Yang (Hymenoptera: Eulophidae) in China. Biocontrol Sci Technol. 27(3):1–10. [Google Scholar]
- Xiong M, Zhou QS, Zhang YZ.. 2019. The complete mitochondrial genome of Encyrtus infelix (Hymenoptera: Encyrtidae). Mitochondrial DNA Part B. 4(1):114–115. [Google Scholar]
- Yuan ML, Zhang QL, Guo ZL, Wang J, Shen YY.. 2015. The complete mitochondrial genome of Corizus tetraspilus (Hemiptera: Rhopalidae) and phylogenetic analysis of Pentatomomorpha. PLOS One. 10(6):e0129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Gao FL, Li WX, Jakovlić I, Zou H, Zhang J, Wang GT.. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. [DOI] [PubMed] [Google Scholar]
- Zhao YN, Wang FZ, Zhang XY, Zhang SU, Guo SL, Zhu GP, Liu Q, Li M.. 2016. Transcriptome and expression patterns of chemosensory genes in antennae of the parasitoid wasp Chouioia cunea. PLOS One. 11(2):e0148159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW192646. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA691215, SRP301245, and SRS8004285, respectively.