Abstract
Onobrychis viciifolia is mainly distributed in Europe and has been widely cultivated in North and Northwest of China. The complete chloroplast genome was sequenced using the Illumina Hiseq X-Ten platform. The genome lacks an inverted repeat (IR) region, containing 76 protein-coding genes, 30 tRNAs genes and 4 rRNAs. The overall GC content is 34.6%. A phylogenetic tree based on the whole chloroplast genomes of 14 species indicated that Onobrychis viciifolia belonged to the tribe Hydysareae in IRLC group of the subfamily Papilionoideae (Leguminosae), and it was sister to the genus Hedysarum.
Keywords: Chloroplast genome, Onobrychis viciifolia
Onobrychis viciifolia is a deep-rooted perennial legume with erect or sub-erect hollow stems growing up to a height of 100 cm (Frame et al. 1998). As a high quality forage crop, it has been cultivated in Northwest China for a long period (Xu 1998). Previous studies reported that Onobrychis viciifolia has good tolerance to drought-stress, salt-stress (Beyaz et al. 2017; Beyaz 2019), a high nutritive value, high voluntary intake and palatability to grazing animals (Bhattarai et al. 2016), but wild resources has been barely witnessed. A good knowledge in genomic information of this species would contribute to the study on its molecular studies, geographical distribution, diversity, breeding and fodder production.
The fresh leaves of Onobrychis viciifolia was collected in Yunwu Mountains National Natural Reserve, Ningxia, China (106°23′15″E, 36°15′38″N), and the voucher specimen was deposited in the herbarium of South China Botanical Garden, Chinese Academy of Sciences (IBSC, collection #: L.Duan 2018014). We extracted the total genomic DNA with CTAB approach (Doyle 1987), the genomic libraries were prepared and sequenced using the Illumina Hiseq X-Ten platform (Illumina Inc. San Diego, CA). The resultant sequences were filtered following Yao et al. (2016), the adaptor-free reads were then assembled with SPAdes 3.11 (Bankevich et al. 2012). We annotated the assembly of complete chloroplast (cp) genomes using the Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. 2004) and deposited the genomes in GenBank (accession number: MW007721).
About 1.09 Gb raw reads of Onobrychis viciifolia were obtained, with coverage of 918× and 121,932 bp in length. The cp genome lacked inverted repeat (IR) region. The genome contained 76 protein-coding genes (CDS), 30 transfer RNA genes(tRNA), 4 ribosomal RNA genes (rRNA), within which 16 genes (ndhA, ndhB, rpl2, rpl16, petD, petB, clpP, rps12, atpF, rpoC1, trnG-UCC, trnA-UGC, trnI-GAU, trnL-UAA, trnV-UAC and trnK-UUU) had one intron, 2 gene (rps12, ycf3) has two introns, respectively. Overall GC content of the whole genome was 34.6%.
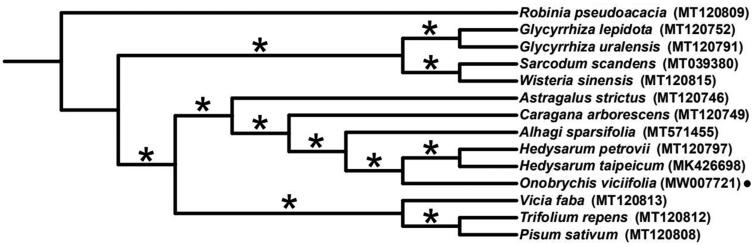
To infer the phylogenetic relationships among this species and its related taxa, whole cp genomes of 13 Papilionoideae species were downloaded from GenBank, which were aligned with that of Onobrychis viciifolia by applying MAFFT v.7 (Katoh and Standley 2013). Based on the alignment, a aximum-likelihood (ML) tree was constructed using IQ-TREE v.1.6 (Nguyen et al. 2015). The result (Figure 1) showed that Onobrychis viciifolia, Hedysarum petrovii, Hedysarum taipeicum and Alhagi sparsifolia constituted a well-supported tribe Hydysareae [as in Duan et al. (2020)], serving as the sister of the Hedysarum genus. This tribe was a member of the inverted repeat-lacking clade (IRLC), which in turn belonged to the clade of Hologalegina in papilionoid legumes as suggested by previous studies (Wojciechowski et al. 2004; Schrire 2005).
Figure 1.
Maximum likelihood (ML) phylogenetic tree based on 14 chloroplast genomes of Fabaceae. The position of Onobrychis viciifolia is indicated with black dot. The bootstrap values of 100% are shown on branches with asterisks.
Funding Statement
This study was supported by the grants from the Technology Key Research and Development Project of Guangxi [Grant no. Guike AB18221054] and Science and technology program of Guangdong Province, China [2019B121202006].
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The voucher specimen was deposited in the herbarium of South China Botanical Garden, Chinese Academy of Sciences (IBSC, collection #: L.Duan 2018014) (http://herbarium.scbg.cas.cn/).
To infer the phylogenetic relationships among this species and its related taxa, whole cp genomes of 13 Papilionoideae species were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/).
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW007721. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA690644, SRR13385163, and SAMN17255786, respectively (https://www.ncbi.nlm.nih.gov/sra/?term=SRR13385163).
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai S, Coulman B, Biligetu B.. 2016. Sainfoin (Onobrychis viciifolia scop.): renewed interest as a forage legume for western Canada. Can J Plant Sci. 96(5):748–756. [Google Scholar]
- Beyaz R. 2019. Biochemical responses of sainfoin shoot and root tissues to drought stress in in vitro culture. Legume Res. 42(2):173–177. [Google Scholar]
- Beyaz R, Sancak C, Yildiz M.. 2017. Morphological and biochemical responses of sainfoin (Onobrychis viciifolia Scop.) ecotypes to salinity. Legume Res. 41(2):253–258. [Google Scholar]
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Duan L, Loc PK, Zhang Z-R, Chen H-F.. 2020. The complete chloroplast genome of ornamental liana Sarcodum scandens (Fabaceae.). Mitochondrial DNA Part B. 5(2):1427–1428. [Google Scholar]
- Frame J, Charlton JFL, Laidlaw AS.. 1998. Temperate forage legumes. Wallingford (UK): CAB International. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrire BD. 2005. Trbie Millettieae. In: Lewis G, Schrire B, Mackinder B, Lock M, editors. Legumes of the world (pp 367–387). Kew: Royal Botanic Gardens. [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ.. 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot. 91(11):1846–1862. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]
- Xu LR. 1998. Onobrychis. In: Cui HB, editor. Flora Reipublicae Popularis Sinicae. Beijing: Science Press; Vol. 42(2), p. 218–221. [Google Scholar]
- Yao X, Tan YH, Liu YY, Song Y, Yang JB, Corlett RT.. 2016. Chloroplast genome structure in Ilex (Aquifoliaceae). Sci Rep. 6(1):28559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The voucher specimen was deposited in the herbarium of South China Botanical Garden, Chinese Academy of Sciences (IBSC, collection #: L.Duan 2018014) (http://herbarium.scbg.cas.cn/).
To infer the phylogenetic relationships among this species and its related taxa, whole cp genomes of 13 Papilionoideae species were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/).
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW007721. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA690644, SRR13385163, and SAMN17255786, respectively (https://www.ncbi.nlm.nih.gov/sra/?term=SRR13385163).