ABSTRACT
Background: The Tn916-Tn1545 family of Integrative Conjugative Elements (ICE) are mobile genetic elements (MGEs) that play a role in the spread of antibiotic resistance genes. The Tn916 harbors the tetracycline resistance gene tet(M) and it has been reported in various bacterial species. The increase in the levels of tetracycline resistance among oral streptococci is of great concern primarily due to the abundance of these species in the oral cavity and their ability to act as reservoirs for antibiotic resistance genes.Methods: In the current study, we screened 100 Norwegian clinical oral streptococcal isolates for the presence and diversity of the Tn916-Tn1545 family. In addition, we investigated the transferability the elements, and the associated transfer frequencies.Results: We observed that 21 isolates harboured the Tn916-Tn1545 family and that two of these elements were the novel Tn6815 and Tn6816. The most prevalent member of the Tn916 -Tn1545 family observed in the Norwegian clinical oral streptococcal isolates was the wild type Tn916.Conclusion: The detection of other members of this family of ICE and varying transfer frequencies suggests high versatility of the Tn916 element in oral streptococci in Norway.
KEYWORDS: Integrative Conjugative Elements (ICEs), oral streptococci, mobile genetic elements, antibiotic resistance, conjugation, tetracycline resistance, ddPCR, Tn6815, Tn6816
Introduction
The oral cavity is the second highest bacterial populated area of the human body [1]. With more than 110 approved species, streptococci are the most abundant species in the oral microbial communities [2]. In general, oral streptococci are classified as commensal bacteria and constitute part of the oral microbiota of virtually all humans. Some strains, however, have been shown to have pathogenic abilities, causing invasive infections such as infective endocarditis, septicemia, and pneumonia in neutropenic individuals [3,4]. In addition to the possible invasive pathogenicity, oral streptococci are gathering significance as they have been implicated in being reservoirs of antibiotic resistance genes [5–9], and carry substantial resistance genes as part of the ‘oral resistome’ [10]. Sequence analysis of evolutionary important genes in oral streptococci suggests that the presence of antibiotic resistance genes in these species are due, at least in part, to Horizontal Gene Transfer (HGT) [11]. One of the mechanisms of HGT involves the transfer of mobile genetic elements (MGEs) such as Integrative Conjugative Elements (ICE).
ICE constitute a very large and diverse group of MGEs of which the Tn916-Tn1545 family is one of the largest members. The members of this family are by definition, self-transferable genetic elements that can exist as circular intermediates or as part of a chromosome [12–14]. Although very diverse, the elements in the Tn916-Tn1545 family have Tn916 in the backbone structure with the similar conjugative transfer, transcriptional regulation and recombination modules. Whereas Tn916 has the tet(M) gene in the accessory function module, other antibiotic genes have been observed in other members of this family such as the presence of tet(S) in Tn6000 [15,16] and Tn916S [16,17]; erm(B) in Tn3872 [18]; tet(M) and mef(E) in Tn2009 [19]; and tet(M), erm(B) and mef(E) in Tn2017 [20]. Tn916, which was first isolated from Enterococcus faecalis DS16 [21], is a well-documented prototype of the Tn916-Tn1545 family. It is the smallest member of the Tn916-Tn1545 family and has been found in 35 different bacterial genera [22,23]. The wide spread of Tn916, is in part, attributed to the presence of integrase and excision genes, which enable the ICE to excise (cut) itself from one location and integrate itself in another location. The transcription regulation module of Tn916 tightly regulates the excisionase and integrase, which are located at the 3´end of the element. This module consists of orf5, orf6, orf7, orf8, orf9, orf10 and orf12, and the importance of this module is reflected by being highly conserved [24]. It has been proposed that transcription starts from a promoter that is upstream of orf12 and runs through orf12, tet(M) and the downstream Open Reading Frames (ORFs) [25]. The transcription of these ORFs not only leads to the self-mobilization of the element but also increases the levels of tet(M), which enables the bacteria to survive in the presence of tetracyclines.
Tetracyclines, like all other antibiotics in Norway, are regulated and controlled by prescription. In 2018, tetracyclines were the second most prescribed group of antibiotics in Norway, accounting for 26% of all defined daily doses for systemic antibacterial agents [26]. Although the use of antibiotics continues to decrease in Norway, cases of antibiotic resistant bacterial isolates continue to appear [26]. This increase, coupled with the higher number of antibiotic resistance related infections highlight the need to understand the underlying factors that promote the spread and stability of tetracycline in bacterial populations.
Recent approaches to study the oral microbiome have re-enforced the abundance of Streptococcus ssp. in the oral cavity and the high prevalence of antibiotic resistance genes. A study by Almeida et al. 2020 [27] found that 72% of the samples isolated from the oral microbiota had at least one antibiotic resistance gene, whilst a study by Christensen and Sørensen found that more than 90% of the saliva samples analyzed carried two antibiotic resistance genes [28]. In another study, analysis of 342 clinical oral streptococci found that 44% were resistant to tetracycline, whereas 23.1% were resistant to a combination of erythromycin, tetracycline and ofloxacin [29].
The growing number of studies investigating the prevalence of resistance genes in oral streptococci illustrate the role these species play in the spread of antibiotic resistance genes. There are, however, few studies that have investigated the diversity and transferability of the Tn916-Tn1545 family. In this study, we investigated the presence of the tet(M) carrying ICE belonging to the Tn916-Tn1545 family in antibiotic resistant oral streptococci collected in Norway. We analyzed the diversity and transferability of these ICEs to determine the role that the Tn916-Tn1545 family plays in the spread of these resistance determinants.
Material and methods
Bacterial strains
The bacterial strains used in this study were part of the oral streptococci collection strains obtained from Norwegian hospitals and submitted to the National Competence Service for the Identification of Antibiotic Resistance (K-RES). The 100 clinical strains used in this study consisted of Streptococcus mitis (n = 42), Streptococcus oralis (n = 22), Streptococcus sanguinis (n = 7), Streptococcus salivarius (n = 7), Streptococcus anginosus (n = 7), Streptococcus gordonii (n = 2), Streptococcus constellatus (n = 2), Streptococcus intermedius (n = 1), Streptococcus mutans (n = 1) and nine unclassified streptococcal species. In addition, the type strains S. oralis ATCC 35037, S. mitis ATCC 49456, S. sanguinis ATCC 10556, S. gordonii ATCC 10558, S. pneumoniae ATCC 49616, Bacillus subtilis BS34A (NZ_LN680001.1) and B. subtilis BS49 (NZ_LN649259.1) were included in this study as experimental controls.
Construction of kanamycin resistant oral streptococcal strains
The type strains S. oralis ATCC 35037, S. mitis ATCC 49456, S. sanguinis ATCC 10556 and S. gordonii ATCC 10558 were subjected to mutagenesis using the EZ-Tn5 < kan-2> transposons kit (Lucigen, Middleton, WI) according to the manufacturer’s instruction. In brief, the competent oral streptococcal cells were prepared as described by Smith et al. [30]. The EZ-Tn5 < kan-2> transposon was introduced into the competent cells by electroporation using the MicroPulser Electroporator (Bio-Rad, Pleasanton, CA, USA) according to manufacturer´s instructions. The kanamycin resistant isolates S. oralis SOK10, S. mitis SMK7, S. sanguinis Sg10, S. mutans M8 and S. gordonii G3 were used as recipients in the Tn916- Tn1545 family transferability assay.
Assessment of Minimal Inhibitory Concentrations (MIC) of Tn916 carrying isolates
The MIC to tetracycline, erythromycin, penicillin, gentamicin, and clindamycin of the clinical oral streptococcal isolates were determined using the E-test according to the manufacturer’s instructions (BioMérieux SA, Marcy l’Etoile, France). In short, a single colony was used to prepare a fresh overnight culture in BHI broth. Bacterial cultures with a turbidity of 0.5 McFarland were inoculated onto the Mueller-Hinton-F agar supplemented with 5% horse blood, prior to the addition of the respective antibiotic strips. The agar plates were incubated in an aerobic atmosphere at 37°C for 24 hours. The isolate S. pneumoniae ATCC 49616 were used as a control strain. The results were interpreted according to the standards set by the European Committee for Antimicrobial Susceptibility Testing (EUCAST) (www.eucast.org) and the Clinical and Laboratory Standards Institute (CLSI) (https://clsi.org/).
Characterization of the Tn916-Tn1545 family found in oral streptococci
The genomic DNA used in the conventional Polymerase Chain Reaction (PCR) and ddPCR was extracted using the QIAcube automated system (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. To investigate the presence of the Tn916-Tn1545 family in oral streptococcal strains, PCR was performed as previously described [31–33]. The presence of the tet (M) gene and the integrase and excisionase regions (Int and Xis genes) were determined using the primers listed in Table 1. The PCR reaction was performed in a volume of 25 µl which contained 12.5 µl DreamTaq 2X (Thermofisher Scientific), 0.5 µM of each specific primer, 8 µl of water and 2.5 µl DNA template. Subsequently, the samples that tested positive for both these targets were subjected to Long PCR using Long A primers (amplify the region between 38 bp and 9,884 bp of Tn916) and Long B primers (amplify position 9,824 bp to 17,947 bp of Tn916). These primers (listed in Table 1) were designed to detect and amplify two large fragments in Tn916, Tn6002, Tn6003, Tn1545, Tn3878, Tn2009, Tn2010, Tn2017, Tn6084 and Tn6079 [34]. In brief, the long PCRs were performed using Platinum SuperFI (Thermofisher Scientific) in a total volume of 25 µl. The PCR reaction mix contained 12.5 µl Platinum SuperFI mix, 5 µl Enhancer, 1 µl of each primer (10 pmol/µl), 3.5 µl water and 2 µl DNA template. The PCR reactions were run according to the manufacturer’s instructions. The two long PCR amplicons were subjected to restriction fragment length polymorphism (RFLP) by enzymatic digestion with HincII as previously described by Ciric et al. 2012 [34] .
Table 1.
Details of primers and probes used
| Target | Forward primer | Reverse primer | Probe sequence and label (chlorophore) | Amplicon size | Annealing temperature | Reference |
|---|---|---|---|---|---|---|
| tet(M) | GTR AYG AAC TTT ACC GAA TC | ATC GYA GAA GCG GRT CAC | N/A | 633bp | 55°C | [31] |
| Int and Xis genes | CGC CAAAGG GTC TTG TAT ATG | GCT GTA GGT TTT ATC AGC TTT TGC | N/A | 650bp | 58°C | [50] |
| Long A (position 38–9,884 bp) | GGA CTT ATC CAC TTT ATC AAG G | AAA CAG AAG CAG TGA GAA GA | N/A | 9806bp | 58°C | [51,52] |
| Long B (Position 9,824–17,947 bp) | GAA AAC TTT AGT GAT TGG TGG | CTG TAG GAA GAT ACT TCA CG | N/A | 8123bp | 58°C | [53] |
| Circular intemidate (conventional PCR) | CGT GAA GTA TCT TCC TAC A |
AC CTT GAT AAA GTG TGA TAA | N/A | 166bp | 56°C | [54] |
| Circular intemidate (ddPCR) | CGT GAA GTA TCT TCC TAC A | GAC CTT GAT AAA GTG TGA TAA | AAT ACT CGA AAG CAC ATA GAA TAA GGC FAM/HEX |
167bp | 56°C | [41] |
| Int and Xis (ddPCR) | ATA CTC CCA TAC AGT CAA TAG TCC | AGT TCC ACC CCT GCA TGG | CCG TCG CAGGCA ATG AGT ATG GCT FAM |
88bp | 56°C | [41] |
Next generation sequencing of the Tn916-Tn1545 family found in oral streptococci
The extracted streptococcal DNA for the NGS was obtained using the modified Marmur extraction procedure as described by Salva-Serra et al. [35]. Based in their RFLP pattern, we selected six oral streptococcal isolates for NGS using the Illumina Nextseq 550 platform and the Pacific Biosciences Sequel instrument (PacBio) applying Sequel Polymerase v3.0, SMRT cells v3 LR and Sequencing chemistry v3.0. The Illumina Nextseq 550 was conducted at the Genomics Support Center, Tromso whereas as the PacBio sequencing was performed at the NorSeq (Oslo). Hybrid assemblies were generated using Canu [36]. The assemblies/contigs were re-ordered using Abacas 1.3.1 [37] with Tn916 (U09422.1) as a guiding reference. The protein coding regions in the assemblies were predicted using PROKKA version 1.12 [38] and the similarities and differences between the assembled elements and the Tn916-Tn1545 family were assessed using Easy fig [39]. BLAST was used to align the assembled elements to the wild type Tn916.
Circularization ratio of the Tn916-Tn1545 family in oral streptococci
The circularization ratio (CR) of the Tn916-Tn1545 family was determined by investigating the presence of the circular intermediate (CI) in the selected isolates. The presence of CI was determined by conventional PCR as previously described [40]. The primers used in this PCR are designed to produce an amplicon only when the element has been excised from the chromosome and the left and right junctions of the element join to form the CI. The CR was determined by ddPCR as previously described by Lunde et al. [41] and expressed as a ratio of CI to the number of the Tn916-Tn1545 family elements that were present in a bacterial population.
Transferability and conjugation frequency of the Tn916-Tn1545 family in oral streptococci
The in vivo transferability and conjugation frequency of the Tn916-Tn1545 family was determined by filter mating experiments as previously described by Roberts et al. [42]. In brief, the donor and recipient isolates were cultivated on BHI agar plates with selection (according to the species selective markers, as listed in Table 2). The overnight cultures were prepared from single colonies and incubated at 37°C for 20 hours prior to mixing the donor and the recipient species in a 1:1 ratio. The mixed cultures were centrifuged at 1,000 g for 5 minutes and the supernatant was discarded. The cells were suspended in 1 ml of BHI broth and then 100 µl of the cell suspension was plated on a cellulose nitrate filter (0.45 µM, Sartorius Stedim Biotec; Germany). The conjugation experiments were conducted in three biological replicates and the generated transconjugants were screened phenotypically (on selective agar) and genotypically (by PCR) to verify the presence of the Tn916-Tn1545 family elements. The conjugation frequency of the Tn916-Tn1545 like elements was accessed both inter- and intra-species.
Table 2.
Bacterial strains used in the inter- and intra-species transfer of the Tn916-Tn1545 family to clinical and reference oral streptococci
| Strain | Background and relevant phenotype | Reference |
|---|---|---|
| Bacillus subtilis BS49 | Laboratory strain::Tn916, TetR | [55] |
| S. sanguinis Ssg41 | Clinical isolate:: Tn916, TetR | [41] |
| S. oralis SO52 | Clinical isolate:: Tn916, TetR | [41] |
| S. mitis SM28 | Clinical isolate:: Tn6815, TetR | [41] |
| S. oralis SO32 | Clinical isolate, GentaR | This study |
| S. gordonii G3 | ATCC 10558::Tn5, KanR | This study |
| S. mutans M8 | ATCC 25175::Tn5, KanR | This study |
| S. mitis SMK7 | ATCC 49456::Tn5, KanR | This study |
| S. oralis SOK10 | ATCC 35,037::Tn5, KanR | This study |
| S. sanguinis Sg10 | ATCC 10556::Tn5, KanR | This study |
Results
Distribution of the tet(M) gene and Int and Xis genes in oral streptococcal isolates
The 100 oral streptococcal isolates used in this study were screened for the presence of the Tn916-Tn1545 family using primers that detect the tet(M) gene and the integrase/excisionase regions (Int and Xis genes) of these elements. A total of 24 isolates harbored tet(M) whereas 38 isolates carried the integrase/excisionase. The most dominant species in this study was S. mitis (n = 42) and the prevalence of tet(M) in these isolates was 26.2% (n = 11). S. oralis and S. sanguinis had a tet (M) prevalence of 41% (9/22) and 28.6% (2/7), respectively. Of the seven S. salivarius, seven S. anginosus and three S. gordonii isolates that were tested, only one isolate of each species had tet(M). In S. constellatus, only one of the two studied isolates were found to harbor the tetracycline resistance gene. The tet(M) gene was not detected in the tested S. intermedius and S. mutans isolates. In total, 21% (21/100) tested positive for both the tet(M) gene and the integrase/excisionase region and these were considered to harbor Tn916-Tn1545 like elements. The species distribution of both tet(M) and the integrase/excisionase regions is illustrated in Figure 1.
Figure 1.

A graph illustrating the species distribution of the oral streptococci isolates included in this study. The isolates that tested PCR negative for the presence of tet(M), Int and Xis genes are shown in green whereas isolates that were PCR positive for tet(M), Int and Xis genes are illustrated in red. The blue bars indicate isolates that only tested positive for Int and Xis. S.sp*; other streptococcal species including S. anginosus, S. constellatus and S. intermediate and member of the S. milleri group
Assessment of minimal inhibitory concentrations of Tn916 carrying isolates
The antibiotic resistance profile of the 21 isolates harboring the Tn916-Tn1545 family were assessed by determining the MIC to tetracycline, erythromycin, penicillin, gentamicin and clindamycin, and the result are shown in Table 3. The MIC to tetracycline ranged from 0.190 to 96 g/ml. All isolates that carried the tet(M) gene were phenotypically resistant to tetracycline with the exception of isolate S. oralis SO04 where the tet(M) gene was silent and the detected MICs were below the resistance threshold (≤ 2 g/ml) as determined by EUCAST. The MIC values to erythromycin ranged from 0.023 to >256 g/ml and of the 21 isolates, five were classified as resistant to erythromycin with MIC values ranging from 3 to > 256 g/ml. The MIC to penicillin, gentamycin and clindamycin where found to be 0.016 to 1.5 g/ml, 0.16 to 32 g/ml and clindamycin 0.094 to > 256 g/ml, respectively. Although most strains were resistant to more than two antibiotics, only 16% (n = 4) of the tested isolates were resistant to four of the five tested antibiotics. The isolate S. mitis SM28 was the only isolate found to be phenotypically resistant to penicillin with a MIC of 6 g/ml.
Table 3.
Antibiotic resistance profile of oral streptococcal isolates harboring the Tn916-Tn1545 family collected in Norway
| Isolate | Tetracycline g/ml |
Erythromycin g/ml |
Penicillin g/ml |
Gentamycin g/ml |
Clindamycin g/ml |
|---|---|---|---|---|---|
| S. sanguinis SS33 | 24 (R) | 0.047 (S) | 0.190 (S) | 6 (IE) | 0.125 (S) |
| S. sanguinis SS41 | 32 (R) | 0.016 (S) | 0.047 (S) | 1.5 (IE) | 0.160 (S) |
| S. oralis SO1 | 32 (R) | >256 (R) | 0.016 (S) | 16 (IE) | >256 (R) |
| S. oralis SO4 | 0.190 (S) | 0.125 (S) | 0.023 (S) | 12 (IE) | 0.190 (S) |
| S. oralis SO30 | 24 (R) | 0.047 (S) | 0.094 (S) | 8.00 (IE) | 0.094 (S) |
| S. oralis SO44 | 24 (R) | 3 (R) | 1 (I) | 24 (IE) | 0.094 (S) |
| S. oralis SO47 | 32 (R) | 0.047 (S) | 0.125 (S) | 32 (IE) | 0.094 (S) |
| S. oralis SO52 | 32 (R) | 0.094 (S) | 0.064 (S) | 24 (IE) | 0.125 (S) |
| S. oralis SO62 | 2 (S) | 6 (R) | 0.047 (S) | 24 (IE) | 0.75 (R) |
| S. oralis SO67 | 64 (R) | 0.047 (S) | 0.125 (S) | 12 (IE) | 0.094 (S) |
| S. oralis SO69 | 96 (R) | 6 (R) | 1.500 (I) | 32 (IE) | 0.125 (S) |
| S. oralis SO90 | 2 (S) | 0.125(S) | 0.064 (S) | 12 (IE) | 0.32 (S) |
| S. mitis SM2 | 48(R) | >256 (R) | 0.094 (S) | 0.16 (IE) | >256 (R) |
| S. mitis SM28 | 64 (R) | >256 (R) | 6 (R) | 6 (IE) | >256 (R) |
| S. mitis SM29 | 32 (R) | 0.047 (S) | 0.047 (S) | 1.5 (IE) | 0.064 (S) |
| S. mitis SM74 | 2 (S) | 0.064 (S) | 0.380 (I) | 24 (IE) | 0.094 (S) |
| S. mitis SM81 | 48 (R) | 0.032 (S) | 0.190 (S) | 6 (IE) | 0.125 (S) |
| S. gordonii SG71 | 24 (R) | 0.047 (S) | 0.016 (S) | 4 (IE) | 0.064 (S) |
| S. anginosus SA46 | 12 (R) | 0.094 (S) | 0.047 (S) | 12 (IE) | 0.094 (S) |
| S. salivarius SSv51 | 96 (R) | 0.064 (S) | 0.023 (S) | 12 (IE) | 0.125 (S) |
| S. constellatus SC99 | 16 (R) | 0.023 (S) | 0.094 (S) | 3 (IE) | 0.047 (S) |
S = Susceptible; I = Intermediate; R = Resistant; IE = Intrinsic Resistance.
Characterization of the Tn916-Tn1545 family found in oral streptococci
The Tn916-Tn1545 family elements were characterized by amplifying two fragments that covered a large portion of the 18 kb wild type Tn916; Long A (position 38 bp to 9,844 bp) and Long B (position 9,824 to 17,947 bp). When subjected to RFLP analysis, the large amplicons indicated that 15 out of the 21 isolates harbored the wild type Tn916. The RFLP pattern of isolates S. oralis SO62, S. oralis SO67, S. mitis SM74, S. oralis SO90, S. mitis SM28 and S. constellatus SC99 varied from the wild type Tn916 (as shown in Supplementary Figure S1). This suggested that these isolates potentially harbor other members of the Tn916-Tn1545family and were thus selected for further analysis with NGS.
Next generation sequencing of the Tn916-Tn1545 family found in oral streptococci
Analysis of the NGS resulted in the assembly of complete ICE in all of the isolates with the exception of isolate S. oralis SO62. The assembled elements are available under the Bioproject 660235 on NCBI. Sequence comparisons of the wild type Tn916 with the entire length of Tn916-Tn1545 like elements found high similarity in the isolates S. mitis SM74 and S. oralis SO90 of 98.8% and 98.8%, respectively. Analysis of the S. oralis SO67 assembled genome, showed the presence of two Tn916 elements. The elements were completely identical and showed 99.8% sequence identity to Tn916 with 99% query coverage (17,994/18,032). The resulting variations in the RFLP patterns compared to the wild type Tn916 were found to be due to SNPs and in silico digestion of the sequenced elements confirmed the obtained RFLP pattern.
In S. mitis SM28, an insertion of 5,265 bp was observed respectively as shown in Figure 2. Sequence comparison of the assembled element in S. mitis SM28 showed 99.3% similarity with ICESpn22664 (accession number HG799489.1) [43] with a coverage of 98%. The element was also found to harbor the erm(B), orf3, hin_1 and tnpA (a transposase) genes which are inserted within orf9 and are located downstream of tet(M). The Blast results of this region showed high similarity (99.89%) to Tn917 (accession number M11180.2). This suggested that Tn6815 is a composite element in which Tn917 has been inserted into position 14,525 bp of Tn916 and extends to position 19,790 bp. Downstream of position 19,790 bp, the element was identical to the 3´end of Tn916. This element has been given a new name, Tn6815, and registered in the Transposon registry [44]. The sequence of the novel Tn6815 is available on NCBI under the accession number LR828204.1
Figure 2.
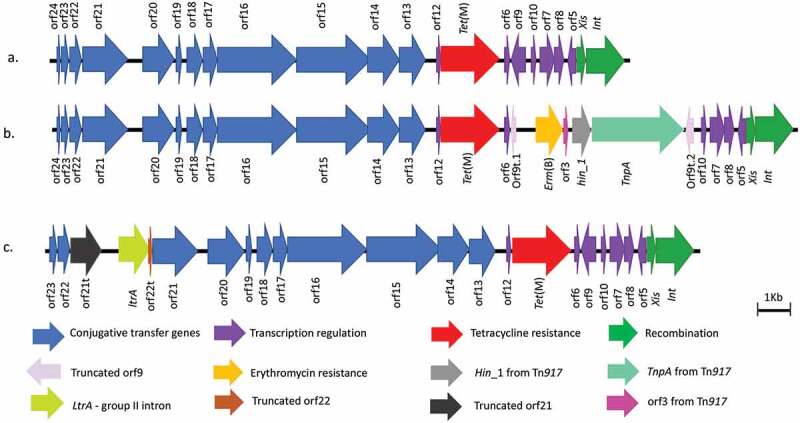
A schematic representation of the structure of Tn916 and the Tn916-Tn1545 family elements. The image in A. is Tn916 from E. faecalis DS16 (accession number U09422.1); B. illustrates Tn6815 from S. mitis SM28 (accession number LR828204.); and C. Tn6816 from S. constellatus SC99 (accession number LR828206.1). The element Tn6815 carries the erm(B) hin_1, orf3 and TnpA Transposase from Tn917 inserted between two truncated regions of orf9 (orft.1 and orft.2). The element in S. constellatus SC99 has been designated Tn6816 and it carries group II intron reverse transcriptase/maturase, truncated orf21 and orf22 and a complete copy of orf21 inserted upstream of orf20
BlastN alignment between LR828204 (Tn6815) and U09422 showed that Tn916 covered 77% of Tn6815 with two matches going from 1 to 14,524 to the left of the 5,266 bp insertion and 19,790 to 23,299 right of the insertion. The 5´end of the alignment ranged from 1 to 14,524 in both elements and was 99% identical whereas the 3´end after the insertion was 98% identical to Tn916 and corresponded to positions 14,519 bp to 18,032 bp in Tn916. The Tn6815 carried two truncated orf9 segments which have been designated orf9t.1 and orft.2. The orf9t.1 was 136 bp and lacked 218 bp of orf9 and the orf9t.2 lacked the first 131 bp of the 5´end but had the remaining 222 bp of the 3´end. The right side of the alignment started at 14,519 (which is 131 bp within orf9) and covered the rest of Tn916. The Tn6815 also carried a 5 bp repeat of orf9 on either side of the 5,266 bp insertion. BlastN analysis showed that the insertion was 99% identical to Tn917 (accession number M11180.2) and that three ranges were present in Tn6815. The first range started at 90 bp and went to 5,355 bp, which is the entire sequence of Tn917. The second range was from 92 bp to 162 bp (corresponded to 15,988 bp to 16,058 bp in Tn6815) and the third range was from 1,550 bp to 1,623 bp which corresponded to 14,524 bp to 14,597 bp in Tn6815. These last two ranges were 96% identical (68/71bp similar) indicating that there was a repeated segment with Tn917.
In S. constellatus SC99, an element harboring a 1,663 bp insertion was observed. The insertion was located 401bp before the end orf21 and corresponded to the ltrA gene, a group II intron reverse transcriptase/maturase and part of peptidase P60. The ltrA gene was followed by 91bp of the 3´end of orf22 (lacking 295 bp of orf22) and a complete copy of orf21. BlastN analysis indicated that the element had two copies of orf21and orf22. The first copy of orf21 was truncated and only 1,061 bp long. It has been designated orf21t and was located upstream of the inserted ltrA and downstream of the first and complete copy of orf22. The second copy of orf21 was complete and located downstream of ltrA. Sequence comparison of this insertion showed 99.9% similarity and 100% coverage with the ltrA gene found in the Enterococcus avium strain 352 chromosome (accession number CP034169.1). As no matches of 99% or more were obtained from sequence comparison analysis, this element has been designated a new name, Tn6816, and registered in the Transposon registry [44]. The sequence of the novel Tn6816 is available on NCBI under the accession number LR828206.1
Circularization ratio of the Tn916-Tn1545 family in oral streptococci
The presence of CI was first determined by conventional PCR and CR was determined by ddPCR. In the absence of selective pressure, the conventional PCR results indicated the presence of the CI molecules in all the tested isolates with the exception of S. mitis SM74. The quantification of the CI molecules in the bacterial population by ddPCR was found to range from 0% in two isolates to 0.2% per element as indicated in Figure 3. Interestingly, the three highest levels of CI observed in this study were from S. mitis isolates. This coupled with the fact that S. mitis is one of the most abundant streptococcus species in the oral cavity warranty the need to further investigate the evolution of Tn916 in S. mitis.
Figure 3.
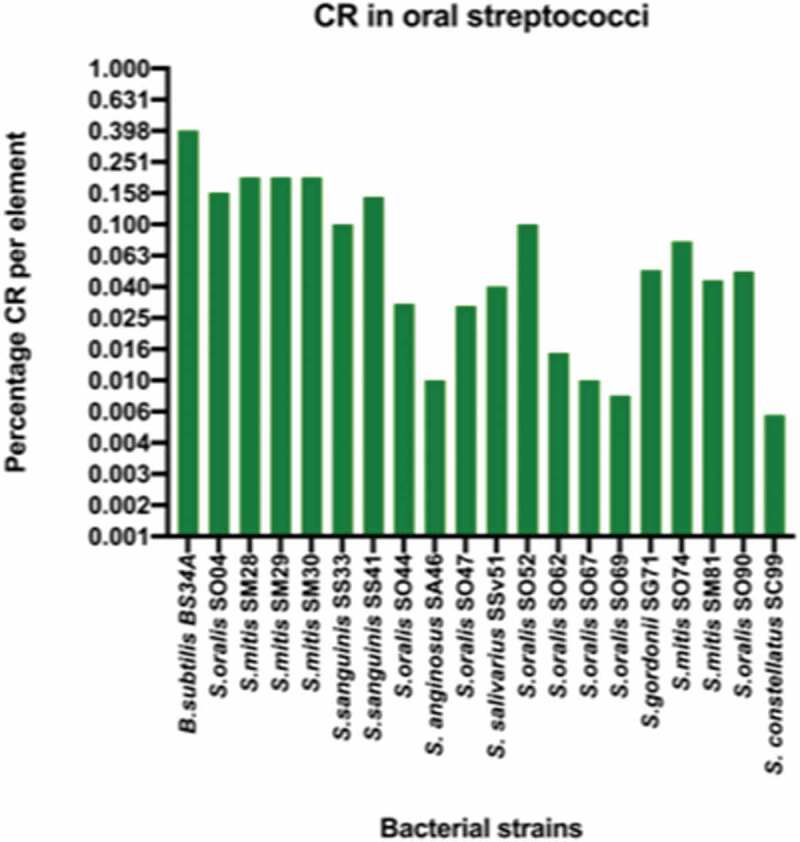
A graph illustrating the percentage of circularization rate (CR) in oral streptococci. B. subtilis BS34A was used as a positive control for the excision of Tn916 and shows the highest levels of CR. The clinical oral streptococci isolates have CR that ranges from zero to 0.25 percentage CR per element
Transferability and conjugation frequency of the Tn916-Tn1545 family in oral streptococci
The in vivo transferability of the Tn916-Tn1545 family was investigated by filter mating experiments in which both laboratory and clinical isolates were used as donor strains. The strain B. subtilis BS49 was included in this study as a control strain for the transfer of Tn916 as this element is known to be transferable with a CR of 9.7% [41], and exists as two copies [45]. To evaluate the transferability of Tn916-Tn1545 elements in the clinical setting, we used clinical isolates harboring the wild type Tn916 as donor strains in the conjugation experiments. Of the 30 filter mating experiments, 28 yielded transconjugants with frequencies that are summarized in Table 4. The bacterial strain B. subtilis BS49 was able to transfer Tn916 to five out of the six recipients. Interestingly, the isolate in which no transfer was detected (S. oralis SO32) is a clinical isolate whereas the other recipients are laboratory strains. In the successful conjugation experiments, the transfer frequency of Tn916 from B. subtilis BS49 ranged from 6.0 (± 4.03) × 10−9 to 1.5 × 10−1 (±0.25). The frequencies of transfer ranged between to 4.0 (± 3.0) × 10−7 to 3.5 (± 6.0) × 10° per recipient when the clinical isolate S. sanguinis Ssg41 harboring Tn916 was used as a donor. The transfer of Tn916 from the clinical isolate S. oralis SO52 was observed to ranged from 8.4 × 10−3 (±0.01) to 4.7 (± 4.1) × 10−1. The transfer frequency of Tn6815 ranged between 7.4(± 9.7 × 10−6 and 5.8 (± 3.6) × 10−2 whereas for Tn6516, the transfer frequencies ranged between below detection in S. oralis SO32 to 1.5 (± 2.7) × 10−1 in S. sanguinis Sg10.
Table 4.
Transfer frequencies of the Tn916-Tn1545 family in oral streptococci per recipients
| Donor (element) | S. oralis SOK10 | S. mitis SMK7 | S. sanguinis Sg10 | S. gordonii G3 | S. mutans M8 | S. oralis SO32 |
|---|---|---|---|---|---|---|
|
Bacillus subtilis BS49; TetR ::Tn916 |
4.7 (± 8.1) × 10−6 |
6.0 (± 4.03) × 10−9 |
2.9 (± 4.9) × 10−5 |
1.5 (±0.25) × 10−1 | 3.2 (± 4.1) × 10−6 |
Not detected |
| S. sanguinis Ssg41; Clinical isolate TetR :: Tn916 | 3.5 (± 6.0) × 10° |
1.7 (± 1.8) × 10−1 |
1.0 (±0.09) × 10−6 |
1.0 (± 1.7) × 10−3 | 3.0 (± 4.6) × 10−6 |
4.0 (± 3.0) × 10−7 |
| S. oralis SO52; Clinical isolate TetR :: Tn916 | 3.1 (± 2.3) × 10−2 |
2.1 (± 1.2) × 10−2 |
6.2 (±0.09) × 10−2 |
4.7 (± 4.1) × 10−1 | 6.2 (±0.09) × 10−2 |
8.4 (±0.01) × 10−3 |
| S. mitis SM28; Clinical isolate TetR ::Tn6815 | 2.9 (± 3.1) × 10−3 |
2.5(± 3.9) × 10−3 |
7.4 (± 9.7) × 10−6 |
1.4 (± 3.5) × 10−3 | 3.3 (± 5.4) × 10−2 |
5.8 (± 3.6) × 10−2 |
| S. constellatus SC99; Clinical isolate :: TetR Tn6816 | 2.2 (± 1.9) × 10−3 |
1.2 (± 2.1) × 10−2 |
1.5 (± 2.7) × 10−1 |
1.0 (± 1.6) × 10−1 |
1.8 (± 2.0) × 10−3 |
Not detected |
TetR – Tetracycline resistant
The random selection of twenty colonies from the resulting transconjugants per filter mating confirmed the presence of both the tet(M) and the Int and Xis genes of Tn916 by conventional PCR. In addition, growth of the transconjugants on BHI agar plates containing both selection makers, i.e. tetracycline and kanamycin, verified the transfer of the Tn916-Tn1545 family to the recipient cells. Sequence analysis of the tet (M) promoter region in all the selected clinical isolates, which is known to influence the rate of transfer, showed nucleotide changes as shown in Figure 4.
Figure 4.
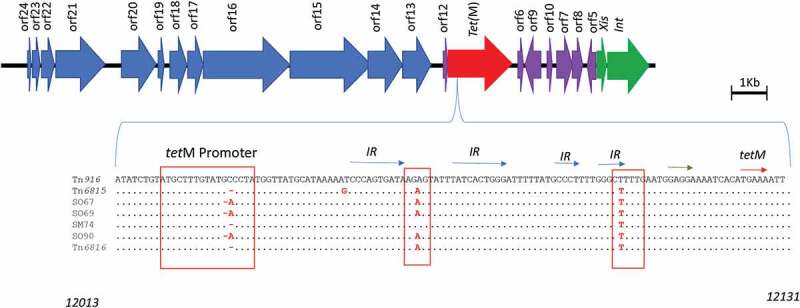
A nucleotide alignment of the promoter region in Tn916 and six oral streptococci clinical isolates highlighting single nucleotide changes in the clinical isolates. The dots indicate nucleotides that are similar to the reference Tn916 (KM615585), whereas the red nucleotides indicate SNPs. The red boxes indicate seemingly conserved SNPs, the blue arrows; inverted repeats (IR), the green arrow; the ribosomal protein binding site, and the red arrow; the start of the tetracycline resistance gene tetM. The numbers in the bottom right and left corners of the image indicate the nucleotide position in the reference E. faecalis DS16 (U09422.1). Tn6815 and Tn6816 are novel transposons that have been isolates from the clinical isolates S. mitis SM28 and S. constellatus SC99
Discussion
Oral streptococci are the most abundant bacterial species in the oral cavity and are regarded as commensal bacteria. Streptococcus species can, however, cause serious infections if they enter the blood stream. There has been an increase in the number of antibiotic resistant oral streptococci thus the aim of this study was to determine the prevalence of the Tn916-Tn1545 family in a collection of oral streptococci from Norway. The presence of tet(M) was detected in 24% the 100 oral streptococcal isolates. This rate of carriage is within the previously reported prevalence of 21% and 29.6% of tet(M) in oral streptococci [46,47]. The above-mentioned studies were from 1984 and 2017 respectively, therefore it is interesting that despite the time span, different geographic locations and changes in usage of antibiotics, the carriage of tet(M) appears to be similar. Of the 100 investigated isolates, the Tn916-Tn1545 family was detected in 21% of the isolates with the highest prevalence per species occurring in S. oralis (41%). S. mitis was the most prevalent species in this study (41%), however only 26.2% of the isolates harbored the Tn916-Tn1545 family. This raises the question of whether the elements have species preference or not, a notation that requires further study. Analysis of the diversity of Tn916-Tn1545 in our study, revealed a total of three different elements with the most occurring being Tn916. The other two elements are new members in the Tn916-Tn1545 family, i.e. Tn6815 and Tn6816. Other studies have documented a diversity of four elements in a collection of 48 oral streptococci where the most frequent occurring was Tn3872 [34].
The presence of both tetracycline and erythromycin resistance genes in Tn6815 and ltrA in Tn6816 is an indication of the role that these elements play in the elasticity of the bacterial genome and HGT. Although the diversity of these elements in our study seems low, it should be noted that based on the inclusion criteria set for further analysis for the Tn916-Tn1545 family excluded the analysis of 14 oral streptococci that tested positive only for the integrase/excisionase regions and not tet (M). It can be speculated that including these samples in this study might have increased the diversity of the detected elements.
Based on the RFLP analysis, the most prevalent form of the Tn916-Tn1545 family observed was the prototype Tn916, which was present in 15 isolates. Interestingly, despite carrying the same element, the 15 isolates had different MIC and lower levels of the CI when compared to the wildtype of B. subtilis BS34. As in many other clinical isolates, the variations in the level of resistance to tetracycline may be attributed to several factors such as the presence of other tetracycline resistance genes in the chromosome and/or the strength of the tet promoter [25]. Analysis of the tet(M) promoter regions in the sequenced isolates (Figure 4) illustrates some of the SNPs that we identified. The fact that these SNPs were observed in nearly all the analyzed clinical isolates, and since it has been hypothetized that their presence is due to beneficial evolutionary advantages requires further analysis and investigations .
Although all but one of the Tn916 like elements found in this study harbored only one resistance gene, the antibiotic resistance profile indicated that five isolates were resistant to erythromycin and that four of the tested isolates were resistant to four of the five tested antibiotics. This may suggest the presence of other MGE or the presence of other resistance genes within the bacterial chromosomes and further highlights the role that oral streptococci play in the spread of antibiotic resistance among bacteria.
The six isolates that were identified as having Tn916-like elements based on the observed difference in the RFLP were further investigated by NGS. The results from the NGS showed that in three of the six isolates (isolates S. oralis SO67, S. mitis SM74 and S. oralis SO90), the observed differences in the RFLP are due to SNPs and/or mutations that result in the introduction of a new HincII digestion target site. According to the sequences of these isolates, they were more than 99% similar to the prototype Tn916. These observations highlighted the need to consider the suitability of the using the RFLP method as a way of distinguishing Tn916 from Tn916-like elements. In the isolate S. oralis SO67, assembly of the genome, suggested the presence of two identical copies of Tn916 in two different chromosomal locations. According to our knowledge, this is the first time that two identical copies of Tn916 have been identified in two different positions in the same chromosome of an oral streptococcal clinical isolate.
S. mitis is one of the most predominant oral Streptococcus species in the oral cavity. The isolate S. mitis SM28 was found to harbor the novel Tn6815 which contained the erm(B) gene inserted downstream to tet(M). The presence of erm(B) and tet(M) genes rendered S. mitis SM28 the only isolate found to have two resistance genes in the same element in this study. Tn6815 harbored two truncated copies of orf9. In spite of the absence of a complete orf9, Tn6815 was able to form CI and excise from the genome suggesting that an alternative mechanism is regulating the excision process, as has been suggested to occur in other Tn916-like elements [23]. The 5,265 bp insertion downstream from the tet(M) gene resulted in an element that was 99% identical to an ICE that has been previously isolated from S. pneumonia [43]. In contrast with the other previously identified elements that carry insertions downstream from the tet(M) gene such as Tn2009 and Tn3872 [19], the Tn6815 in S. mitis SM28 was transferable to other oral streptococcal strains (Table 4). In addition to carrying both the tet(M) and the erm(B) genes, S. mitis SM28 was also shown to be resistant to penicillin, which is usually the first drug of choice for oral streptococcal infections. The ability of Tn6815 to transfer to other species, further supported previous findings that oral streptococci play an active role in the dissemination of resistance genes by being reservoirs of antibiotic resistant genes.
The formation of the CI is the first step in the transfer of the Tn916-Tn1545 elements, hence the presence of the CI and CR may indicate the functionality of the ICE to disseminate associated genes, including resistance genes. To show that the element is transferable, we conducted in vivo inter- and intra-species conjugation assays. Interestingly, we were able to show both inter- and intra-species transfer of Tn916-Tn1545 elements with transfer frequencies ranging from 6.0 (± 4.03) × 10−9 to 3.5 (± 6.0) × 10° (Table 4). Conjugation frequencies of Tn916 to S. gordonii, S. salivarius and S. sanguinis have previously been reported to range from 10−8 to 10−5 [48,49]. Although some of the observed conjugation frequencies reported in this study were comparable to those previously reported, 20/28 of the observed conjugation frequencies were considered higher than what has been previously reported [48,49]. Interestingly all the higher observed conjugation frequencies where observed when clinical isolates where used as donor suggesting that clinical isolates may be more efficient in spreading Tn916 despite having a lower CR. In spite of the bacterial isolate B. subtilis BS49 having two copies of Tn916 and having higher CR than the clinical oral streptococci in this study, it was observed that the B. subtilis BS49 yielded the lowest number of transconjugants compared to the studied oral streptococci. In this study, we observed varying transfer frequencies of the Tn916-Tn1545 elements within the same recipients when different donors were used in the filter mating and vice versa. These findings suggested that other factors than the donor, recipient, number of elements and/or the number of CI influence the rate at which the Tn916-Tn1545 family elements are transferred within and between species.
The clinical oral Streptococcus isolates were all found to have a low CR. Sequence analysis of the tet(M) promoter region in all the selected clinical isolates revealed the presence of some SNPs (Figure 4). As this region has been shown to influence the transcription and transfer of the element, the observed SNPs might be responsible for the observed low levels of CR.
The presence of the Tn916-Tn1545 family in commensal oral streptococci raises concern as it highlights the role that these abundant species may play in the increasing rates of antibiotic resistance. In this study, 21% of the investigated oral streptococci have been found to harbor a diverse elements of the Tn916-Tn1545 family, with the highest prevalence occurring among S. mitis isolates. One of these S. mitis isolates was found to harbor a new element, i.e. Tn6815 that had two resistance genes, i.e tet(M) and erm(B). Tn6815 is transferable to other species with a relatively high transfer frequency. These findings underscore the role of S. mitis plays in the spread of antibiotic resistance within and across bacterial species. There is a need to further investigate the factors that contribute to the stability of the Tn916-Tn1545 elements in oral streptococci.
Supplementary Material
Acknowledgments
The current study was supported by the Department of Clinical Dentistry (IKO), UiT The Arctic University of Norway. Our gratitude goes to the National Advisory Unit for Detection of Antimicrobial Resistance (K-RES), University Hospital of North Norway for providing the majority of the strains used in this study, and to Bjørg Christina Haldorsen, Berit Tømmerås and Ida Sofie Furuholmen for the excellent technical support. The publication charges for this article have been funded by a grant from the publication fund of UiT The Arctic University of Norway.
Disclosure statement
The authors have no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].John R. Tagg, Philip A. Wescombe, John D. F. Hale, Jeremy P. Burton. (2019). Chapter 7 Streptococcus. In: Lactic Acid Bacteria: Microbiological and Functional Aspects (5th ed.). CRC Press (Boca Raton). 10.1201/9780429057465. [DOI] [Google Scholar]
- [3].Bruckner L, Gigliotti F. Viridans group streptococcal infections among children with cancer and the importance of emerging antibiotic resistance. Semin Pediatr Infect Dis.2006;17:153-60. [DOI] [PubMed] [Google Scholar]
- [4].Shenep JL. Viridans-group streptococcal infections in immunocompromised hosts. Int J Antimicrob Agents. 2000;14(2):129–12. [DOI] [PubMed] [Google Scholar]
- [5].Julius A, Subbiah U, Elango S.. Designing universal primer for the identification of erythromycin and tetracycline resistance genes in oral Streptococci. Indian J Public Health Res Dev. 2019;10(11):2838. [Google Scholar]
- [6].Kim S-M, Kim HC, Lee S-WS. Characterization of antibiotic resistance determinants in oral biofilms. J Microbiol. 2011;49(4):595–602. [DOI] [PubMed] [Google Scholar]
- [7].Roberts AP, Mullany P. Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev Anti Infect Ther. 2010;8(12):1441–1450. [DOI] [PubMed] [Google Scholar]
- [8].Roberts AP, Mullany P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):856–871. [DOI] [PubMed] [Google Scholar]
- [9].Seville LA, Patterson AJ, Scott KP, et al. Distribution of tetracycline and erythromycin resistance genes among human oral and fecal metagenomic DNA. Microbial Drug Resist. 2009;15(3):159–166. [DOI] [PubMed] [Google Scholar]
- [10].Sukumar S, Roberts AP, Martin FE, et al. Metagenomic insights into transferable antibiotic resistance in oral bacteria. J Dent Res. 2016;95(9):969–976. [DOI] [PubMed] [Google Scholar]
- [11].Richards VP, Palmer SR, Pavinski Bitar PD, et al. Phylogenomics and the dynamic genome evolution of the genus Streptococcus. Genome Biol Evol. 2014;6(4):741–753. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155(5):376–386. [DOI] [PubMed] [Google Scholar]
- [13].Osborn AM, Böltner D. When phage, plasmids, and transposons collide: genomic islands, and conjugative-and mobilizable-transposons as a mosaic continuum. Plasmid. 2002;48(3):202–212. [DOI] [PubMed] [Google Scholar]
- [14].Salyers AA, Shoemaker NB, Stevens AM, et al. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59(4):579–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brouwer MS, Mullany P, Roberts AP. Characterization of the conjugative transposon Tn 6000 from Enterococcus casseliflavus 664.1 H1 (formerly Enterococcus faecium 664.1 H1). FEMS Microbiol Lett. 2010;309(1):71–76. [DOI] [PubMed] [Google Scholar]
- [16].Roberts AP, Davis IJ, Seville L, et al. Characterization of the ends and target site of a novel tetracycline resistance-encoding conjugative transposon from Enterococcus faecium 664.1 H1. J Bacteriol. 2006;188(12):4356–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lancaster H, Roberts AP, Bedi R, et al. Characterization of Tn916S, a Tn916-like element containing the tetracycline resistance determinant tet (S). J Bacteriol. 2004;186(13):4395–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McDougal LK, Tenover FC, Lee LN, et al. Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42(9):2312–2318. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Del Grosso M, d’Abusco AS, Iannelli F, et al. Tn2009, a Tn916-like element containing mef (E) in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2004;48(6):2037–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Del Grosso M, Camilli R, Libisch B, et al. New composite genetic element of the Tn916 family with dual macrolide resistance genes in a Streptococcus pneumoniae isolate belonging to clonal complex 271. Antimicrob Agents Chemother. 2009;53(3):1293–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Franke AE, Clewell D. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of” conjugal” transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145(1):494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ciric L, Mullany P, Roberts AP. Antibiotic and antiseptic resistance genes are linked on a novel mobile genetic element: tn6087. J Antimicrob Chemother. 2011;66(10):2235–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Reynolds LJ, Anjum MF, Roberts AP. Detection of a novel, and likely ancestral, Tn916-like element from a human saliva metagenomic library. Genes (Basel). 2020;11(5):548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17(6):251–258. [DOI] [PubMed] [Google Scholar]
- [25].Su Y, He P, Clewell D. Characterization of the tet (M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob Agents Chemother. 1992;36(4):769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].2018 NN-V . Usage of antimicrobial agents a nd occurrence of antimicrobial resistance in Norway. Tromso/Oslo; 2018. Report No.: 1502–2307 [Google Scholar]
- [27].Almeida V, Azevedo J, Leal HF, et al. Bacterial diversity and prevalence of antibiotic resistance genes in the oral microbiome. Plos One. 2020;15(9):e0239664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Christensen TM, Sørensen TK. Presence and levels of antibiotic resistance genes in saliva from dental students in Tromsø. Investigation of cfxA and erm (B) in Saliva Samples. UiT Norges arktiske universitet; 2016. [Google Scholar]
- [29].Davidovich NV, Galieva A, Davydova N, et al. Spectrum and resistance determinants of oral streptococci clinical isolates. Klin Lab Diagn. 2020;65(10):632–637. . [DOI] [PubMed] [Google Scholar]
- [30].Smith HE, Wisselink HJ, Vecht U, et al. High-efficiency transformation and gene inactivation in Streptococcus suis type 2. Microbiology. 1995;141(1):181–188. [DOI] [PubMed] [Google Scholar]
- [31].Bertrand S, Huys G, Yde M, et al. Detection and characterization of tet (M) in tetracycline-resistant Listeria strains from human and food-processing origins in Belgium and France. J Med Microbiol. 2005;54(12):1151–1156. . [DOI] [PubMed] [Google Scholar]
- [32].Doherty N, Trzcinski K, Pickerill P, et al. Genetic diversity of the tet (M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob Agents Chemother. 2000;44(11):2979–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Guardabassi L, Dijkshoorn L, Collard J-M, et al. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J Med Microbiol. 2000;49(10):929–936. [DOI] [PubMed] [Google Scholar]
- [34].Ciric L, Ellatif M, Sharma P, et al. Tn916-like elements from human, oral, commensal streptococci possess a variety of antibiotic and antiseptic resistance genes. Int J Antimicrob Agents. 2012;39(4):360. . [DOI] [PubMed] [Google Scholar]
- [35].Salvà-Serra F, Svensson-Stadler L, Busquets A, et al. A protocol for extraction and purification of high-quality and quantity bacterial DNA applicable for genome sequencing: a modified version of the Marmur procedure. 2018.
- [36].Koren S, Walenz BP, Berlin K, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Assefa S, Keane TM, Otto TD, et al. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics. 2009;25(15):1968–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. [DOI] [PubMed] [Google Scholar]
- [39].Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Manganelli R, Romano L, Ricci S, et al. Dosage of Tn916 circular intermediates in Enterococcus faecalis. Plasmid. 1995;34(1):48–57. [DOI] [PubMed] [Google Scholar]
- [41].Lunde TM, Roberts AP, Al-Haroni M. Determination of copy number and circularization ratio of Tn 916-Tn 1545 family of conjugative transposons in oral streptococci by droplet digital PCR. J Oral Microbiol. 2019;11(1):1552060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Roberts AP, Mullany P, Wilson M. Gene transfer in bacterial biofilms. Methods Enzymol.2001;336:60-5. [DOI] [PubMed] [Google Scholar]
- [43].Croucher NJ, Hanage WP, Harris SR, et al. Variable recombination dynamics during the emergence, transmission and ‘disarming’of a multidrug-resistant pneumococcal clone. BMC Biol. 2014;12(1):49. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tansirichaiya S, Rahman MA, Roberts AP. The transposon registry. Mob DNA. 2019;10(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Browne HP, Anvar SY, Frank J, et al. Complete genome sequence of BS49 and draft genome sequence of BS34A, Bacillus subtilis strains carrying Tn916. FEMS Microbiol Lett. 2015;362(3):1–4. [DOI] [PubMed] [Google Scholar]
- [46].Horaud T, Delbos F. Viridans streptococci in infective endocarditis: species distribution and susceptibility to antibiotics. Eur Heart J. 1984;5(suppl_C):39–44. [DOI] [PubMed] [Google Scholar]
- [47].Sun J-Q, Li L, Zhao K, et al. Molecular epidemiology of tetracycline resistance among viridians group streptococci isolated from various clinical specimens. 2017.
- [48].Roberts AP, Cheah G, Ready D, et al. Transfer of Tn916-like elements in microcosm dental plaques. Antimicrob Agents Chemother. 2001;45(10):2943–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fitzgerald GF, Clewell D. A conjugative transposon (Tn919) in Streptococcus sanguis. Infect Immun. 1985;47(2):415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Marra D, Scott JR. Regulation of excision of the conjugative transposon Tn916. Mol Microbiol. 1999;31(2):609–621. [DOI] [PubMed] [Google Scholar]
- [51].Wang H, Roberts AP, Mullany P. DNA sequence of the insertional hot spot of Tn916 in the Clostridium difficile genome and discovery of a Tn916-like element in an environmental isolate integrated in the same hot spot. FEMS Microbiol Lett. 2000;192(1):15–20. [DOI] [PubMed] [Google Scholar]
- [52].Poyart C, Quesne G, Acar P, et al. Characterization of the Tn916-like transposon Tn3872 in a strain of Abiotrophia defectiva (Streptococcus defectivus) causing sequential episodes of endocarditis in a child. Antimicrob Agents Chemother. 2000;44(3):790–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- [54].Manganelli R, Ricci S, Pozzi G. Conjugative transposon Tn916: evidence for excision with formation of 5ʹ-protruding termini. J Bacteriol. 1996;178(19):5813–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Roberts A, Browne H, Anvar S, et al. Complete genome sequence of BS49 and draft genome sequence of BS34A, Bacillus subtilis strains carrying Tn916. FEMS Microbiol Lett. 2014;362(3):1–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


