ABSTRACT
Genomic aberrations are frequently found in anaplastic thyroid cancer (ATC). However, the functional genes in aberrantly genomic regions are largely unclear. In this study, we identified a long noncoding RNA (lncRNA) HOTAIRM1, whose encoding gene was amplified and expression was upregulated in ATC compared with papillary thyroid cancer and normal thyroid. Increased genomic copy number and expression of HOTAIRM1 were both correlated with poor survival of ATC patients. Functional assays revealed that HOTAIRM1 promoted proliferation, inhibited apoptosis, and promoted migration and invasion of ATC cells in vitro, and promoted ATC tumour growth and metastasis in vivo. HOTAIRM1 was found to bind ILF3, repress the binding between ILF3 and precursor miR-144 (pre-miR-144), block the effects of ILF3 on stabilizing pre-miR-144, and therefore downregulate pre-miR-144. Intriguingly, HOTAIRM1 was also found to directly bind primary miR-144 (pri-miR-144), repress the binding between pri-miR-144 and DROSHA, block the processing of pri-miR-144 by DROSHA, and therefore upregulate pri-miR-144 and downregulate pre-miR-144. Thus, HOTAIRM1 remarkably downregulated pre-miR-144 and further downregulated miR-144. Knockdown of ILF3 and DROSHA abolished the effects of HOTAIRM1 on pre-miR-144 and miR-144. The expression of miR-144 was downregulated and reversely correlated with HOTAIRM1 in ATC. Via repressing miR-144 biogenesis, HOTAIRM1 upregulated MET and activated AKT signalling. miR-144 overexpression reversed the oncogenic roles of HOTAIRM1 in ATC. Altogether, these findings identified a genomic copy number amplified and highly expressed lncRNA HOTAIRM1, which exerted oncogenic roles via repressing miR-144 biogenesis in ATC. Our data suggested HOTAIRM1 as a potential prognostic biomarker and therapeutic target for ATC.
KEYWORDS: Anaplastic thyroid cancer, copy number variation, long noncoding RNA, progression, microRNA, biogenesis
Introduction
Thyroid cancer is the most common endocrine malignancy [1]. According to 2020 cancer statistics data in USA, the incidence of thyroid cancer ranks the tenth in both sexes and the fifth in female [2]. Based on the histopathological characteristics, well-differentiated papillary thyroid cancer (PTC) is the major subtype of thyroid cancer [1]. However, most thyroid cancer related deaths are caused by anaplastic thyroid cancer (ATC), which is undifferentiated and the most aggressive subtype of thyroid cancer [3]. Although intensive and multimodal therapeutic approaches including surgery, systemic chemotherapy, radiotherapy, molecule-targeted treatment, and immunotherapy have been used to fight against ATC, the prognosis of ATC is still very poor with overall survival of less than one year after diagnosis [3,4,5]. Therefore, it is urgent to understand the underlying molecular mechanisms driving the initiation and progression of ATC.
Recent genomic and transcriptomic analyses of ATC have identified more molecular aberrations compared with those in PTC [6,7]. In addition to the co-mutated genes with PTC, such as BRAF, RAS, AKT1 and TERT, several genes were identified to be predominant in ATC, such as the mutation of TP53 and ATM, the loss of CDKN2A, the amplification of TWIST1, CCNE1 and CDK6 [6,8]. Genomic copy number variations in ATC are also more common than those in PTC [6,8]. Different studies have identified various genomic copy number aberrations, including the loss of 8p, 9p21.3, q36.1, the amplification of 8q, 19q12, 19q13, q21.2, and so on [6,8]. However, the protein-encoding genes localized in these aberrant copy number regions are mostly cancer-unrelated. The potential cancer-related genes in these regions need to be further investigated.
Genomic and transcriptomic high throughput sequencings have found that less than 2% of human genome encode for proteins, with about 21,000 protein-encoding genes [9]. The number of genes encoding long noncoding RNAs (lncRNAs) is about 58,000 [10]. Therefore, we hypothesized that these aberrant genomic copy number regions in ATC may contain cancer-related lncRNAs. lncRNA is a class of novel transcripts with more than 200 nucleotides in length and limited protein-coding potential [11]. Many lncRNAs have been identified as cancer-related [12,13,14]. In addition to the aberrant expressions of various lncRNAs in different types of cancers, many lncRNAs have been also demonstrated to play critical regulatory roles in cancer growth, metastasis, drug-resistance, and so on [15,16,17,18].
microRNA (miRNA) is a class of small non-coding RNA with 19–25 nucleotides in length [19]. Aberrant expressions and functions of miRNAs have been intensively investigated in a variety of cancers [20,21]. Similar to lncRNAs, miRNAs have been also demonstrated to play important roles in various cancer cell biological behaviours [22,23,24]. The biogenesis of miRNAs is a multiple-step process [25,26,27]. The long primary transcripts (pri-miRNAs) are transcribed from genomic DNA. In the nucleus, the pri-miRNAs are processed by DROSHA to generate precursor miRNAs (pre-miRNAs). The pre-miRNAs are exported to the cytoplasm, where they are processed by DICER to generate mature miRNAs.
Via analysing the previously reported lncRNA expression microarray results and genomic sequencing results of ATC tissues [8,28], we identified an lncRNA HOTAIRM1 whose expression was upregulated in ATC. HOTAIRM1 is localized in chromosome 7p15.2, which is a copy number amplified region in ATC. In this study, we further measured the expression levels and genomic copy number of HOTAIRM1 in ATC. In addition, we found that HOTAIRM1 drove ATC progression via repressing miR-144 biogenesis.
Results
The expression of HOTAIRM1 is upregulated and the genomic copy number of HOTAIRM1 is amplified in ATC
To investigate the expression levels and genomic copy number of HOTAIRM1 in ATC, we collected 20 normal thyroid tissues, 20 PTC tissues, and 20 ATC tissues. HOTAIRM1 expression was measured by RT-qPCR and the results revealed that the expression of HOTAIRM1 was markedly increased in ATC tissues compared with normal thyroid and PTC tissues (Fig. 1A). Next, the genomic copy number of HOTAIRM1 was measured in these normal thyroid, PTC, and ATC tissues by qPCR. Consistent with HOTAIRM1 expression, the genomic copy number of HOTAIRM1 was markedly increased in ATC tissues compared with normal thyroid and PTC tissues (Fig. 1B). Furthermore, the expression of HOTAIRM1 was significantly positively correlated with the genomic copy number of HOTAIRM1 in ATC tissues (Fig. 1C), suggesting that the genomic amplification of HOTAIRM1 contributes to the upregulation of HOTAIRM1 expression in ATC. Kaplan-Meier survival analysis of the correlation between HOTAIRM1 expression, HOTAIRM1 genomic copy number and survival of ATC patients revealed that higher expression of HOTAIRM1 and higher genomic copy number of HOTAIRM1 were both correlated with poor survival (Fig. 1D and E). The expressions of HOTAIRM1 in normal thyroid follicular epithelial cell (Nthy-ori 3–1), PTC cells (TPC-1 and HTH83), and ATC cells (8505 C, C643, and FRO) were measured by RT-qPCR and the results revealed that HOTAIRM1 was also markedly increased in ATC cells compared with normal thyroid follicular epithelial cell and PTC cells (Fig. 1F). In accordance with HOTAIRM1 expression, the genomic copy number of HOTAIRM1 was increased in ATC cells compared with normal thyroid follicular epithelial cell and PTC cells (Supplementary Fig. 1A).
Figure 1.
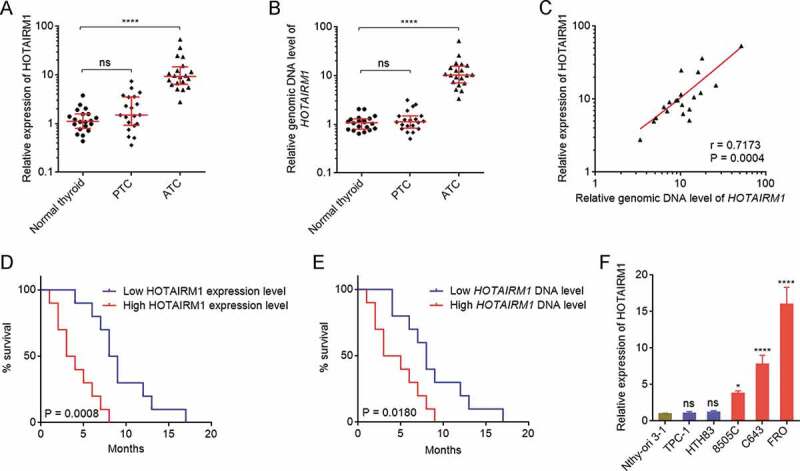
The expression and genomic copy number of HOTAIRM1 in ATC. (A) HOTAIRM1 expressions in 20 normal thyroid tissues, 20 PTC tissues, and 20 ATC tissues were measured by RT-qPCR. GAPDH was used as endogenous control. Three randomly selected thyroid tissues were used as the reference samples to calculate the relative expressions. (B) Genomic copy numbers of HOTAIRM1 in 20 normal thyroid tissues, 20 PTC tissues, and 20 ATC tissues were measured by qPCR. LINE1 was used as endogenous control. Three randomly selected thyroid tissues were used as the reference samples to calculate the relative genomic copy numbers. For A-B, data were shown as median with interquartile range. ns, not significant, ****P < 0.0001 by Kruskal-Wallis test, followed by Dunn’s multiple comparisons test. (C) The correlation between HOTAIRM1 expression and genomic copy number of HOTAIRM1 in 20 ATC tissues. r = 0.7173, P = 0.0004 by Spearman correlation analysis. (D) Kaplan-Meier survival analyses of the correlation between HOTAIRM1 expression level and overall survival of ATC patients. n = 20, P = 0.0008 by log-rank test. HOTAIRM1 median expression level was used as cut-off. (E) Kaplan-Meier survival analyses of the correlation between HOTAIRM1 genomic copy number level and overall survival of ATC patients. n = 20, P = 0.0180 by log-rank test. HOTAIRM1 median copy number level was used as cut-off. (F) HOTAIRM1 expressions in normal thyroid follicular epithelial cell (Nthy-ori 3–1), PTC cells (TPC-1 and HTH83), and ATC cells (8505 C, C643, and FRO) were measured by RT-qPCR. GAPDH was used as endogenous control. Data were shown as mean ± standard deviation of three independent experiments. ns, not significant, *P < 0.05, ****P < 0.0001 by one-way ANOVA, followed by Dunnett’s multiple comparisons test
HOTAIRM1 has four different isoforms (Supplementary Fig. 1B). RT-qPCR results revealed that the isoform HOTAIRM1-204 which lacks exon 2 was the major isoform in ATC tissues and ATC cells (Supplementary Fig. 1 C and D). Therefore, in this study we focused on HOTAIRM1-204. The coding potential of HOTAIRM1 was calculated using three in silico tools, namely coding potential assessment tool (CPAT), coding potential calculator (CPC), and PhyloCSF. The CPAT score of HOTAIRM1 was 0.0046758, and the CPC score of HOTAIRM1 was −0.745026. These low scores suggested that HOTAIRM1 was a noncoding RNA. PhyloCSF tracks from UCSC (http://genome.ucsc.edu/) showed no regions with high codon conservation, which further supported the noncoding nature of HOTAIRM1. Subcellular fractions revealed that HOTAIRM1 was mainly localized in the nucleus of ATC cells (Supplementary Fig. 1E).
HOTAIRM1 promotes proliferation, migration, and invasion, and represses apoptosis of ATC cells
Given the increased expression of HOTAIRM1 in ATC, we next investigated the potential roles of HOTAIRM1 in ATC. We overexpressed HOTAIRM1 in 8505 C and C643 cells via stable infection of HOTAIRM1 overexpressing lentivirus (Fig. 2A and B). Cell Counting Kit-8 (CCK-8) assays revealed that 8505 C and C643 cells overexpressing HOTAIRM1 had faster cell proliferation rates than control 8505 C and C643 cells (Fig. 2C and D). 5-ethynyl-2ʹ-deoxyuridine (EdU) incorporation assays revealed that 8505 C and C643 cells overexpressing HOTAIRM1 had more EdU positive cells than control 8505 C and C643 cells (Fig. 2E), supporting that overexpression of HOTAIRM1 promotes proliferation of ATC cells. Annexin V-PI staining and flow cytometry revealed that 8505 C and C643 cells overexpressing HOTAIRM1 had less apoptotic cells than control 8505 C and C643 cells (Fig. 2F). Transwell migration assays revealed that 8505 C and C643 cells overexpressing HOTAIRM1 had stronger migratory ability than control 8505 C and C643 cells (Fig. 2G). Transwell invasion assays revealed that 8505 C and C643 cells overexpressing HOTAIRM1 had stronger invasive ability than control 8505 C and C643 cells (Fig. 2H). In summary, these findings demonstrated that overexpression of HOTAIRM1 promotes proliferation, inhibits apoptosis, and promotes migration and invasion of ATC cells.
Figure 2.
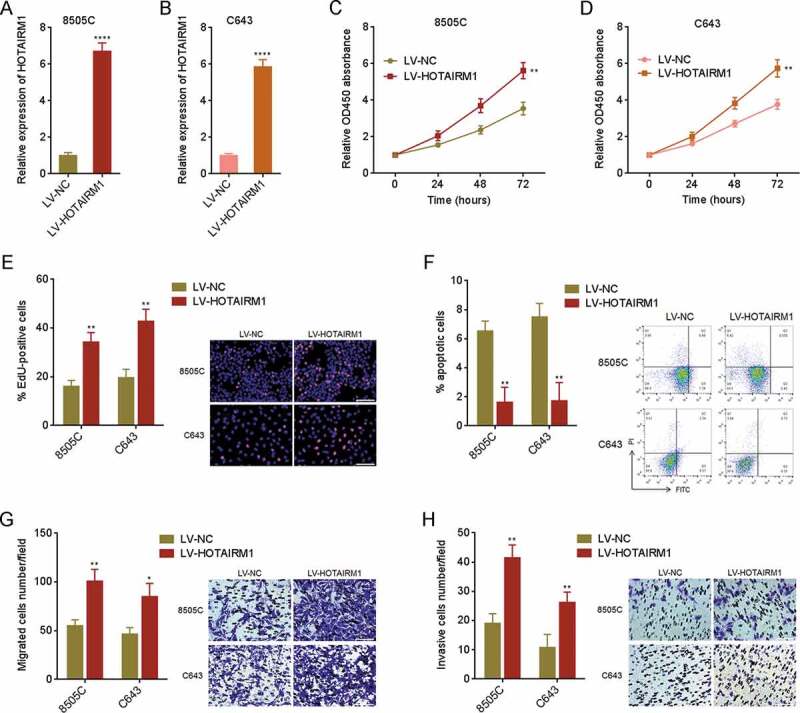
The roles of HOTAIRM1 in proliferation, apoptosis, migration, and invasion of ATC cells. (A) HOTAIRM1 expressions in HOTAIRM1 overexpressed and control 8505 C cells were measured by RT-qPCR. GAPDH was used as endogenous control. (B) HOTAIRM1 expressions in HOTAIRM1 overexpressed and control C643 cells were measured by RT-qPCR. GAPDH was used as endogenous control. (C) Cell proliferation of HOTAIRM1 overexpressed and control 8505 C cells were measured by CCK-8 assays. Absorbance values at 450 nm were measured to track cell proliferation. (D) Cell proliferation of HOTAIRM1 overexpressed and control C643 cells were measured by CCK-8 assays. Absorbance values at 450 nm were measured to track cell proliferation. (E) Cell proliferation of HOTAIRM1 overexpressed and control 8505 C and C643 cells were measured by EdU incorporation assays. Red colour indicates EdU positive and proliferative cells. Scale bar = 100 μm. (F) Cell apoptosis of HOTAIRM1 overexpressed and control 8505 C and C643 cells were measured by Annexin-V-FITC/PI staining, followed by flow cytometry. (G) Cell migration of HOTAIRM1 overexpressed and control 8505 C and C643 cells were measured by transwell migration assays. Scale bar = 100 μm. (H) Cell invasion of HOTAIRM1 overexpressed and control 8505 C and C643 cells were measured by transwell invasion assays. Scale bar = 100 μm. Data were shown as mean ± standard deviation of three independent experiments. *P < 0.05, **P < 0.01, ****P < 0.0001 by Student’s t-test
Knockdown of HOTAIRM1 inhibits proliferation, migration, and invasion, and promotes apoptosis of ATC cells
To further investigate the influence of HOTAIRM1 on proliferation, apoptosis, migration, and invasion of ATC cells, HOTAIRM1 was knocked down in FRO cells via stable infection of two independent HOTAIRM1 interfering lentiviruses (Fig. 3A). CCK-8 assays revealed that FRO cells with HOTAIRM1 knockdown had slower cell proliferation rates than control FRO cells (Fig. 3B). EdU incorporation assays revealed that FRO cells with HOTAIRM1 knockdown had less EdU positive cells than control FRO cells (Fig. 3C), supporting that knockdown of HOTAIRM1 inhibits proliferation of ATC cells. Annexin V-PI staining and flow cytometry revealed that FRO cells with HOTAIRM1 knockdown had more apoptotic cells than control FRO cells (Fig. 3D). Transwell migration assays revealed that FRO cells with HOTAIRM1 knockdown had weaker migratory ability than control FRO cells (Fig. 3E). Transwell invasion assays revealed that FRO cells with HOTAIRM1 knockdown had weaker invasive ability than control FRO cells (Fig. 3F). In summary, these findings demonstrated that knockdown of HOTAIRM1 inhibits proliferation, induces apoptosis, and inhibits migration and invasion of ATC cells.
Figure 3.
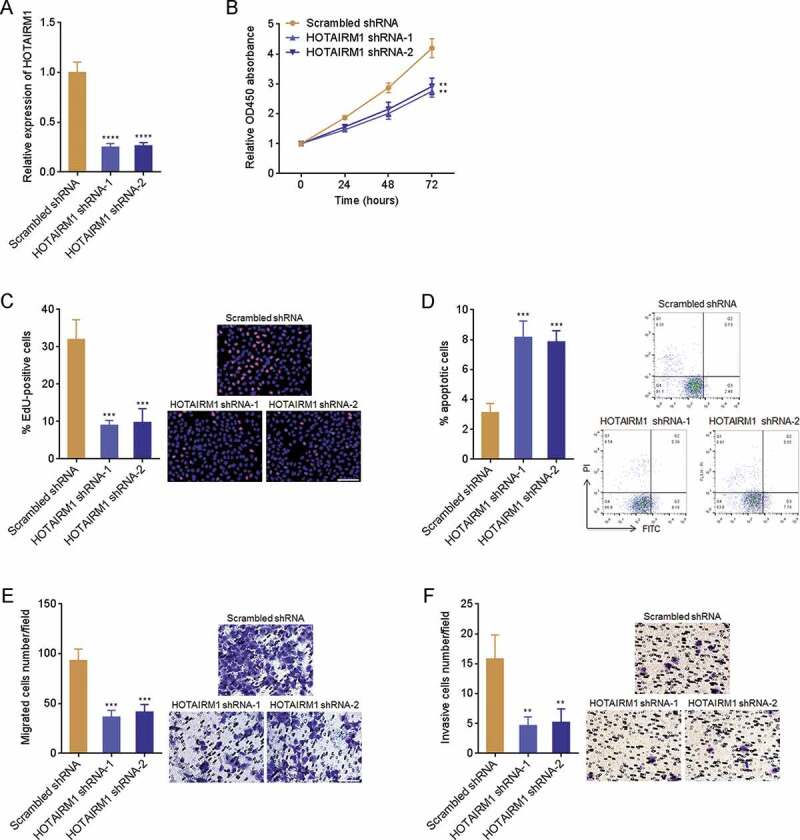
The roles of HOTAIRM1 knockdown in proliferation, apoptosis, migration, and invasion of ATC cells. (A) HOTAIRM1 expressions in HOTAIRM1 depleted and control FRO cells were measured by RT-qPCR. GAPDH was used endogenous control. (B) Cell proliferation of HOTAIRM1 depleted and control FRO cells were measured by CCK-8 assays. Absorbance values at 450 nm were measured to track cell proliferation. (C) Cell proliferation of HOTAIRM1 depleted and control FRO cells were measured by EdU incorporation assays. Red colour indicates EdU positive and proliferative cells. Scale bar = 100 μm. (D) Cell apoptosis of HOTAIRM1 depleted and control FRO cells were measured by Annexin-V-FITC/PI staining, followed by flow cytometry. (E) Cell migration of HOTAIRM1 depleted and control FRO cells were measured by transwell migration assays. Scale bar = 100 μm. (F) Cell invasion of HOTAIRM1 depleted and control FRO cells were measured by transwell invasion assays. Scale bar = 100 μm. Data were shown as mean ± standard deviation of three independent experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001 by one-way ANOVA, followed by Dunnett’s multiple comparisons test
HOTAIRM1 promotes growth and metastasis of ATC in vivo
Given the roles of HOTAIRM1 in proliferation, apoptosis, migration, and invasion of ATC cells in vitro, we further investigated the potential roles of HOTAIRM1 in ATC in vivo. 8505 C cells overexpressing HOTAIRM1 or control were subcutaneously inoculated into nude mice. The tumours derived from 8505 C cells overexpressing HOTAIRM1 showed faster growth and formed larger tumours than those derived from control 8505 C cells (Fig. 4A-C). In addition, FRO cells with HOTAIRM1 knockdown or control were subcutaneously inoculated into nude mice. The tumours derived from HOTAIRM1 depleted FRO cells showed slower growth and formed smaller tumours than those derived from control FRO cells (Fig. 4D-F). To elucidate the influence of HOTAIRM1 on ATC metastasis, luciferase-labelled 8505 C cells overexpressing HOTAIRM1 or control were injected into tail vein of nude mice. Lung metastasis was tracked by bioluminescence imaging. As shown in Fig. 4G, 8505 C cells overexpressing HOTAIRM1 formed more lung metastases than control 8505 C cells. Luciferase-labelled HOTAIRM1 depleted or control FRO cells were injected into tail vein of nude mice, followed by bioluminescence imaging tracking. The bioluminescence imaging results revealed that HOTAIRM1 depleted FRO cells formed less lung metastases than control FRO cells (Fig. 4H). In summary, these findings demonstrated that HOTAIRM1 promotes growth and metastasis of ATC in vivo.
Figure 4.
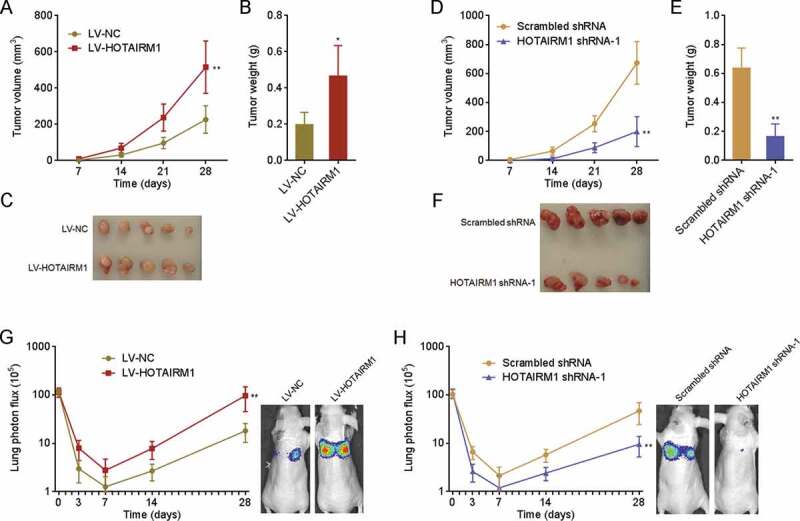
The roles of HOTAIRM1 in ATC growth and metastasis in vivo. (A-C) HOTAIRM1 overexpressed or control 8505 C cells were subcutaneously inoculated into nude mice. Subcutaneous tumours volumes were measured every 7 days (A). At the 28th day after inoculation, subcutaneous tumours were resected, weighed (B), and photographed (C). (D-F) HOTAIRM1 depleted or control FRO cells were subcutaneously inoculated into nude mice. Subcutaneous tumours volumes were measured every 7 days (D). At the 28th day after inoculation, subcutaneous tumours were resected, weighed (E), and photographed (F). (G) Luciferase-labelled HOTAIRM1 overexpressed or control 8505 C cells were injected into tail vein of nude mice, followed by bioluminescence imaging to track lung metastasis. Represent images of mice at the 28th day after injection were shown. (H) Luciferase-labelled HOTAIRM1 depleted or control FRO cells were injected into tail vein of nude mice, followed by bioluminescence imaging to track lung metastasis. Represent images of mice at the 28th day after injection were shown. Data were shown as mean ± standard deviation of five mice in each group. *P < 0.05, **P < 0.01 by Mann-Whitney test
HOTAIRM1 downregulates pre-miR-144 and mature miR-144 via binding ILF3
Given the oncogenic roles of HOTAIRM1 in ATC in vitro and in vivo, we next investigated the molecular mechanisms underlying the oncogenic roles of HOTAIRM1 in ATC. One of the major mechanisms of lncRNAs frequently reported is to bind proteins [29]. Therefore, we searched the potential proteins bound to HOTAIRM1 using in silico tool RNA-Protein Interaction Prediction (RPISeq) (http://pridb.gdcb.iastate.edu/RPISeq/). Notably, we focused on ILF3, which had a high prediction score of 0.97. ILF3 is a well-known nucleus-localized double strand RNA binding protein [30]. The predominantly nuclear localization of HOTAIRM1 in ATC cells supported the potential interaction between HOTAIRM1 and ILF3 (Supplementary Fig. 1E). RNA pull-down assays using in vitro transcribed biotin-labelled HOTAIRM1 identified the significant enrichment of ILF3 in HOTAIRM1 group (Fig. 5A). UV crosslinking RNA immunoprecipitation (RIP) assays with ILF3 antibody identified the specific enrichment of HOTAIRM1 in ILF3 group (Fig. 5B). Therefore, the results of RNA pull-down and RIP assays suggested that HOTAIRM1 interacts with ILF3. ILF3 was reported to bind pre-miR-144, increase pre-miR-144 stability, and therefore upregulate the expressions of pre-miR-144 and mature miR-144 in human chronic myelogenous leukaemia cells [31]. miR-144 has tumour suppressive roles in several cancers [32,33]. More importantly, our previously findings have demonstrated that miR-144 also functions as a tumour suppressor in ATC [34,35]. Therefore, we further explored whether HOTAIRM1 modulated miR-144 via binding ILF3. RIP assays found that ILF3 also bound pre-miR-144 in ATC cells (Fig. 5C). Furthermore, RIP assays revealed that ILF3 bound less pre-miR-144 in HOTAIRM1 overexpressed 8505 C cells than control 8505 C cells (Fig. 5D). RIP assays also revealed that ILF3 bound more pre-miR-144 in HOTAIRM1 depleted FRO cells than control FRO cells (Fig. 5E). These data demonstrated that HOTAIRM1 represses the binding between ILF3 and pre-miR-144. In accordance with the influence of HOTAIRM1 on the binding between ILF3 and pre-miR-144, pre-miR-144 and mature miR-144 were markedly downregulated in 8505 C cells overexpressing HOTAIRM1 compared with control 8505 C cells (Fig. 5F). pre-miR-144 and mature miR-144 were markedly upregulated in HOTAIRM1 depleted FRO cells compared with control FRO cells (Fig. 5G). To elucidate whether ILF3 mediated the repressive influence of HOTAIRM1 on pre-miR-144 and miR-144, we silenced ILF3 in 8505 C cells via transfection of ILF3 small interfering RNA (siRNA). At the condition of ILF3 silencing, the downregulation of pre-miR-144 and miR-144 in 8505 C cells overexpressing HOTAIRM1 were diminished (Fig. 5H). The upregulation of pre-miR-144 and miR-144 in HOTAIRM1 depleted FRO cells were also diminished in the condition of ILF3 silencing (Fig. 5I). In summary, these findings demonstrated that HOTAIRM1 downregulates pre-miR-144 and mature miR-144 partially via binding ILF3 and relieving the protective roles of ILF3 on pre-miR-144 and miR-144.
Figure 5.
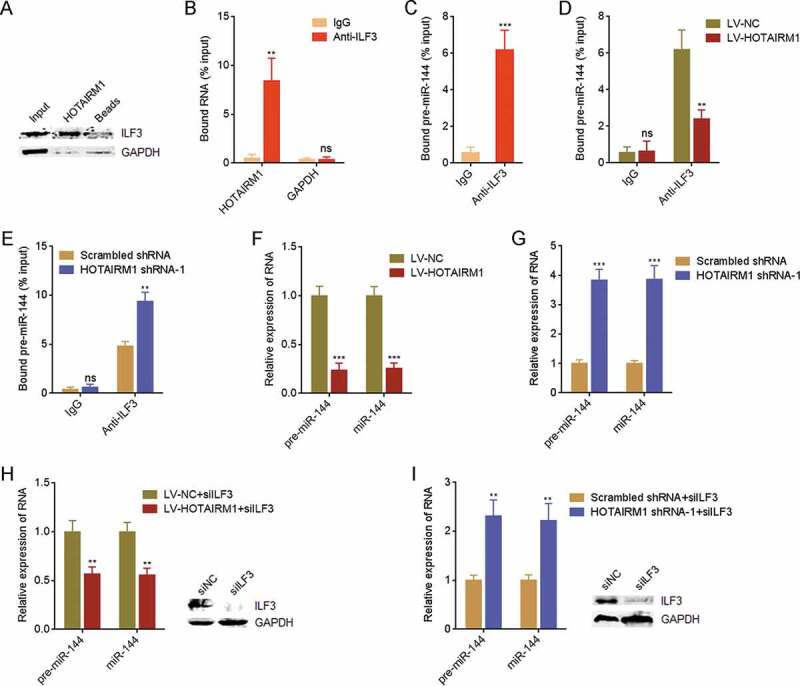
HOTAIRM1 binds ILF3 and downregulates pre-miR-144 and miR-144. (A) RNA pull-down assays using in vitro transcribed biotin-labelled HOTAIRM1 were performed in 8505 C cells, followed by western blot to measure interacted proteins. GAPDH protein was used as negative control. (B) RIP assays with ILF3 antibody were performed in 8505 C cells, followed by RT-qPCR to measure interacted transcripts. GAPDH mRNA was used as negative control. (C) RIP assays with ILF3 antibody were performed in 8505 C cells, followed by RT-qPCR to measure bound pre-miR-144. Non-specific IgG was used as negative control. (D) RIP assays with ILF3 antibody were performed in HOTAIRM1 overexpressed or control 8505 C cells, followed by RT-qPCR to measure bound pre-miR-144. (E) RIP assays with ILF3 antibody were performed in HOTAIRM1 depleted or control FRO cells, followed by RT-qPCR to measure bound pre-miR-144. (F) pre-miR-144 and miR-144 expressions in HOTAIRM1 overexpressed and control 8505 C cells were measured by RT-qPCR. RNU6-2_11 was used as endogenous control for pre-miR-144. U6 was used as endogenous control for miR-144. (G) pre-miR-144 and miR-144 expressions in HOTAIRM1 depleted and control FRO cells were measured by RT-qPCR. RNU6-2_11 was used as endogenous control for pre-miR-144. U6 was used as endogenous control for miR-144. (H) After transient transfection of ILF3 siRNA pool into HOTAIRM1 overexpressed or control 8505 C cells, pre-miR-144 and miR-144 expressions were measured by RT-qPCR. RNU6-2_11 was used as endogenous control for pre-miR-144. U6 was used as endogenous control for miR-144. (I) After transient transfection of ILF3 siRNA pool into HOTAIRM1 depleted or control FRO cells, pre-miR-144 and miR-144 expressions were measured by RT-qPCR. RNU6-2_11 was used as endogenous control for pre-miR-144. U6 was used as endogenous control for miR-144. Data were shown as mean ± standard deviation of three independent experiments. ns, not significant, **P < 0.01, ***P < 0.001 by Student’s t-test
HOTAIRM1 represses the biogenesis of pre-miR-144 and mature miR-144 via binding pri-miR-144
The data in Fig. 5H and I suggested that ILF3 silencing could not completely abolish the influence of HOTAIRM1 on pre-miR-144 and mature miR-144, which suggested that other mechanisms also mediated the modulation of pre-miR-144 and miR-144 by HOTAIRM1. pri-miR-144 is transcribed from genomic DNA and processed by DROSHA to generate pre-miR-144. Then, pre-miR-144 is exported to cytoplasm and processed by DICER to generate mature miR-144. Therefore, we investigated whether HOTAIRM1 modulated pri-miR-144. Our results showed that pre-miR-144 was increased in 8505 C cells overexpressing HOTAIRM1 compared with control 8505 C cells (Fig. 6A). ILF3 silencing did not modulate the effects of HOTAIRM1 on pri-miR-144 (Fig. 6B). Furthermore, pri-miR-144 was reduced in HOTAIRM1 depleted FRO cells compared with control FRO cells (Fig. 6C). The converse influences of HOTAIRM1 on pri-miR-144 and pre-miR-144 implied that HOTAIRM1 might regulate the processing of pri-miR-144. Notably, potential binding between HOTAIRM1 and pri-miR-144 was predicted by in silico tool IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp). As shown in Fig. 6D, 594–614 nucleotides of HOTAIRM1 were predicted to bind pri-miR-144 at 18–35 nucleotides upstream of pre-miR-144. HOTAIRM1 antisense probes were used to enrich HOTAIRM1 and bound transcripts in 8505 C cells, followed by RT-qPCR to measure the enrichment of interacted transcripts. The RT-qPCR results showed that pri-miR-144 was markedly enriched in HOTAIRM1 antisense probes group (Fig. 6E). RNA pull-down assays using in vitro transcribed biotin-labelled HOTAIRM1 showed that pri-miR-144 was markedly enriched in HOTAIRM1 group, which was abolished by the mutation of 594–614 nucleotides of HOTAIRM1 (Fig. 6F). These data suggested that HOTAIRM1 binds pri-miR-144. To elucidate whether the interaction between HOTAIRM1 and pri-miR-144 modulated the processing of pri-miR-144 by DROSHA, RIP assays with DROSHA antibody revealed that DROSHA bound less pri-miR-144 in 8505 C cells overexpressing HOTAIRM1 than control 8505 C cells (Fig. 6G). RIP assays also revealed that DROSHA bound more pri-miR-144 in HOTAIRM1 depleted FRO cells than control FRO cells (Fig. 6H). These data demonstrated that HOTAIRM1 represses the binding of pri-miR-144 to DROSHA. Next, we investigated the levels of pre-miR-144 and miR-144 at the condition of both ILF3 and DROSHA silencing. At the condition of both ILF3 and DROSHA silencing, the downregulation of pre-miR-144 and miR-144 in 8505 C cells overexpressing HOTAIRM1 were abolished (Fig. 6I). The upregulation of pre-miR-144 and miR-144 in HOTAIRM1 depleted FRO cells were also abolished in the condition of both ILF3 and DROSHA silencing (Fig. 6J). Collectively, these findings demonstrated that HOTAIRM1 binds ILF3, reduces the binding of ILF3 to pre-miR-144, therefore relieves the protective roles of ILF3 on pre-miR-144 stability, and decreases pre-miR-144; furthermore, HOTAIRM1 also directly binds pri-miR-144, reduces the binding of DROSHA to pri-miR-144, represses the biogenesis of pre-miR-144 (Fig. 6K). Therefore, HOTAIRM1 markedly downregulates pre-miR-144 and mature miR-144.
Figure 6.
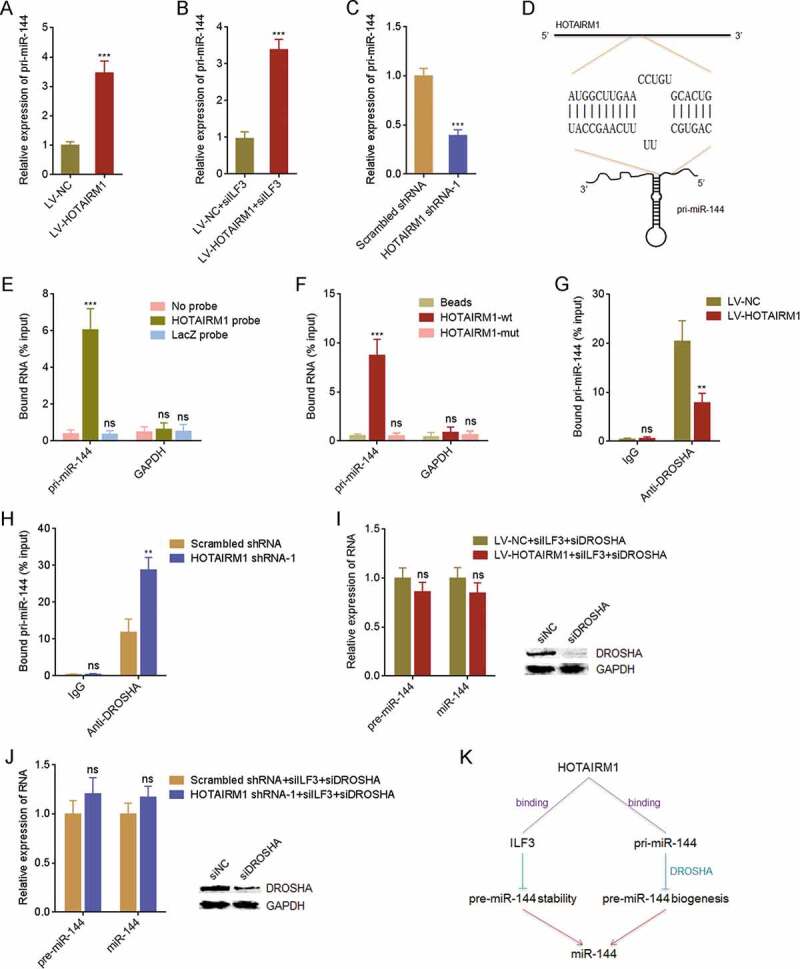
HOTAIRM1 binds pri-miR-144 and represses pre-miR-144 and miR-144 biogenesis. (A) pri-miR-144 expressions in HOTAIRM1 overexpressed and control 8505 C cells were measured by RT-qPCR. GAPDH was used as endogenous control. (B) After transient transfection of ILF3 siRNA pool into HOTAIRM1 overexpressed or control 8505 C cells, pri-miR-144 expression was measured by RT-qPCR. GAPDH was used as endogenous control. (C) pri-miR-144 expressions in HOTAIRM1 depleted and control FRO cells were measured by RT-qPCR. GAPDH was used as endogenous control. (D) The binding sites between HOTAIRM1 and pri-miR-144 predicted by IntaRNA. (E) HOTAIRM1 antisense probes were used to enrich transcripts interacted with HOTAIRM1 in 8505 C cells, followed by RT-qPCR to measure the enrichment of pri-miR-144. GAPDH mRNA was used as negative control. (F) RNA pull-down assays using in vitro transcribed biotin-labelled wild-type (wt) or binding sites mutated (mut) HOTAIRM1 were performed in 8505 C cells, followed by RT-qPCR to measure interacted transcripts. GAPDH mRNA was used as negative control. (G) RIP assays with DROSHA antibody were performed in HOTAIRM1 overexpressed or control 8505 C cells, followed by RT-qPCR to measure bound pri-miR-144. (H) RIP assays with DROSHA antibody were performed in HOTAIRM1 depleted or control FRO cells, followed by RT-qPCR to measure bound pri-miR-144. (I) After transient transfection of ILF3 and DROSHA siRNA pools into HOTAIRM1 overexpressed or control 8505 C cells, pre-miR-144 and miR-144 expressions were measured by RT-qPCR. RNU6-2_11 was used as endogenous control for pre-miR-144. U6 was used as endogenous control for miR-144. (J) After transient transfection of ILF3 and DROSHA siRNA pools into HOTAIRM1 depleted or control FRO cells, pre-miR-144 and miR-144 expressions were measured by RT-qPCR. RNU6-2_11 was used as endogenous control for pre-miR-144. U6 was used as endogenous control for miR-144. (K) A model depicting the influences of HOTAIRM1 on miR-144. Data were shown as mean ± standard deviation of three independent experiments. ns, not significant, **P < 0.01, ***P < 0.001 by Student’s t-test
The expression of miR-144 is downregulated and reversely correlated with HOTAIRM1 in ATC tissues
Given the markedly repressive roles of HOTAIRM1 on miR-144, we next investigated whether the modulation also existed in vivo. The expressions of miR-144 in the same 20 normal thyroid tissues, 20 PTC tissues, and 20 ATC tissues used in Fig. 1 were measured by RT-qPCR and the results revealed that the expression of miR-144 was markedly reduced in ATC tissues compared with normal thyroid and PTC tissues (Fig. 7A). Furthermore, the expression of miR-144 was markedly negatively correlated with the expression of HOTAIRM1 in ATC tissues (Fig. 7B), supporting the repressive roles of HOTAIRM1 on miR-144 in vivo.
Figure 7.
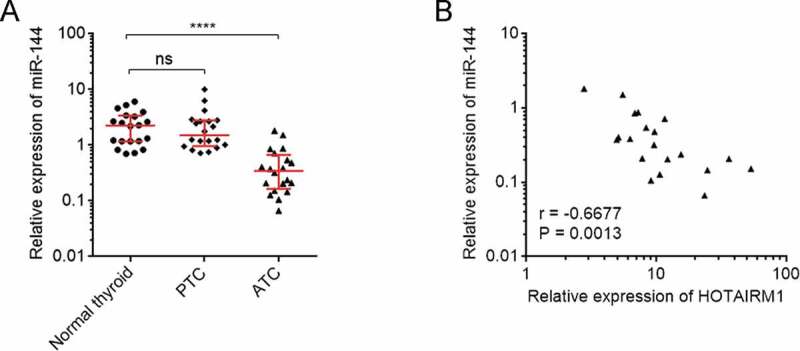
The correlation between miR-144 and HOTAIRM1 expression in ATC tissues. (A) miR-144 expressions in 20 normal thyroid tissues, 20 PTC tissues, and 20 ATC tissues were measured by RT-qPCR. U6 was used as endogenous control. Three randomly selected thyroid tissues were used as the reference samples to calculate the relative expressions. Data were shown as median with interquartile range. ns, not significant, ****P < 0.0001 by Kruskal-Wallis test, followed by Dunn’s multiple comparisons test. (B) The correlation between miR-144 and HOTAIRM1 expressions in 20 ATC tissues. r = −0.6677, P = 0.0013 by Spearman correlation analysis
HOTAIRM1 upregulates MET via repressing miR-144
MET, which is a receptor tyrosine kinase and a critical oncogenic molecule, is revealed to be miR-144 target in several cancers [36,37]. Therefore, we further investigated the influence of HOTAIRM1 on MET. Dual luciferase reporter assays revealed that the luciferase activity of MET 3ʹUTR was increased in 8505 C cells overexpressing HOTAIRM1 compared with control 8505 C cells, and the increase was abolished by overexpression of miR-144 (Fig. 8A). Conversely, the luciferase activity of MET 3ʹUTR was reduced in HOTAIRM1 depleted FRO cells compared with control FRO cells (Fig. 8B). In accordance with the luciferase activity, MET expression was increased in 8505 C cells overexpressing HOTAIRM1 compared with control 8505 C cells, and the increase was abolished by overexpression of miR-144 (Fig. 8C). MET expression was reduced in HOTAIRM1 depleted FRO cells compared with control FRO cells (Fig. 8D). As a receptor tyrosine kinase, MET activates downstream PI3K-AKT signalling [36]. Therefore, we further investigated the influence of HOTAIRM1 on AKT signalling. The phosphorylation level of AKT was increased in 8505 C cells overexpressing HOTAIRM1 compared with control 8505 C cells (Fig. 8E). Conversely, the phosphorylation level of AKT was reduced in HOTAIRM1 depleted FRO cells compared with control FRO cells (Fig. 8F). Furthermore, the expressions of MET in the same 20 normal thyroid tissues, 20 PTC tissues, and 20 ATC tissues used in Fig. 1 were measured by RT-qPCR and the results revealed that the expression of MET was consistently increased in ATC tissues (Fig. 8G). The expression of MET was negatively correlated with the expression of miR-144 and positively correlated with the expression of HOTAIRM1 in ATC tissues (Fig. 8H and I). In summary, these findings demonstrated that HOTAIRM1 upregulates MET and activates AKT signalling via repressing miR-144.
Figure 8.
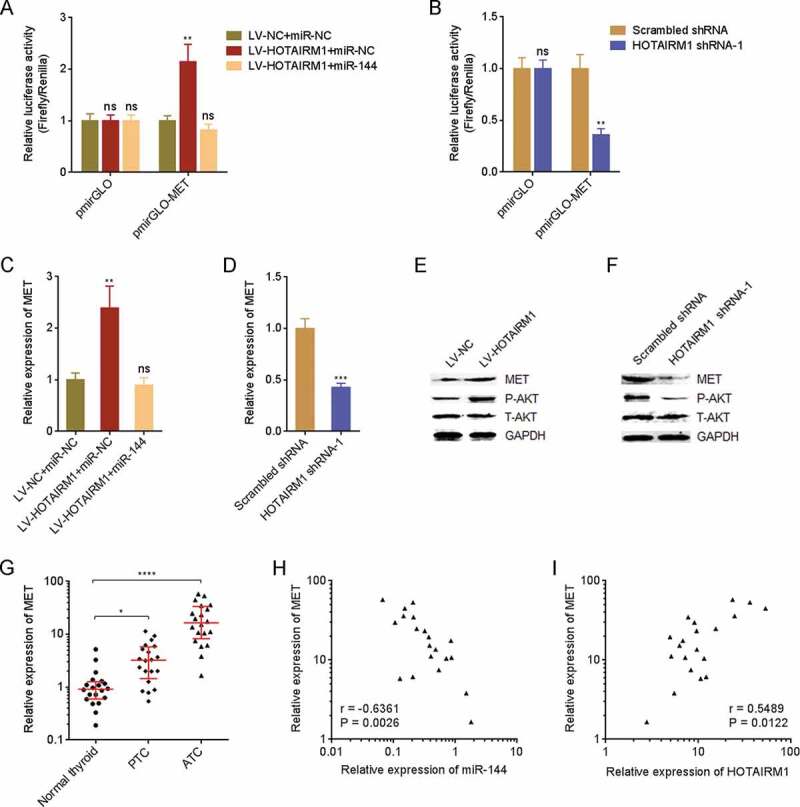
HOTAIRM1 upregulates MET via repressing miR-144. (A) Luciferase reporter containing MET 3ʹUTR (pmirGLO-MET) was co-transfected with miR-144 mimics into HOTAIRM1 overexpressed or control 8505 C cells. 48 hours after transfection, the luciferase activities were measured by dual luciferase reporter assays. Results were shown as relative ratio of firefly luciferase activity to renilla luciferase activity. (B) pmirGLO-MET or empty reporter pmirGLO was transfected into HOTAIRM1 depleted or control FRO cells. 48 hours after transfection, the luciferase activities were measured by dual luciferase reporter assays. Results were shown as relative ratio of firefly luciferase activity to renilla luciferase activity. (C) After transient transfection of miR-144 mimics or miRNAs negative control (miR-NC) into HOTAIRM1 overexpressed or control 8505 C cells, MET expression was measured by RT-qPCR. GAPDH was used as endogenous control. (D) MET expressions in HOTAIRM1 depleted and control FRO cells were measured by RT-qPCR. GAPDH was used as endogenous control. For A-D, data were shown as mean ± standard deviation of three independent experiments. ns, not significant, **P < 0.01, ***P < 0.001 by one-way ANOVA followed by Dunnett’s multiple comparisons test (A and C) or Student’s t-test (B and D). (E) MET protein levels and AKT phosphorylation levels in HOTAIRM1 overexpressed and control 8505 C cells were measured by western blot. (F) MET protein levels and AKT phosphorylation levels in HOTAIRM1 depleted and control FRO cells were measured by western blot. (G) MET expressions in 20 normal thyroid tissues, 20 PTC tissues, and 20 ATC tissues were measured by RT-qPCR. GAPDH was used as endogenous control. Three randomly selected thyroid tissues were used as the reference samples to calculate the relative expressions. Data were shown as median with interquartile range. *P < 0.05, ****P < 0.0001 by Kruskal-Wallis test, followed by Dunn’s multiple comparisons test. (H) The correlation between MET and miR-144 expressions in 20 ATC tissues. r = −0.6361, P = 0.0026 by Spearman correlation analysis. (I) The correlation between MET and HOTAIRM1 expressions in 20 ATC tissues. r = 0.5489, P = 0.0122 by Spearman correlation analysis
miR-144 abolished the roles of HOTAIRM1 in proliferation, apoptosis, migration, and invasion of ATC cells
To elucidate whether the oncogenic roles of HOTAIRM1 in ATC were dependent on the repression of miR-144, we transiently overexpressed miR-144 in HOTAIRM1 overexpressed 8505 C cells via transfection of miR-144 mimics. CCK-8 assays revealed that the faster cell proliferation rate of HOTAIRM1 overexpressed 8505 C cells was reversed by miR-144 overexpression (Fig. 9A). EdU incorporation assays revealed that the increased EdU-positive cell proportion of HOTAIRM1 overexpressed 8505 C cells was reversed by miR-144 overexpression (Fig. 9B). Annexin V-PI staining and flow cytometry revealed that the reduced apoptotic cell proportion of HOTAIRM1 overexpressed 8505 C cells was reversed by miR-144 overexpression (Fig. 9C). Transwell migration assays revealed that the increased migratory ability of HOTAIRM1 overexpressed 8505 C cells was reversed by miR-144 overexpression (Fig. 9D). Transwell invasion assays revealed that the increased invasive ability of HOTAIRM1 overexpressed 8505 C cells was reversed by miR-144 overexpression (Fig. 9E). In summary, these findings demonstrated that the roles of HOTAIRM1 in promoting proliferation, inhibiting apoptosis, and promoting migration and invasion of ATC cells are dependent on the negative modulation of miR-144.
Figure 9.
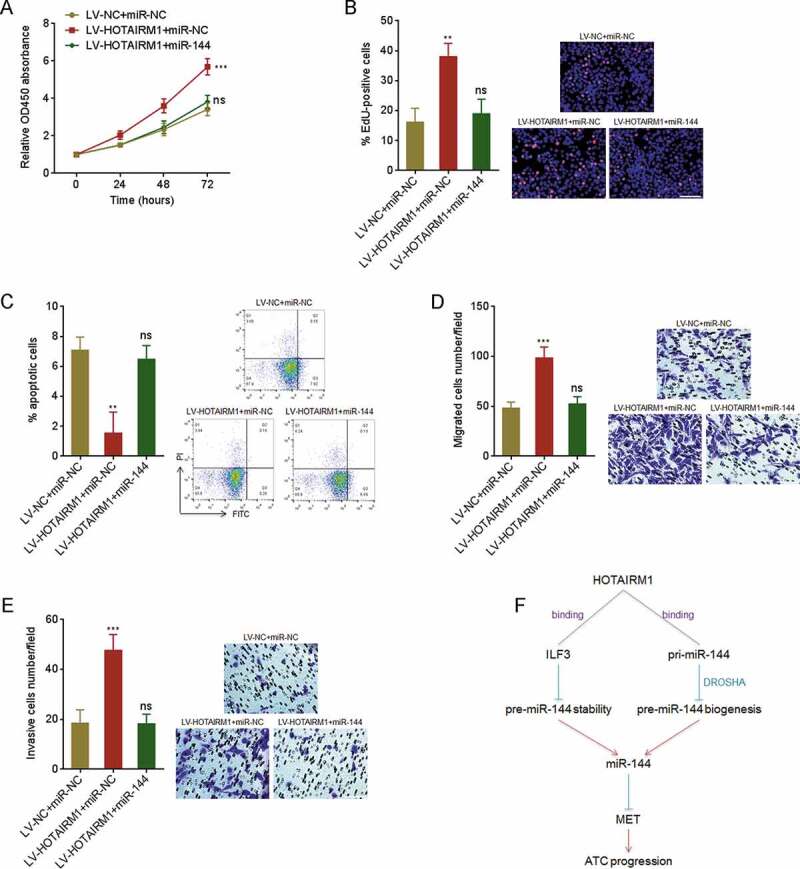
The roles of HOTAIRM1 in proliferation, apoptosis, migration and invasion of ATC cells are dependent on the inhibition of miR-144. (A) After transient transfection of miR-144 mimics or miRNAs negative control (miR-NC) into HOTAIRM1 overexpressed or control 8505 C cells, cell proliferation was measured by CCK-8 assays. Absorbance values at 450 nm were measured to track cell proliferation. (B) After transient transfection of miR-144 mimics or miR-NC into HOTAIRM1 overexpressed or control 8505 C cells, cell proliferation was measured by EdU incorporation assays. Red colour indicates EdU positive and proliferative cells. Scale bar = 100 μm. (C) After transient transfection of miR-144 mimics or miR-NC into HOTAIRM1 overexpressed or control 8505 C cells, cell apoptosis was measured by Annexin-V-FITC/PI staining, followed by flow cytometry. (D) After transient transfection of miR-144 mimics or miR-NC into HOTAIRM1 overexpressed or control 8505 C cells, cell migration was measured by transwell migration assays. Scale bar = 100 μm. (E) After transient transfection of miR-144 mimics or miR-NC into HOTAIRM1 overexpressed or control 8505 C cells, cell invasion was measured by transwell invasion assays. Scale bar = 100 μm. (F) Schematic of the roles of HOTAIRM1 in ATC progression via repressing miR-144. Data were shown as mean ± standard deviation of three independent experiments. ns, not significant, **P < 0.01, ****P < 0.0001 by one-way ANOVA, followed by Dunnett’s multiple comparisons test
Discussion
ATC is the most lethal and uncommon subtype of thyroid cancer [3]. Unlike other common malignancies, the studies and knowledge of ATC are relative lacking [38]. The molecular mechanisms underlying the initiation and development of ATC are just beginning to be investigated. Wang et al. identified 11 upregulated and 7 downregulated lncRNAs in two ATC tissues compared with PTC tissues using microarray [28]. Ravi et al. investigated genomic copy number levels in 10 ATC tissues compared with normal thyroid tissues [8]. Combining these two analyses, we identified a genomic copy number amplified and expression upregulated lncRNA HOTAIRM1 in ATC. To further confirm the expression levels and genomic copy number of HOTAIRM1 in ATC, we collected 20 ATC tissues, 20 PTC tissues and 20 normal thyroid tissues. Our data found that the expression levels and copy number of HOTAIRM1 were both significantly increased in ATC tissues compared with PTC tissues and normal thyroid tissues. The expression levels and copy number of HOTAIRM1 were significantly positively associated in ATC tissues. Our data suggested that increased genomic copy number of HOTAIRM1 contributed to HOTAIRM1 expression upregulation in ATC. Furthermore, our findings demonstrated that increased expression levels and copy number of HOTAIRM1 were both correlated with poor survival of ATC patients. Therefore, we identified HOTAIRM1 as an upregulated and prognosis-related lncRNA in ATC. Whether the genomic amplification of HOTAIRM1 is a local event or part of large segment amplification event needs further investigation. To elucidate whether the aberrant expression levels and copy number of HOTAIRM1 contribute to the outcome of ATC, we further investigated the potential biological roles of HOTAIRM1 in ATC.
HOTAIRM1 is localized in chromosome 7p15.2. HOTAIRM1 has several different isoforms according to Ensembl. In ATC tissues, we identified the isoform HOTAIRM1-204 as predominantly expressed. HOTAIRM1 (HOTAIRM1-204) has two exons and 783 nucleotides in length. In different cancers, HOTAIRM1 displays different expression changes and roles. HOTAIRM1 is reported to be overexpressed and has oncogenic roles in glioblastoma multiforme and pancreatic ductal adenocarcinoma [39,40]. HOTAIRM1 is also reported to be downregulated and has tumour suppressive roles in ovarian cancer and colorectal cancer [41,42]. These previous reports suggested that the roles of HOTAIRM1 in different tumours may be organ-specific. In ATC, using gain-of and loss-of function assays we found that HOTAIRM1 promoted proliferation, inhibited apoptosis, and promoted migration and invasion of ATC cells. Using xenograft assays, we demonstrated that HOTAIRM1 promoted ATC tumour growth and metastasis in vivo. Therefore, our data suggested HOTAIRM1 as an oncogenic lncRNA in ATC.
Different subcellular localizations of lncRNAs influence the molecular mechanisms exerted by lncRNAs. For cytoplasmic lncRNAs, they can competitively bind miRNAs and relieve the repressing roles of miRNAs on targets [43]. Cytoplasmic lncRNAs can also bind mature mRNAs and change the stability and/or translation of interacted mRNAs [44]. For nuclear lncRNAs, they can bind primary transcripts and change the processing of primary transcripts, in particular inducing alternative splicing of mRNAs [45]. For some cytoplasmic and nuclear lncRNAs, they directly bind cytoplasmic and nuclear proteins, respectively, change the subcellular localization, post-translation modification, stability, expression, and/or functions of interacted proteins [46].
In this study, combining bioinformatic prediction and experimental verification, we found that HOTAIRM1 bound IIF3. ILF3, also known as nuclear factor 90 (NF90), is a double-stranded RNA binding protein [47]. Except the effects of binding and stabilizing mRNAs, ILF3 has been reported to bind precursor miRNAs, particular pre-miR-144, and further stabilize and upregulate pre-miR-144 [31]. In this study, we found that the interaction between HOTAIRM1 and ILF3 competitively repressed the binding between ILF3 and pre-miR-144, relieved the stabilization of pre-miR-144 exerted by ILF3, and therefore downregulated pre-miR-144 and mature miR-144. Except miR-144, the effects of HOTAIRM1 on other ILF3 targets need further investigation.
Intriguingly, in addition to the downregulation of pre-miR-144, we also found that HOTAIM1 increased pri-miR-144. Further investigation revealed that HOTAIRM1 directly bound pri-miR-144, repressed the binding between pri-miR-144 and DROSHA, repressed the processing of pri-miR-144 by DROSHA, and therefore increased pri-miR-144 and reduced pre-miR-144. HOTAIRM1 significantly downregulated pre-miR-144 and miR-144 via dual mechanisms. Our data found that depletion of ILF3 partially blocked the effects of HOTAIRM1 on pre-miR-144 and miR-144, and while concurrent depletion of ILF3 and DROSHA nearly completely abolished the effects of HOTAIRM1 on pre-miR-144 and miR-144. These findings supported the dual mechanisms of HOTAIRM1 on the modulation of pre-miR-144 and miR-144. Previous reports have shown that many lncRNAs directly bind mature miRNAs and change the function of interacted miRNAs [48]. In this study, we showed a different functional manner of lncRNA on miRNA biogenesis. The negative correlation between miR-144 and HOTAIRM1 expression in ATC tissues supported the repression of miR-144 by HOTAIRM1. Overexpression of miR-144 reversed the roles of HOTAIRM1 in promoting proliferation, inhibiting apoptosis, and promoting migration and invasion of ATC cells. These data supported that miR-144 was an important mediator of the roles of HOTAIRM1 in ATC.
In summary, we identified HOTAIRM1 as an expression-upregulated and genomic copy number amplified lncRNA in ATC, which drove ATC progression via repressing miR-144 biogenesis. Our findings suggested HOTAIRM1 as a potential prognostic biomarker and therapeutic target for ATC.
Materials and methods
Human Tissue samples
Twenty noncancerous thyroid tissues, twenty PTC tissues, and 20 ATC tissues were acquired at the First Hospital of Shanxi Medical University (Taiyuan, China) during the period of January 2012 to December 2018. All specimens were confirmed by pathological examination. The ethics committee of the First Hospital of Shanxi Medical University approved the acquirement of tissues specimens. All participants signed written informed consents. This study followed the Declaration of Helsinki.
Cell culture
Human normal thyroid follicular epithelial cell line (Nthy-ori 3–1), PTC cell lines (TPC-1 and HTH83), and ATC cell lines (8505 C, C643, and FRO) were obtained from BeNa Culture Collection (Beijing, China). Nthy-ori 3–1 was cultured in F-12 K medium with 10% foetal bovine serum (FBS). TPC-1 was cultured in L15 medium with 10% FBS, HTH83, 8505 C, C643 and FRO were cultured in RPMI-1640 medium with 10% FBS. All cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Plasmids, siRNAs, and transfection
Full-length HOTAIRM1 was PCR-amplified from cDNA with the primers 5ʹ-CGAGCTCAAAAGTTTGCCGGCTTC-3ʹ (forward) and 5ʹ-GCTCTAGACTTTTTTCAATTTTAATACATTTAT-3ʹ (reverse). Next, the PCR products were subcloned to the Sac I and Xba I sites of the pSPT19 plasmid (Roche) to construct pSPT19-HOTAIRM1. pSPT19-HOTAIRM1 with 594–614 nucleotides of HOTAIRM1 deleted was synthesized by GenScript (Nanjing, China), named as pSPT19-HOTAIRM1-mut. The 3ʹUTR of MET was PCR-amplified from cDNA with the primers 5ʹ-CGAGCTCTAGACATTCCTTTGGTTGG-3ʹ (forward) and 5ʹ-GCTCTAGAAAGTGTGTAGTCCTGTTGAT-3ʹ (reverse). Next, the PCR products were subcloned into the Sac I and Xba I sites of the pmirGLO plasmid (Promega) to construct pmirGLO-MET. siRNA pools targeting ILF3 or DROSHA were purchased from Dharmacon (ON-TARGETplus Human ILF3 siRNA, Cat. L-012442-00-0010 and ON-TARGETplus Human DROSHA siRNA, Cat. L-016996-00-0010, respectively). miR-144 mimics and negative control (NC) were purchased from GenePharma (Shanghai, China). Transfection and co-transfection of plasmids, siRNAs, and miR-144 mimics were performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s guidelines.
Lentivirus transduction and stable cell lines construction
HOTAIRM1 overexpressing lentivirus was purchased from GenePharma. To construct ATC cells stably overexpressing HOTAIRM1, 8505 C and C643 cells were infected with HOTAIRM1 overexpressing lentivirus and selected with 2 µg/ml puromycin for four weeks. Two pairs of cDNA oligonucleotides to inhibit HOTAIRM1 expression were designed and synthesized by GenePharma. After annealing, double-strand oligos were inserted into the shRNA lentivirus expressing plasmid pLV6/EF-1aF/Puro (GenePharma). The constructed plasmid was co-transfected with pGag/Pol, pRev and pVSV-G (GenePharma) into HEK-293 FT cells to produce the lentivirus inhibiting HOTAIRM1. Lentiviruses were harvested at 48 hours after transfection and filtered through 0.45 μm PVDF filters. Scrambled shRNA lentivirus was used as negative control. Recombinant lentiviruses were concentrated 100-fold using ultracentrifugation (2 hours at 50,000 g). The lentivirus-containing pellet was dissolved in RPMI-1640 medium, aliquoted and stored at −80°C. FRO cells were infected with the shRNA lentivirus and selected with 2 µg/ml puromycin for four weeks to construct HOTAIRM1 stably depleted FRO cells. The shRNAs sequences were: for HOTAIM1 shRNA-1, 5ʹ-GATCCGGCAAAGGCCGATTTGGAGTTTCAAGAGAACTCCAAATCGGCCTTTGCCTTTTTTG-3ʹ (forward) and 5ʹ-AATTCAAAAAAGGCAAAGGCCGATTTGGAGTTCTCTTGAAACTCCAAATCGGCCTTTGCCG-3ʹ (reverse); for HOTAIM1 shRNA-2, 5ʹ-GATCCGCCGTTCAATGAAAGATGAATTCAAGAGATTCATCTTTCATTGAACGGCTTTTTTG-3ʹ (forward) and 5ʹ-AATTCAAAAAAGCCGTTCAATGAAAGATGAATCTCTTGAATTCATCTTTCATTGAACGGCG-3ʹ (reverse); for scrambled shRNA, 5ʹ-GATCCGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAACTTTTTTG-3ʹ (forward) and 5ʹ-AATTCAAAAAAGTTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAACG-3ʹ (reverse).
RNA extraction, reverse transcription (RT), and quantitative polymerase chain reaction (qPCR)
RNA extraction was performed using the TRIzol reagent (Invitrogen). The extracted RNA was used to carry out reverse transcription (RT) using the M-MLV Reverse Transcriptase (Invitrogen). For the quantitation of HOTAIRM1 and MET expression, quantitative polymerase chain reaction (qPCR) was performed using the SYBR® Premix Ex Taq™ II (TAKARA) on StepOnePlus Real-Time PCR System (Applied Biosystems) with the primers 5ʹ-AAGATGAACTGGCGAGAGGT-3ʹ (forward) and 5ʹ-CAGGAATGAGTAACACGGAG-3ʹ (reverse) for HOTAIRM1, 5ʹ-AATCAACCCGCCCCACACAC-3ʹ (forward) and 5ʹ-ACCCCCTCCCCCATAAATCC-3ʹ (reverse) for exon 2–3 of HOTAIRM1, 5ʹ-GATTGATTGCTGGTGTTGT-3ʹ (forward) and 5ʹ-CATTTCTGTAGTTGGGCTTA-3ʹ (reverse) for MET, 5ʹ-GTCGGAGTCAACGGATTTG-3ʹ (forward) and 5ʹ-TGGGTGGAATCATATTGGAA-3ʹ (reverse) for GAPDH. For the quantitation of pri-miR-144 expression, qPCR was performed using the TaqManTM Pri-miRNA Assay (Cat. 4,427,012, Applied Biosystems) on the StepOnePlus Real-Time PCR System (Applied Biosystems). For the quantitation of pre-miR-144 expression, qPCR was performed using the miScript SYBR Green PCR Kit (QIAGEN) and miScript Precursor Assay (QIAGEN) on the StepOnePlus Real-Time PCR System (Applied Biosystems). For the quantitation of mature miR-144 expression, qPCR was performed using the TaqManTM Advanced miRNA Assay (Cat. A25576, Applied Biosystems) on the StepOnePlus Real-Time PCR System (Applied Biosystems).
DNA extraction and qPCR
Genomic DNA was extracted from tissues using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) following the manufacturer’s guidelines. Genomic copy number was measured by qPCR assays which were performed using the SYBR® Premix Ex Taq™ II (TAKARA) on StepOnePlus Real-Time PCR System (Applied Biosystems) with the primers 5ʹ-TGGACAAATGGCTGATGGTG-3ʹ (forward) and 5ʹ-GGAAGTAGGGTGATGAGGAA-3ʹ (reverse) for HOTAIRM1, 5ʹ-AAAGCCGCTCAACTACATGG-3ʹ (forward) and 5ʹ-TGCTTTGAATGCGTCCCAGAG-3ʹ (reverse) for LINE1.
Nuclear and cytoplasmic RNA fractionation
Nuclear/cytoplasmic fractionation was performed using the PARIS Kit (Invitrogen) according to the manufacturer’s guidelines. HOTAIRM1 levels in nuclear and cytoplasmic cellular fractions were detected by RT-qPCR as described above.
Cell proliferation, apoptosis, migration, and invasion assays
Cell Counting Kit-8 (CCK-8) and 5-ethynyl-2ʹ-deoxyuridine (EdU) incorporation assays were conducted to detect cell proliferation. CCK-8 assay was undertaken using the Cell Counting Kit-8 (Cat. CK04, Dojindo) following the manufacturer’s guidelines. EdU incorporation assay was undertaken using the Cell-Light EdU Apollo643 In Vitro Kit (Cat. C10310-2, RiboBio, Guangzhou, China). Cell apoptosis was detected using the FITC Annexin V Apoptosis Detection Kit I (BD Biosciences). Cell migration and invasion were detected by transwell migration and invasion assays, respectively. 4 × 104 indicated ATC cells re-suspended in RPMI-1640 medium without FBS were plated onto the upper chamber of transwell inserts (pore size 8 μm; Corning). For migration and invasion assays, the transwell inserts were coated without or with Matrigel, respectively. RPMI-1640 medium with 20% FBS were added into the bottom chamber. After culture for 48 hours, the cells remaining in the upper chamber were removed. The cells on the bottom chamber were fixed, stained, and counted using light microscope (Zeiss).
Animal experiment in vivo
Athymic nude mice were purchased from Model Animal Research Centre of Nanjing University (Nanjing, China) and were maintained in a pathogen-free facility in Shanxi Medical University. 2 × 106 indicated ATC cells were subcutaneously injected into the flanks of nude mice. Tumours volumes were measured using a caliper and calculated following the formula 0.5 × L× S2 (L, long axes; S, short axes) every 7 days. After breeding for 28 days, the mice were sacrificed and the tumours were resected and weighed. To monitor ATC lung metastasis, indicated ATC cells were infected with luciferase expressing lentivirus (GeneChem, Shanghai, China) and selected with 1000 µg/ml neomycin for two weeks to construct luciferase stably labelled ATC cells. A total of 1 × 106 luciferase stably labelled ATC cells were injected into the tail vein of nude mice. At the indicted time after injection, lung metastases were monitored by bioluminescence imaging using the IVIS@ Lumina II system (Caliper Life Sciences). Animal experiments were approved by the animal care committee of the First Hospital of Shanxi Medical University (Taiyuan, China).
RNA-protein interaction assays
RNA-protein interaction was detected by RNA pull-down and UV crosslinking RIP assays. For RNA pull-down assay, HOTAIRM1 was in vitro transcribed from pSPT19-HOTAIRM1 and biotinylated using the Biotin RNA Labelling Mix (Roche) and Sp6 RNA polymerase (Roche). After purification using RNase-free DNase I (Roche) and RNeasy Mini Kit (QIAGEN), 3 µg of in vitro transcribed biotin-labelled HOTAIRM1 were incubated with 1 mg of whole-cell lysate from 8505 C cells for 1 hour at 25°C. Streptavidin agarose beads (Invitrogen) were used to enrich biotin-labelled transcripts and interacted proteins. The enriched protein was detected by western blot. For UV crosslinking RIP assay, UV crosslinking was undertaken at 400 mJ/cm2 using XLE-1000 UV crosslinker (Spectroline). Immunoprecipitation was conducted using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) and primary antibodies against ILF3 (5 µg per reaction; ab131004, Abcam) or DROSHA (5 µg per reaction; #3410, Cell Signalling Technology). The enriched RNA was detected by RT-qPCR as described above.
Western blot
Whole-cell lysates were acquired from indicated ATC cells using RIPA Lysis Buffer (Beyotime, Jiangsu, China) added with PMSF (Beyotime) and phosphatase inhibitors cocktail (Sigma). Protein concentrations were determined using the Enhanced BCA Protein Assay Kit (Beyotime). The samples were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto PVDF membranes (Millipore). After blocking in 0.5% non-fat dry milk, the membranes were incubated with primary antibodies against ILF3 (1:2000; ab131004, Abcam), DROSHA (1:1000; #3410, Cell Signalling Technology), MET (1:1000; #8198, Cell Signalling Technology), Phospho-AKT (1:2000; #4060, Cell Signalling Technology), AKT (1:1000; #4685, Cell Signalling Technology), or GAPDH (1:10,000; T0004, Affinity). After three washes using TBST, the membranes were incubated with IRdye 800-conjugated goat anti-rabbit IgG or IRdye 700-conjugated goat anti-mouse IgG and scanned by an Odyssey infrared scanner (Li-Cor).
RNA-RNA interaction assays
To detect the RNA interacted with HOTAIRM1, RNA pull-down and Chromatin isolation by RNA purification (ChIRP) assays were conducted. For RNA pull-down assay, wild-type or mutated HOTAIRM1 was in vitro transcribed from pSPT19-HOTAIRM1 or pSPT19-HOTAIRM1-mut respectively as described above. 3 µg of in vitro transcribed biotin-labelled HOTAIRM1 were incubated with 1 mg of whole-cell lysate from 8505 C cells for 1 hour at 25°C. Then, AMT (Sigma) was added (20 µg/ml) and incubated for another 10 min, followed by irradiation for 30 min under UV365 nm bulbs. After crosslinking, streptavidin agarose beads (Invitrogen) were added to enrich biotin-labelled transcripts and interacted RNAs. The enriched RNA was detected by RT-qPCR as described above. ChIRP assay was conducted in 8505 C cells using the EZ-Magna ChIRP RNA Interactome Kit (Millipore) to enrich interacted RNA according to the manufacturer’s guidelines. The sequences of oligonucleotide probes complementary to HOTAIRM1 were: 1, 5ʹ-cggggatttaaatgccacta-3ʹ; 2, 5ʹ-agaacgcagcttttgctctt-3ʹ; 3, 5ʹ-gttcaggcaaaacagacctc-3ʹ; 4, 5ʹ-caaacacccacatttcaacc-3ʹ; 5, 5ʹ-ccaggaatgagtaacacgga-3ʹ; 6, 5ʹ-aggcagaattggacagtcta-3ʹ; 7, 5ʹ-cggcatgttcaaagtcttca-3ʹ. The enriched RNA was detected by RT-qPCR as described above.
Dual luciferase reporter assays
pmirGLO or pmirGLO-MET was co-transfected with miR-144 mimics or miR-NC into indicated ATC cells. After incubation for another 48 hours, the luciferase activities were detected by the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer’s guidelines.
Statistical analysis
GraphPad Prism 6.0 Software was used to conduct statistical analyses. The detailed statistical analyses methods were indicated in figure legends for each analysis. P < 0.05 was defined as statistically significant.
Supplementary Material
Funding Statement
This work was supported by the Natural Science Foundation of Shanxi Province (201801D121339 to J.L.), National Natural Science Foundation of China (81773150 to L.Z.), Program for the Outstanding Innovative Teams of Higher Learning Institutions of Shanxi (OIT 2017 to L.Z.), and Key Research Program of Shanxi Province in Social Development (201903D321147 to J.L.).
Disclosure statement
The authors declare no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Cabanillas ME, McFadden DG, Durante C.. Thyroid cancer. Lancet. 2016;388:2783–2795. [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- [3].Molinaro E, Romei C, Biagini A, et al. Anaplastic thyroid carcinoma: from clinicopathology to genetics and advanced therapies. Nat Rev Endocrinol. 2017;13:644–660. [DOI] [PubMed] [Google Scholar]
- [4].Saini S, Tulla K, Maker AV, et al. Therapeutic advances in anaplastic thyroid cancer: a current perspective. Mol Cancer. 2018;17:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bible KC, Ryder M. Evolving molecularly targeted therapies for advanced-stage thyroid cancers. Nat Rev Clin Oncol. 2016;13:403–416. [DOI] [PubMed] [Google Scholar]
- [6].Yoo SK, Song YS, Lee EK, et al. Integrative analysis of genomic and transcriptomic characteristics associated with progression of aggressive thyroid cancer. Nat Commun. 2019;10:2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Naoum GE, Morkos M, Kim B, et al. Novel targeted therapies and immunotherapy for advanced thyroid cancers. Mol Cancer. 2018;17:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ravi N, Yang M, Gretarsson S, et al. Identification of targetable lesions in anaplastic thyroid cancer by genome profiling. Cancers (Basel). 2019;11(3):402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yan X, Hu Z, Feng Y, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. [DOI] [PubMed] [Google Scholar]
- [12].Berger AC, Korkut A, Kanchi RS, et al. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell. 2018;33:690–705 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mondal T, Juvvuna PK, Kirkeby A, et al. Sense-antisense lncRNA pair encoded by locus 6p22.3 determines neuroblastoma susceptibility via the USP36-CHD7-SOX9 regulatory axis. Cancer Cell. 2018;33:417–434e7. [DOI] [PubMed] [Google Scholar]
- [14].Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that Interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33:706–720 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hu WL, Jin L, Xu A, et al. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. 2018;20:492–502. [DOI] [PubMed] [Google Scholar]
- [16].Yuan JH, Liu XN, Wang TT, et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19:820–832. [DOI] [PubMed] [Google Scholar]
- [17].Chen F, Chen J, Yang L, et al. Extracellular vesicle-packaged HIF-1alpha-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21:498–510. [DOI] [PubMed] [Google Scholar]
- [18].Zhu XT, Yuan JH, Zhu TT, et al. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. Febs J. 2016;283:3739–3754. [DOI] [PubMed] [Google Scholar]
- [19].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- [20].Yuan JH, Yang F, Chen BF, et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011;54:2025–2035. [DOI] [PubMed] [Google Scholar]
- [21].Sin-Chan P, Mumal I, Suwal T, et al. A C19MC-LIN28A-MYCN oncogenic circuit driven by hijacked super-enhancers is a distinct therapeutic vulnerability in ETMRs: a lethal brain tumor. Cancer Cell. 2019;36:51–67 e7. [DOI] [PubMed] [Google Scholar]
- [22].Yan W, Wu X, Zhou W, et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol. 2018;20:597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jiang Q, Isquith J, Zipeto MA, et al. Hyper-editing of cell-cycle regulatory and tumor suppressor RNA promotes malignant progenitor propagation. Cancer Cell. 2019;35:81–94 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hanniford D, Ulloa-Morales A, Karz A, et al. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020;37:55–70 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Clancy JW, Zhang Y, Sheehan C, et al. An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat Cell Biol. 2019;21:856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ramirez-Moya J, Wert-Lamas L, Riesco-Eizaguirre G, et al. Impaired microRNA processing by DICER1 downregulation endows thyroid cancer with increased aggressiveness. Oncogene. 2019;38:5486–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].O’Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang Y, Hardin H, Chu YH, et al. Long non-coding RNA expression in anaplastic thyroid carcinomas. Endocr Pathol. 2019;30:262–269. [DOI] [PubMed] [Google Scholar]
- [29].Li JK, Chen C, Liu JY, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer. 2017;16:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Castella S, Bernard R, Corno M, et al. Ilf3 and NF90 functions in RNA biology. Wiley Interdiscip Rev RNA. 2015;6:243–256. [DOI] [PubMed] [Google Scholar]
- [31].Nussbacher JK, Yeo GW. Systematic discovery of RNA binding proteins that regulate MicroRNA levels. Mol Cell. 2018;69:1005–1016 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kooshkaki O, Rezaei Z, Rahmati M, et al. MiR-144: a new possible therapeutic target and diagnostic/prognostic tool in cancers. Int J Mol Sci. 2020;21:2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Guo Y, Ying L, Tian Y, et al. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. Febs J. 2013;280:4531–4538. [DOI] [PubMed] [Google Scholar]
- [34].Liu J, Feng L, Zhang H, et al. Effects of miR-144 on the sensitivity of human anaplastic thyroid carcinoma cells to cisplatin by autophagy regulation. Cancer Biol Ther. 2018;19:484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu F, Zhang J, Qin L, et al. Circular RNA EIF6 (Hsa_circ_0060060) sponges miR-144-3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging (Albany NY). 2018;10:3806–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang P, Yang Z, Ye T, et al. lncTUG1/miR-144-3p affect the radiosensitivity of esophageal squamous cell carcinoma by competitively regulating c-MET. J Exp Clin Cancer Res. 2020;39:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sun L, Bian G, Meng Z, et al. MiR-144 inhibits uveal melanoma cell proliferation and invasion by regulating c-Met expression. PLoS One. 2015;10:e0124428. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [38].Saini S, Maker AV, Burman KD, et al. Molecular aberrations and signaling cascades implicated in the pathogenesis of anaplastic thyroid cancer. Biochim Biophys Acta. 2019;1872:188262. [DOI] [PubMed] [Google Scholar]
- [39].Li Q, Dong C, Cui J, et al. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res. 2018;37:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Luo Y, He Y, Ye X, et al. High expression of long noncoding RNA HOTAIRM1 is associated with the proliferation and migration in pancreatic ductal adenocarcinoma. Pathol Oncol Res. 2019;25(4):1567–1577. [DOI] [PubMed] [Google Scholar]
- [41].Chao H, Zhang M, Hou H, et al. HOTAIRM1 suppresses cell proliferation and invasion in ovarian cancer through facilitating ARHGAP24 expression by sponging miR-106a-5p. Life Sci. 2020;243:117296. [DOI] [PubMed] [Google Scholar]
- [42].Ren T, Hou J, Liu C, et al. The long non-coding RNA HOTAIRM1 suppresses cell progression via sponging endogenous miR-17-5p/B-cell translocation gene 3 (BTG3) axis in 5-fluorouracil resistant colorectal cancer cells. Biomed Pharmacother. 2019;117:109171. [DOI] [PubMed] [Google Scholar]
- [43].Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. [DOI] [PubMed] [Google Scholar]
- [44].Mo S, Zhang L, Dai W, et al. Antisense lncRNA LDLRAD4-AS1 promotes metastasis by decreasing the expression of LDLRAD4 and predicts a poor prognosis in colorectal cancer. Cell Death Dis. 2020;11:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yin J, Luo W, Zeng X, et al. UXT-AS1-induced alternative splicing of UXT is associated with tumor progression in colorectal cancer. Am J Cancer Res. 2017;7:462–472. [PMC free article] [PubMed] [Google Scholar]
- [46].Yu Y, Yan R, Chen W, et al. Long non-coding RNA SLC26A4-AS1 exerts antiangiogenic effects in human glioma by upregulating NPTX1 via NFKB1 transcriptional factor. Febs J. 2020 doi: 10.1111/febs.15325. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- [47].Vrakas CN, Herman AB, Ray M, et al. RNA stability protein ILF3 mediates cytokine-induced angiogenesis. Faseb J. 2019;33:3304–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li Y, Yan G, Zhang J, et al. LncRNA HOXA11-AS regulates calcium oxalate crystal-induced renal inflammation via miR-124-3p/MCP-1. J Cell Mol Med. 2020;24:238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


