Abstract
Pantala flavescens is the world’s most abundant and widely distributed dragonfly and with its outstanding migratory capacity an important model system to study insect migration at the evolutionary base of winged insects. We here report on the first complete mitochondrial genome (mitogenome) of P. flavescens sampled from a population in Rufiji River, Tanzania. The mitogenome is 14,853 bp long with an AT-biased base composition (72.7% A + T) and encodes a typical set of 13 protein-coding genes (PCGs), 22 tRNAs, and two rRNAs. The control region (CR) (171 bp) is the shortest reported in any anisopteran odonate, so far. Phylogenetic analyses support the placement of P. flavescens within the Libellulidae.
Keywords: Mitochondrial genome, Odonata, Pantala flavescens, A + T rich control region, migratory insect
The migratory odonate Pantala flavescens is described as the most successful odonate species in terms of abundance and geographic distribution (e.g. Dijkstra and Clausnitzer 2014). This status is likely achieved by its remarkable migratory capacity with long, multigenerational migration routes of up to 18,000 km. This is the longest known distance compared to other migratory insects (Hobson et al. 2012). The exceptional ability for long-distance migration (possibly related to passive dispersal mediated by human transport or wind) explains why P. flavescens is even present on one of the world’s most remoted islands, the Easter Island, Chile (Samways and Osborn 1998). The analysis of whole mitogenomes from migratory as well as resident P. flavescens populations would be a promising approach to further study the genetic pathways linked to the migratory behavior in this species and would substantially extend insights gained from previous single marker gene analyses (Troast et al. 2016; Alvial et al. 2019). We here report on the first mitogenome of P. flavescens from a migratory population in Tanzania.
A standard phenol–chloroform protocol by Hadrys et al. (1992) was used to extract total genomic DNA from a mid-leg of an individual collected at Rufiji River, Tanzania (−7.854316; 38.428008) in the frame of the BIOTA AFRICA project in 2002. The tissue and DNA samples (PFLRRT32002) are stored in the Institute of Animal Ecology, University of Veterinary Medicine Hannover, Foundation, Germany (contact person is the first author). PCR primers were designed based on transcriptomic data (PRJNA239794) from a migratory P. flavescens specimen from China. Sanger sequencing was conducted at DNA Analysis Facility at Yale University, New Haven, CT and Geneious version 8.1.8 (https://www.geneious.com) was used to assemble the overlapping mitogenome sequences. For mitochondrial genome annotation the MITOS WebServer (Bernt et al. 2013) was used and results were manually checked using BLAST (Altschul et al. 1990). The phylogenetic position of P. flavescens was assessed in the context of all available anisopteran mitogenomes to date (10/2020) mined from GenBank. Protein-coding genes (PCGs) were aligned using MAFFT version 7.017 (Kumar et al. 2016) and subsequently concatenated. Maximum likelihood (ML) analyses were performed using the best-fitting model GTR + F + I + G4 under the Bayesian information criterion detected via ModelFinder (Kalyaanamoorthy et al. 2017) with IQ-tree keeping default settings (Nguyen et al. 2015). Ischnura elegans (Zygoptera, NC_031824) served as outgroup.
The complete circular mitochondrial genome sequence of P. flavescens (GenBank Accession No. MW256717) with a length of 14,853 bp is the most compact known mitogenome among dragonflies (Anisoptera). The standard metazoan gene content of 37 genes is identically arranged as in other odonate mitochondrial genomes (e.g. Simon and Hadrys 2013; Feindt et al. 2016). The overall base frequency is AT-biased (72.7%). Regarding invertebrate mitochondrial start codons, ATT (nad2, atp8), ATA (cox1, nad3, nad4, and atp6), ATC (nad6), ATG (cob, cox2, cox3, and nad4l), and TTG (nad1) standard codons are found. All PCGs possess TAA as complete stop codon except for cox2 and nad5, which possess a single T which is likely completed by post-transcriptional polyadenylation. All tRNA secondary structures exhibited a typical clover-leaf structure. However, trnS1 lacked the dihydrouridine (DHU) arm. The AT rich control region (CR) is 171 bp long and therefore the shortest observed CR among all Anisoptera.
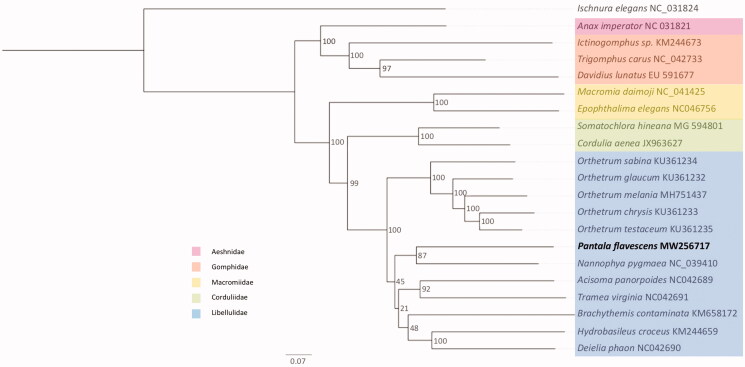
The phylogeny of all available anisopteran mitogenomes is shown in Figure 1. All families within Anisoptera were consistently supported as monophyletic groups if represented by more than one species. Within the Libellulidae, P. flavescens is placed as a sister to Nannophya pygmaea.
Figure 1.
Maximum likelihood (ML) tree for 20 Anisoptera using 13 concatenated mitochondrial protein-coding genes (11,375 bp). Maximum likelihood bootstrap support values are given at each node. The Zygoptera Ischnura elegans served as outgroup and the here presented mitogenome of Pantala flavescens is given in bold.
The first complete mitochondrial genome of P. flavescens from a migratory population from Tanzania paves the way for further comparative approaches, which should include both, other migratory populations (e.g. from Asia or South America) as well as the so far only known non-migratory population from Easter Island, Chile. Such comparative mitogenomic approaches might help to further reconstruct dispersal routes and migration history of this unique odonate model species.
Acknowledgments
We thank Sandra Damm for the tissue sampling as part of the BIOTA AFRICA project funded by the German Federal Ministry of Education and Research.
Funding Statement
This study was supported in part by the Deutscher Akademischer Austauschdienst (DAAD) PROMOS fellowship for FJD to conduct part of his research at The Sackler Institute for Comparative Genomics, American Museum of Natural History, New York City, NY, United States of America supervised by Dr. Rob DeSalle and Bundesministerium für Bildung und Forschung.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW256717.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Alvial IE, Vargas HA, Marinov M, Esquivel C, Araya J, Araya-Donoso R, Vila I, Véliz D.. 2019. Isolation on a remote island: genetic and morphological differentiation of a cosmopolitan odonate. Heredity. 122(6):893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Dijkstra KDB, Clausnitzer V.. 2014. The dragonflies and damselflies of eastern Africa: handbook for all Odonata from Sudan to Zimbabwe. Tervuren, Belgium: Royal Museum for Central Africa. [Google Scholar]
- Feindt W, Osigus H-J, Herzog R, Mason CE, Hadrys H.. 2016. The complete mitochondrial genome of the neotropical helicopter damselfly Megaloprepus caerulatus (Odonata: Zygoptera) assembled from next generation sequencing data. Mitochondrial DNA Part B. 1(1):497–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadrys H, Balick M, Schierwater B.. 1992. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol Ecol. 1(1):55–63. [DOI] [PubMed] [Google Scholar]
- Hobson KA, Anderson RC, Soto DX, Wassenaar LI.. 2012. Isotopic evidence that dragonflies (Pantala flavescens) migrating through the Maldives come from the northern Indian subcontinent. PLOS One. 7(12):e52594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samways M, Osborn R.. 1998. Divergence in a transoceanic circumtropical dragonfly on a remote island. J Biogeography. 25(5):935–946. [Google Scholar]
- Simon S, Hadrys H.. 2013. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol Phylogenet Evol. 69(2):393–403. [DOI] [PubMed] [Google Scholar]
- Troast D, Suhling F, Jinguji H, Sahlén G, Ware J.. 2016. A global population genetic study of Pantala flavescens. PLoS One. 11(3):e0148949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW256717.