ABSTRACT
Sterols are essential lipids for plant growth, and the sterol content is tightly regulated by a fail-safe system consisting of two processes: 1) suppression of excess sterol production by a negative regulator of sterol biosynthesis (HIGR STEROL ESTER 1, HISE1), and 2) conversion of excess sterols to sterol esters by PHOSPHOLIPID STEROL ACYLTRANSFERASE 1 (PSAT1) in Arabidopsis thaliana. The hise1-3 psat1-2 double mutant has a 1.5-fold higher sterol content in leaves than the wild type; this upregulates the expression of stress-responsive genes, leading to disruption of cellular activities in leaves. However, the effects of excess sterols on seeds are largely unknown. Here, we show that excess sterols cause multiple defects in seeds. The seeds of hise1-3 psat1-2 plants had a higher sterol content than wild-type seeds and showed a deeper color than wild-type seeds because of the accumulation of proanthocyanidin. The seed coat in the hise1-3 psat1-2 mutant was abnormally wrinkled. Seed coat formation is accompanied by cell death-mediated shrinkage of the inner integument. In the hise1-3 psat1-2 mutant, transmission electron microscopy showed that shrinkage of the integument was impaired, resulting in a thick seed coat and delayed seed germination. Moreover, psat1-2 and hise1-3 psat1-2 seeds displayed defective imbibition. Taken together, the results suggest that excess sterols impair proper seed coat formation, thereby inhibiting seed germination.
KEYWORDS: Arabidopsis thaliana, seeds, seed coat, HIGH STEROL ESTER 1, PHOSPHOLIPID STEROL ACYL TRANSFERASE 1, sterols, sterol esters
Sterols are essential lipids in plants that function as membrane components and signaling molecules. Sterol production is under strict regulation, as excess sterols are toxic to plants.1,2 We previously identified a negative regulator of sterol biosynthesis, HIGH STEROL ESTER 1 (HISE1; accession At1g60995).2 The rate-limiting sterol biosynthesis enzyme 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR)2 accumulates in the leaves of the Arabidopsis thaliana hise1 mutant.3 HISE1 negatively regulates the protein levels of HMGR to maintain sterol homeostasis.
PHOSPHOLIPID STEROL ACYL TRANSFERASE 1 (PSAT1; accession At1g04010) prevents the overaccumulation of sterols by converting excess sterols into sterol esters, which are then segregated into sterol ester (SE) bodies.2 In previous work from our group, we defined the system involving HISE1 and PSAT1 as a fail-safe regulatory mechanism that maintains sterol homeostasis. Deletion of the genes encoding HISE1 and PSAT1 causes a severe growth defect in Arabidopsis, suggesting that this system is essential for plant growth.2,4 This growth defect is associated with a 1.5-fold higher content of sterols in hise1-3 psat1-2 leaves than in wild-type leaves.2 Thus, the hise1-3 psat1-2 double mutant could be used as a model for investigating the effects of excess sterols on plant growth. In this study, we focused on the phenotype of hise1-3 psat1-2 seeds and showed that the combined deficiency of HISE1 and PSAT1 affects seed development.
We compared dry seeds of hise1-3 psat1-2 double mutants with those of the wild type and hise1-3 and psat1-2 single mutants (Figure 1a). Single mutant seeds were similar in shape to wild-type seeds (Figure 1a–d). The major axis of hise1-3 psat1-2 seeds was slightly longer than that of wild-type and single mutant seeds (Figure 1d); however, the minor axis (Figure 1b) and seed weight (Figure 1c) did not differ between the wild type and hise1-3 psat1-2. These results suggest that hise1-3 psat1-2 seeds were slightly longer than wild-type and single mutant seeds. Furthermore, hise1-3 psat1-2 seeds were deeper in color than wild-type and single mutant seeds (Figure 1a). In Arabidopsis, seed color is derived from proanthocyanidins in the seed coat.5 The proanthocyanidin content of seeds was measured using a simple quantitation method described previously.5 The hise1-3 psat1-2 seeds contained less acetone-soluble proanthocyanidins and more acetone-insoluble proanthocyanidins than wild-type seeds (Figure 1e). The total proanthocyanidin content of hise1-3 psat1-2 seeds was higher than that of wild-type seeds (Figure 1e). These results suggest that hyperaccumulation of proanthocyanidins in hise1-3 psat1-2 seeds was responsible for the deep seed color. The sterol content of seeds was measured using a colorimetric assay with a Cholesterol Quantitation Kit (Sigma-Aldrich). Total lipids were extracted from seeds as described previously.6 The sterol content of hise1-3 psat1-2 seeds was 6-fold higher than that of wild-type and single mutant seeds (Figure 1f), suggesting that hise1-3 psat1-2 seeds have excess amounts of sterols.
Figure 1.
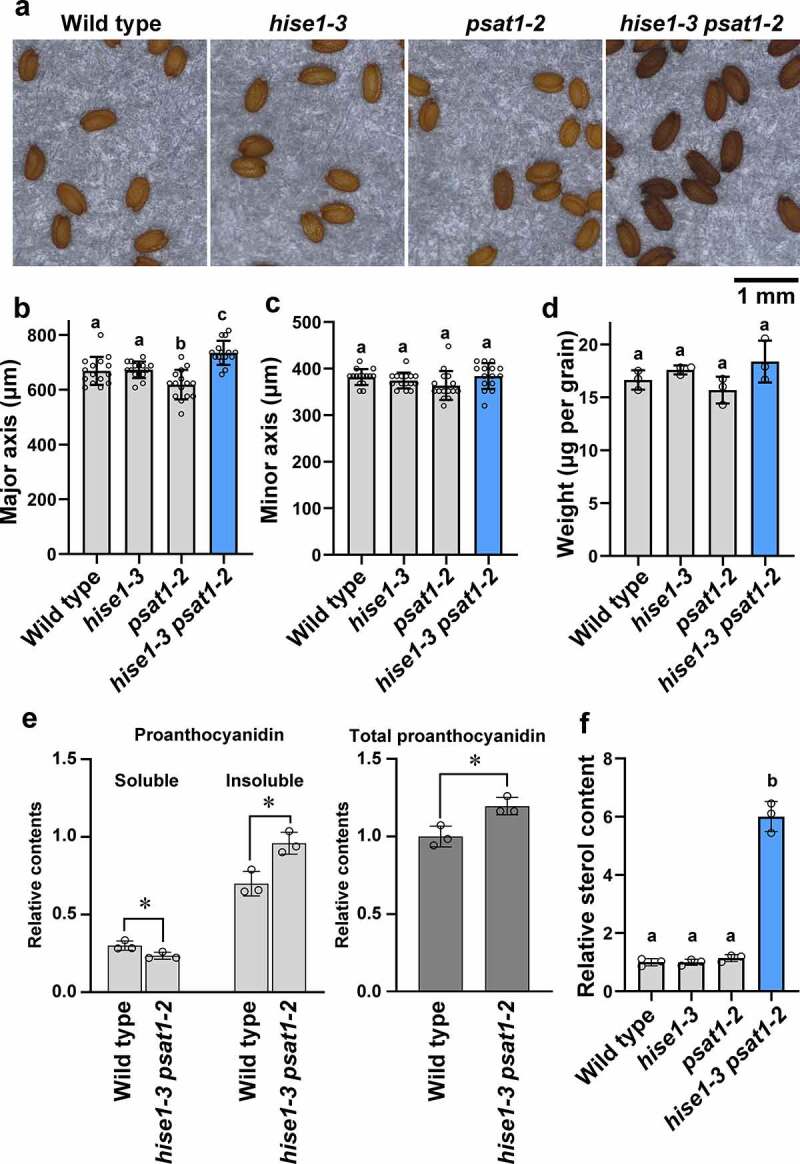
The seeds of an Arabidopsis thaliana double mutant, hise1-3 psat1-2, exhibit deeper color and higher contents of proanthocyanidins than those of the wild type. (a) Photographs of dry seeds of the wild type, hise1-3, psat1-2, and hise1-3 psat1-2. (b, c) Average values of the major axis (b) and minor axis (c) of 15 seed grains of each genotype. Data represent the mean ± standard deviation (SD; n = 15). Different letters indicate significant differences (p < .05; Tukey’s test). (d) Average dry weight of one seed grain. To measure seed weight, 150 seeds were collected and weighed with an electronic balance. The weight of one seed was calculated. Three biological replicates were performed. Data represent the mean ± SD (n = 3). Different letters indicate significant differences (p < .05; Tukey’s test). (e) Relative contents of acetone-soluble (soluble), acetone-insoluble (insoluble), and total proanthocyanidins in wild-type and hise1-3 psat1-2 seeds in three biological replicates. Data represent the mean ± SD (n = 3). Asterisks indicate significant differences between wild-type and hise1-3 psat1-2 seeds (p < .05; two-tailed Student’s t-test). The relative total proanthocyanidin content was calculated as follows: acetone-soluble proanthocyanidin content plus acetone-insoluble proanthocyanidin content was defined as total proanthocyanidin content. The relative total proanthocyanidin content of wild-type seeds was set at 1.0. (f) Relative content of sterols in wild-type, hise1-3, psat1-2, and hise1-3 psat1-2 dry seeds. The sterol content of total lipid extracts from seeds was measured with a Cholesterol Quantitation Kit (Sigma-Aldrich). In this experiment, cholesterol esterase treatment was not performed. The relative content of sterols of wild-type seeds was set at 1.0. Three biological replicates were performed. Data represent the mean ± SD (n = 3). Different letters indicate significant differences (p < .05; Tukey’s test)
To investigate the structure of dry seeds in detail, we performed scanning electron microscopy analysis. The seed coat surface did not differ between the single mutants and the wild type (Figure 2a). However, the seed coat of the hise1-3 psat1-2 double mutant had irregular formations compared with that of the wild type (Figure 2a). The columella of the hise1-3 psat1-2 seed coat exhibited irregular wrinkles (Figure 2a). To examine the seed coat cell layers, we performed transmission electron microscopy analysis of dry seeds. In Arabidopsis, the seed coat is composed of two cell layers in the outer integument (oi1 and oi2) and three cell layers in the inner integument (ii1–3).7 Analysis of dry seeds showed that the wild-type and single mutant seed coat cell layers were shrunken and forming a thin pigment layer (Figure 2b). However, two cell layers in the seed coat of hise1-3 psat1-2 were not shrunken (Figure 2b, hise1-3 psat1-2). We concluded that the two cell layers corresponded to the oi1 and ii1 layers. Analysis of intracellular structures showed that in the hise1-3 psat1-2 mutant, the cells of the oi1 and ii1 layers were irregular compared with those of the perisperm-endosperm envelope (Figure 2c). These results suggest that the irregular formation of cell layers in the seed coat of hise1-3 psat1-2 seeds results in a wrinkled surface of dry seeds, and an excess of sterols may cause the abnormal development of hise1-3 psat1-2 seeds.
Figure 2.
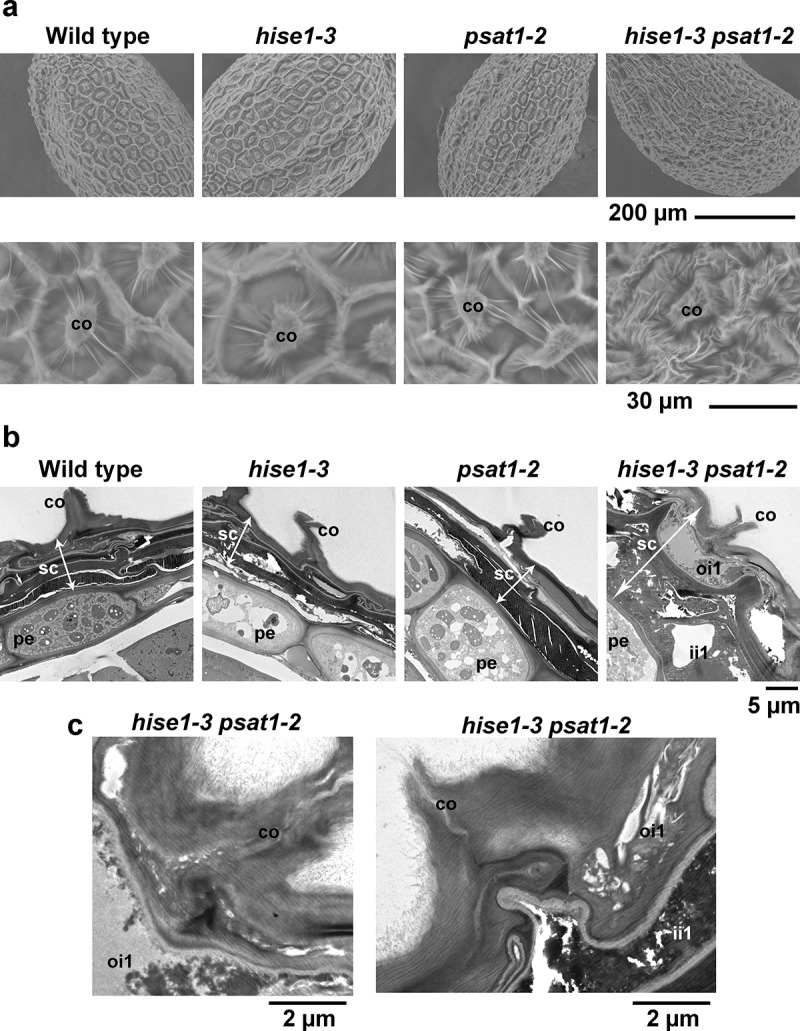
Seeds of the double mutant hise1-3 psat1-2 have an abnormal seed coat. (a) Scanning electron micrographs of the surface of wild-type, hise1-3, psat1-2, and hise1-3 psat1-2 seeds. Six seed grains of each genotype were inspected with similar results. co, columella. (b) Transmission electron micrographs of the seed coat of wild-type, hise1-3, psat1-2, and hise1-3 psat1-2 seeds. co, columella; pe, perisperm endosperm; sc, seed coat; ii1, inner integument 1; oi1, outer integument 1. (c) Transmission electron micrographs of the seed coat of hise1-3 psat1-2 seeds at high magnification. The left panel is the same cell as that of Figure 2(b) (hise1-3 psat1-2). co, columella; ii1, inner integument 1; oi1, outer integument 1
To investigate the effects of excess sterol accumulation on seed viability, we determined the germination rate of wild-type and mutant seeds. The germination rate of wild-type and single mutant seeds was 90% at 3 days after sowing and 100% at 10 days after sowing (Figure 3a), whereas that of hise1-3 psat1-2 seeds was <10% at 3 days after sowing and >90% at 10 days after sowing (Figure 3a). Thus, hise1-3 psat1-2 seeds exhibited delayed germination.
Figure 3.
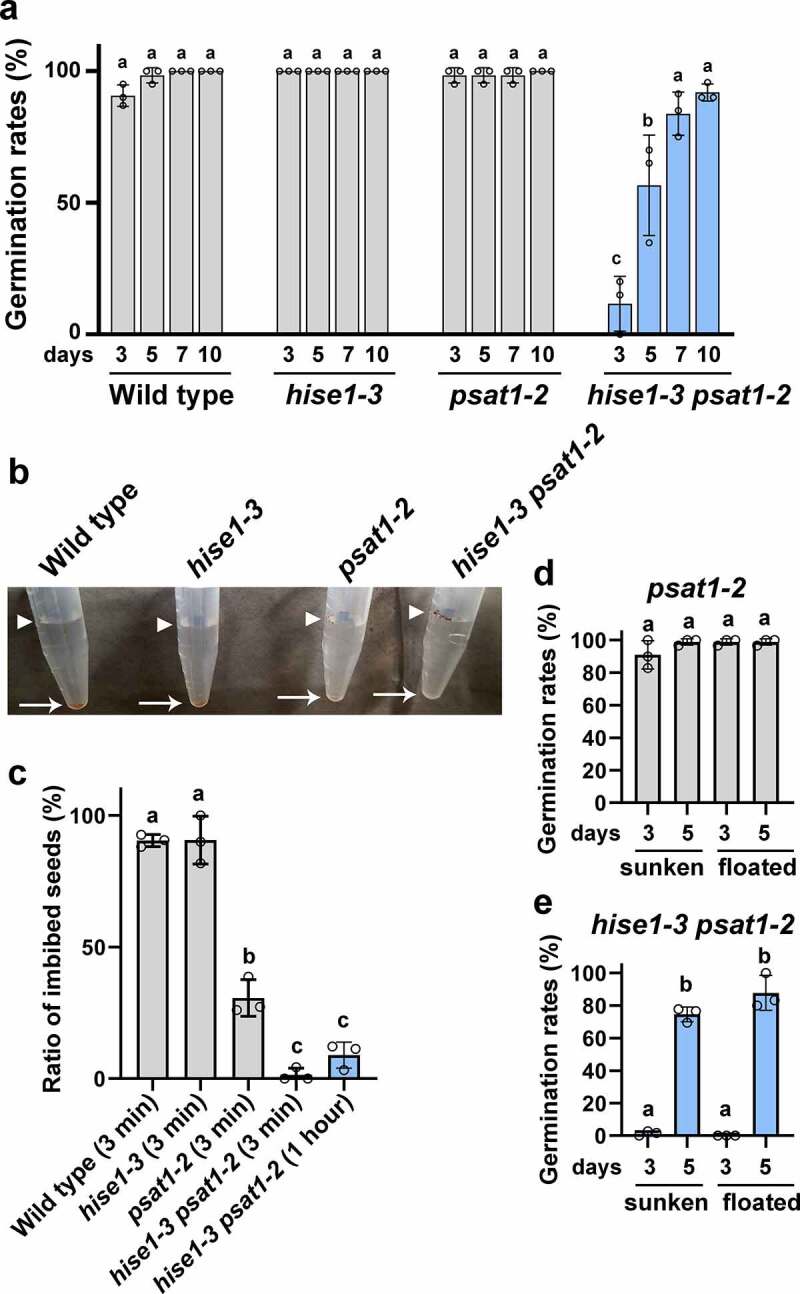
Seeds of psat1-2 and hise1-3 psat1-2 exhibit defective seed imbibition. (a) Rates of seed germination at 3, 5, 7, and 10 days after sowing. Data represent the mean ± SD of three biological replicates (n = 3). Different letters indicate significant differences (p < .05; Tukey’s test). (b) A photograph showing seed imbibition at 3 min after imbibition. Arrowheads indicate un-imbibed seeds, which remained floating. Arrows indicate imbibed seeds that sunk to the bottom of the microtubes. (c) Rates of sunken seeds of wild-type, hise1-3, psat1-2, and hise1-3 psat1-2 at 3 min after imbibition, and rates of sunken seeds of hise1-3 psat1-2 at 1 h after imbibition. Data indicate the mean ± SD of three biological replicates (n = 3). Different letters indicate significant differences (p < .05; Tukey’s test). (d) Germination rates of sunken and floating seeds of psat1-2 at 3 and 5 days after sowing. Seeds of the psat1-2 mutant were divided into sunken and floating groups at 3 min after imbibition, and the germination rates were measured. Three biological replicates were performed. Data represent the mean ± SD (n = 3). Different letters indicate significant differences (p < .05; Tukey’s test). (e) Germination rates of sunken and floating seeds of hise1-3 psat1-2 at 3 and 5 days after sowing. Seeds of the hise1-3 psat1-2 double mutant were divided into sunken and floating groups at 1 h after imbibition, and the germination rates were measured. Three biological replicates were performed. Data represent the mean ± SD (n = 3). Different letters indicate significant differences (p < .05; Tukey’s test)
Next, we investigated the imbibition ability of wild-type and mutant seeds. Wild-type dry seeds initially floated on water, and within 3 min, approximately 90% of seeds had imbibed water and sunk to the bottom of the container (Figure 3b,c). Similar to wild-type seeds, approximately 90% of hise1-3 seeds rapidly sunk to the bottom of the container (Figure 3b,c), suggesting that the imbibition ability of hise1-3 seeds was not impaired. However, >70% of psat1-2 seeds and 99% of hise1-3 psat1-2 seeds floated on water (Figure 3b,c). The proportion of imbibed hise1-3 psat1-2 seeds that remained on the surface of the water did not differ between 3 min and 1 h after imbibition (Figure 3c). These results indicate that the imbibition ability of double mutant seeds was impaired.
To investigate the correlation between seed imbibition and seed germination, psat1-2 seeds were divided into sunken and floating seeds at 3 min after imbibition, and the germination rates were measured. There was no difference between the germination rates of the sunken and floating seeds (Figure 3d), suggesting that psat1-2 seeds with impaired imbibition retain the capacity to germinate. When the hise1-3 psat1-2 seeds were divided into sunken and floating seeds at 1 h after imbibition, there was no difference in the germination rate between the two groups (Figure 3e). This indicates that the delayed germination is present to a similar extent in sunken hise1-3 psat1-2 seeds, which have an intact imbibition ability, and in floating hise1-3 psat1-2 seeds. These results suggest that the germination delay of hise1-3 psat1-2 seeds is derived from the seed coat abnormality and not from the impaired imbibition ability.
In this study, the hise1-3 psat1-2 double mutant showed severe defects in seed coat formation, seed germination, and seed imbibition. Microarray data (DART, http://dandelion.liveholonics.com/dart/index.php; EXPRESSION ATLAS OF ARABIDOPSIS DEVELOPMENT) indicated that HISE1 and PSAT1 genes are expressed during seed development. These results support the hyperaccumulation of sterols in hise1-3 psat1-2 seeds. The present results provide novel insights into the effects of excess sterols on seed physiology, suggesting that excess sterols cause irregular formation of the seed coat (Figure 2), which leads to delayed germination (Figure 3a). Excess sterols may also affect seed imbibition, although this effect does not directly influence seed germination. Overall, the present results suggest that excess sterols reduce the germination ability of dry seeds.
Programmed cell death causes cell shrinkage in seed coat layers.8 A previous study in A. thaliana showed that a member of the Vacuolar Processing Enzyme (VPE) family, δVPE, which is expressed in ii2 and ii3, is involved in programmed cell death in these two layers.9 A δVPE deficient mutant shows delayed programmed cell death in ii2 and ii3.9 Compared with the wild type, hise1-3 psat1-2 seeds showed cell shrinkage in oi2, ii2, and ii3, but not in oi1 and ii1. This suggests that δVPE is functional in hise1-3 psat1-2 seeds and mediates cell death in ii2 and ii3. However, excess sterols in hise1-3 psat1-2 seeds may inhibit programmed cell death in oi1 and ii1 layers in a δVPE-independent manner. Excess sterols disturb cellular activity in leaves and roots.4 Therefore, it is possible that the excess sterols affect cellular activity in the double mutant, thereby altering the programmed cell death process. This abnormality may inhibit cell shrinkage in the hise1-3 psat1-2 seed coat. We suggest that the sterol content is regulated to induce programmed cell death appropriately in the seed coat. In addition, cells in the ii1 layer of the seed coat are involved in proanthocyanidin accumulation.8 This finding together with the present results suggest that the deep coloration and proanthocyanidin hyperaccumulation in hise1-3 psat1-2 seeds are caused by a deficiency in cell shrinkage in the ii1 layer of the hise1-3 psat1-2 seed coat.
Acknowledgments
We thank Kazuo Ebine (NIBB) for technical assistance.
Funding Statement
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) [grant numbers 15H05776 and 22000014 to I.H.-N., 18H02470 and 19H05675 to T.U., and 16K18834 and 19K05809 to T.L.S.]; Leading Initiative for Excellent Young Researchers (LEADER) from the Ministry of Education, Culture, Sports, Science and Technology in Japan (MEXT) [grant number J16HJ00026 to T.L.S.]; Kato Memorial Bioscience Foundation to T.L.S.; the SUNBOR GRANT of Suntory Foundation for Life Science to T.L.S.; the Phytochemical Plant Molecular Science of Strategic Priority Research Promotion Program from Chiba University to T.L.S.; the NIBB Collaborative Research Program (17–301, 18–301, 19-302, 20-301) to T.L.S.; and the Hirao Taro Foundation of KONAN GAKUEN for Academic Research to I.H.-N.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Bouvier-Nave P, Berna A, Noiriel A, Compagnon V, Carlsson AS, Banas A, Stymne S, Schaller H.. Involvement of the phospholipid sterol acyltransferase1 in plant sterol homeostasis and leaf senescence. Plant Physiol. 2010;152(1):1–4. doi: 10.1104/pp.109.145672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimada TL, Shimada T, Okazaki Y, Higashi Y, Saito K, Kuwata K, Oyama K, Kato M, Ueda H, Nakano A.. HIGH STEROL ESTER 1 is a key factor in plant sterol homeostasis. Nat Plants. 2019;5(11):1154–1166. doi: 10.1038/s41477-019-0537-2. [DOI] [PubMed] [Google Scholar]
- 3.Maurey K, Wolf F, Golbeck J. 3-Hydroxy-3-Methylglutaryl coenzyme a reductase activity in ochromonas malhamensis: a system to study the relationship between enzyme activity and rate of steroid biosynthesis. Plant Physiol. 1986;82:523–527. doi: 10.1104/pp.82.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimada TL, Yamaguchi K, Shigenobu S, Takahashi H, Murase M, Fukuyoshi S, Hara-Nishimura I. Excess sterols disrupt plant cellular activity by inducing stress-responsive gene expression. J Plant Res. 2020;133(3):383–392. doi: 10.1007/s10265-020-01181-4. [DOI] [PubMed] [Google Scholar]
- 5.Kitamura S, Matsuda F, Tohge T, Yonekura-Sakakibara K, Yamazaki M, Saito K, Narumi I. Metabolic profiling and cytological analysis of proanthocyanidins in immature seeds of Arabidopsis thaliana flavonoid accumulation mutants. Plant J. 2010;62(4):549–559. doi: 10.1111/j.1365-313X.2010.04174.x. [DOI] [PubMed] [Google Scholar]
- 6.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 7.Kunieda T, Shimada T, Kondo M, Nishimura M, Nishitani K, Hara-Nishimura I. Spatiotemporal secretion of PEROXIDASE36 is required for seed coat mucilage extrusion in Arabidopsis. Plant Cell. 2013;25:1355–1367. doi: 10.1105/tpc.113.110072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haughn G, Chaudhury A. Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci. 2005;10:472–477. doi: 10.1016/j.tplants.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Nakaune S, Yamada K, Kondo M, Kato T, Tabata S, Nishimura M, Hara-Nishimura I. A vacuolar processing enzyme, deltaVPE, is involved in seed coat formation at the early stage of seed development. Plant Cell. 2005;17:876–887. doi: 10.1105/tpc.104.026872. [DOI] [PMC free article] [PubMed] [Google Scholar]


