SUMMARY
Morphological plasticity is a key virulence trait for many fungal pathogens. For the opportunistic fungal pathogen Candida albicans, transitions among yeast, pseudohyphal, and hyphal forms are critical for virulence, because the morphotypes play distinct roles in the infection process. C. albicans morphogenesis is induced in response to many host-relevant conditions and is regulated by complex signaling pathways and cellular processes. Perturbation of either cell-cycle progression or protein homeostasis induces C. albicans filamentation, demonstrating that these processes play a key role in morphogenetic control. Regulators such as cyclin-dependent kinases, checkpoint proteins, the proteasome, the heat shock protein Hsp90, and the heat shock transcription factor Hsf1 all influence morphogenesis, often through interconnected effects on the cell cycle and proteostasis. This review highlights the major cell-cycle and proteostasis regulators that modulate morphogenesis and discusses how these two processes intersect to regulate this key virulence trait.
INTRODUCTION
Candida albicans is a commensal fungus frequently found as a benign member of the human mucosal and skin microbiota. Studies have estimated that as many as 70% of healthy individuals are colonized with C. albicans at any given time (Noble et al., 2017). However, C. albicans is an opportunistic pathogen that can cause superficial infections in immunocompetent individuals, as well as life-threatening systemic disease in immunocompromised patients (Polvi et al., 2015). As such, C. albicans is a leading cause of hospital-acquired infections, particularly infecting patients with indwelling medical devices or compromised immune systems from transplant surgeries or cancer treatments. C. albicans bloodstream infections lead to disseminated candidiasis as the fungus invades the internal organs, with attributable mortality rates often exceeding 40% even with current antifungal therapies (Kullberg and Arendrup, 2015). C. albicans imposes a major burden on human health, causing ~700,000 deaths per year globally because of invasive candidiasis (Bongomin et al., 2017).
The success of C. albicans as a pathogen can be largely attributed to its armamentarium of virulence factors, including its capacity for rapid evolution of resistance to the three classes of antifungals used to treat invasive infections and its ability to form drug-resistant biofilms on medical devices. One of the most influential C. albicans virulence traits is its ability to transition among morphological states: yeast, hyphal, and pseudohyphal forms. In its yeast form, C. albicans persists as round or oval cells that arise from budding, whereas hyphal growth results in the production of tubular cells that remain firmly attached following cytokinesis with no constriction at the site of septation. Pseudohyphal growth is thought to represent an intermediate state between yeast and hyphal filaments and is characterized by chains of ellipsoidal cells with constriction between elongated buds (Carlisle et al., 2009; Noble et al., 2017; Sudbery et al., 2004). Yeast and filamentous forms (hyphae and pseudohyphae) have distinct roles in the infection process: yeasts are crucial for colonization and dissemination through the bloodstream, and filaments enable tissue invasion and deep-seated infection (Saville et al., 2003). Filaments are equipped with additional virulence factors such as adhesins, tissue-degrading proteases, antioxidant proteins, and the peptide toxin candidalysin, which promotes damage to host cells during infection (Noble et al., 2017). As a result of these different contributions to infections, morphogenetic flexibility is critical for C. albicans virulence, and most mutants that are unable to undergo morphogenesis have attenuated virulence in systemic models of infection (Noble et al., 2010). Although C. albicans can undergo other morphological transitions that affect virulence, such as growing as chlamydospores or the yeast-like opaque, gray, and gastrointestinally induced transition (GUT) morphotypes (Noble et al., 2017), this review focuses on the yeast-to-filament transition.
C. albicans transitions to filamentous growth are induced in response to diverse environmental and host-relevant stimuli, which are sensed and transduced through complex, often interconnected, signaling pathways (Sudbery, 2011). One of the most central morphogenetic signaling pathways is the cyclic AMP-protein kinase A (cAMP-PKA) pathway, which is required for filamentation in response to most inducing cues, including serum (Noble et al., 2017; Shapiro et al., 2011; Sudbery, 2011). Inducing cues can activate the guanosine triphosphatase (GTPase) Ras1 and stimulate the activity of the adenylyl cyclase Cyr1, inducing cAMP production. Cyr1 can also be activated directly by cues such as peptidoglycan independent of Ras1. Elevated cAMP levels are important in activating the PKA complex, which consists of two catalytic subunits, Tpk1 and Tpk2, and a regulatory subunit, Bcy1. PKA is thought to phosphorylate the downstream transcription factor Efg1, which activates a transcriptional program governing morphogenesis (Shapiro et al., 2011). Although Efg1 is an integral regulator of filamentation induced through this pathway, it is dispensable for filamentation in response to some cues that depend on cAMP-PKA signaling, implicating additional, unidentified factors downstream of PKA. As an important virulence trait, morphogenesis is tightly regulated by many cellular processes and regulatory circuits (Shapiro et al., 2011).
Maintenance of cellular signaling is crucial for coordinated cellular growth and cell-cycle progression. Eukaryotic cells cycle through four distinct phases: G1, synthesis (S), G2, and mitosis (M) followed by cytokinesis. In addition to the structural differences between yeast and hyphal forms, the morphotypes are distinct in how they progress through the cell cycle. In the yeast form, bud emergence is coordinated with the transition from G1 to S phase and follows the formation of the septin ring. Nuclear division occurs across the mother-bud neck, and cytokinesis results in fully separated rounded cells that disperse after septation (Sudbery, 2011). A similar progression of cell-cycle events occurs in pseudohyphae, but the cells remain attached and elongate because of a prolonged G2 phase (Sudbery, 2011). During hyphal growth, a germ tube forms before the G1-to-S transition and continues to grow, later becoming the site of nuclear division. One daughter nucleus migrates back into the mother cell, but cytokinesis does not result in separation or constriction of the cells. Though the subapical compartment remains in G1, the apical compartment continues progressing through the cell cycle, resulting in elongation of the hyphal tip to form tubular cells (Sudbery, 2011). Progression through the phases of cell cycles in the different forms is coordinated by many proteins, including cyclins, cyclin-dependent kinases (CDKs), and checkpoint proteins (Berman, 2006). Proper cell-cycle progression is integral for regulation of morphogenesis, because depletion of genes that influence cell cycle, including genes involved in DNA replication, chromosome segregation, nuclear division, and DNA repair, induces constitutive filamentation (O’Meara et al., 2015). Filamentation is also induced by pharmacological treatments that cause cell-cycle arrest, reinforcing the connection between these two processes (Berman, 2006; Pérez-Martín et al., 2016).
Regulation of filamentation is inextricably tied to protein homeostasis. Molecular chaperones aid in folding nascent proteins, refolding misfolded proteins, and targeting damaged and terminally misfolded proteins for degradation via the ubiquitin-proteasome pathway (UPP). The molecular chaperone and heat shock protein Hsp90 mediates proteostasis by binding diverse client proteins, regulating their stability and function (Taipale et al., 2010). The heat shock transcription factor Hsf1 also regulates proteostasis via transcriptional regulation of chaperone genes, including HSP90 (Nicholls et al., 2009). In C. albicans, the proteasome, Hsp90, and Hsf1 have been implicated as key regulators of morphogenesis, because their misregulation leads to filamentation in the absence of inducing cues (Hossain et al., 2020; Shapiro et al., 2009; Veri et al., 2018a). This reinforces that proteostasis is critical for maintaining the yeast phase of growth while affecting cell-cycle progression.
It is clear that the cell-cycle and proteostasis machineries contribute to the regulation of filamentation in C. albicans; however, how these processes orchestrate morphogenesis remain elusive. In this review, we highlight the impact of regulators of the cell cycle and proteostasis on C. albicans morphogenesis and explore the connections between these two critical cellular processes.
Cyclins and CDK complexes control cell morphology
During yeast-phase growth, cell morphology is tightly linked to cell-cycle progression through the highly coordinated activity of CDK complexes (Lew and Reed, 1993) (Figure 1). The essential C. albicans CDK Cdc28 is the central hub, with its activity regulated by the temporal binding of cyclins. During G1 of the C. albicans cell cycle, the G1 cyclins Ccn1 and Cln3 are expressed and their association with Cdc28 promotes apical growth and bud elongation (Bachewich and Whiteway, 2005; Chapa y Lazo et al., 2005). As the cell cycle progresses, G1 cyclins are degraded and replaced by the G2 cyclins Clb2 and Clb4, which bind to Cdc28 and suppress polarized growth, driving isotropic bud expansion (Bensen et al., 2005). This coordinated balance between bud elongation and repression results in the formation of fully separated and oval-shaped mother and daughter buds (Sudbery et al., 2004); thus, modulating synthesis or degradation of cyclins dramatically changes cell morphology (Berman, 2006). Cells depleted of Cln3 arrest in G1 and form filaments before re-entering the cell cycle (Bachewich and Whiteway, 2005; Chapa y Lazo et al., 2005). Surprisingly, although loss of Ccn1 does not induce morphogenesis, it does cause a filamentation defect in response to some inducing cues (Loeb et al., 1999). Filamentation is also observed in the absence of G2 cyclins, because the polarized growth promoted by G1 cyclin complexes cannot be fully suppressed (Bensen et al., 2005). Clb2-depleted cells are not viable and arrest with hyperelongated projections, whereas Clb4-depleted cells remain viable and grow constitutively as pseudohyphae (Bensen et al., 2005). In addition, depletion of Cdc28 induces filamentous growth (Umeyama et al., 2006). Overall, opposing activities of CDK complexes ensure maintenance of yeast morphology under normal conditions (Berman, 2006) (Figure 1).
Figure 1. Cyclin-dependent kinase (CDK) complexes control cell-cycle progression and play many critical roles during morphogenesis.
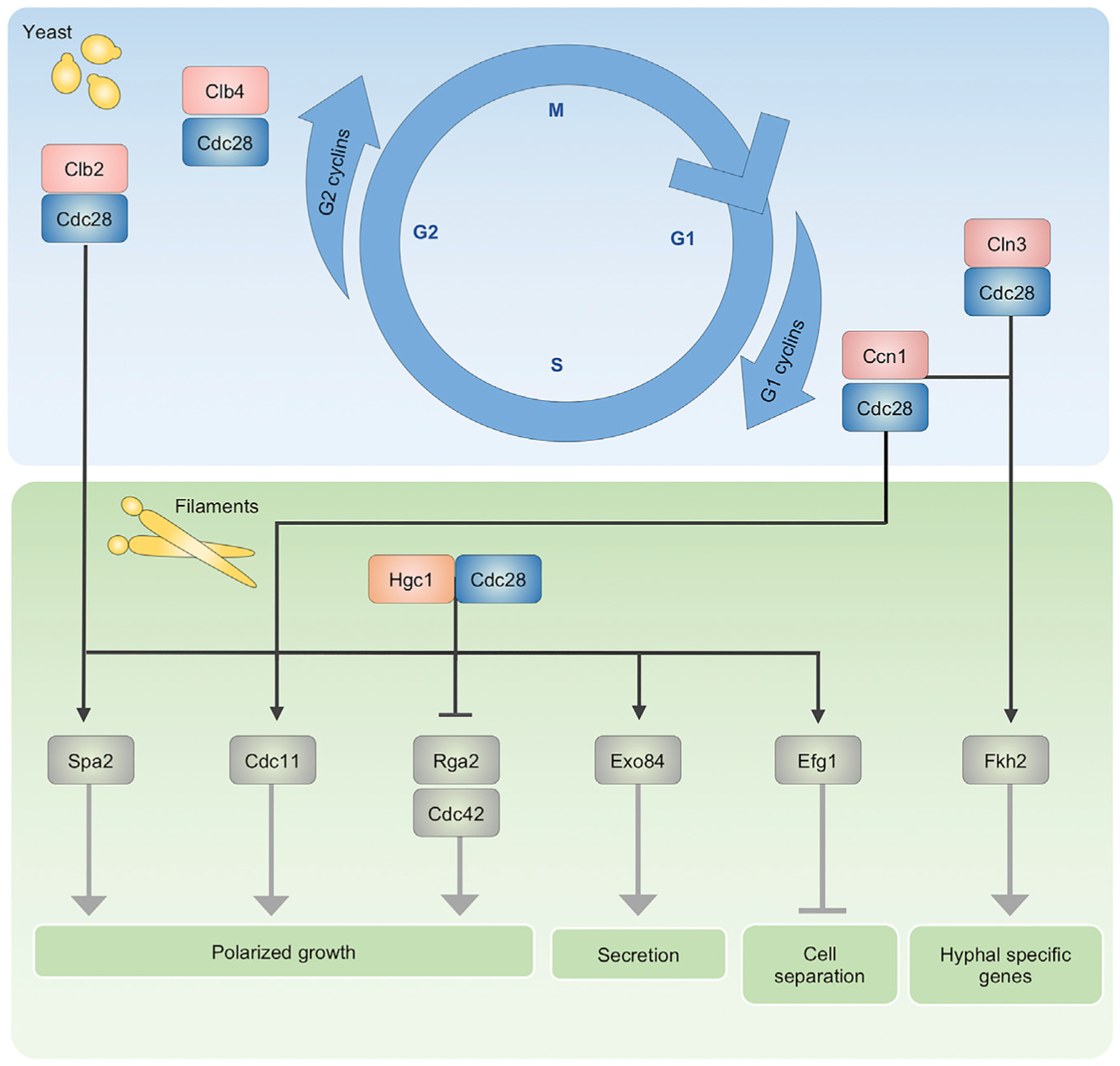
CDK Cdc28 drives the cell cycle and maintains yeast-phase growth through its association with phase-specific cyclins with opposing activities. During early stages of the cell cycle, Cdc28 associates with the G1 cyclins Ccn1 and Cln3; as the cell cycle progresses, these are replaced by the G2 cyclins Clb2 and Clb4. The temporal binding of Cdc28 with these cyclins depends on their coordinated synthesis and degradation. Upon hyphal induction, the hyphal-specific G1 cyclin Hgc1 is expressed and associates with Cdc28 to target various cellular processes that promote morphogenesis, independently or in concert with other CDK complexes. Phosphorylation of the polarisome component Spa2 by Clb2-Cdc28 and Hgc1-Cdc28, the septin Cdc11 by Ccn1-Cdc28 and Hgc1-Cdc28, and the GTPase-activating protein Rga2 by Hgc1-Cdc28 promotes hyphal growth. Phosphorylation of the exocyst subunit Exo84 by Hgc1-Cdc28 regulates polarized secretion at the hyphal tip. The G1 CDK complexes, Ccn1-Cdc28 and Cln3-Cdc28, work together to phosphorylate the transcription factor Fkh2 to enhance expression of hyphal-specific genes.
In addition to their roles in controlling cell-cycle progression and maintaining yeast-phase growth, some CDK complexes are involved in processes required for proper, cell-cycle-independent hyphal development (Figure 1). The G1 cyclin Hgc1 does not regulate the cell cycle, but it is implicated in morphogenesis. HGC1 is a hyphal-specific gene whose expression is induced upon activation of the cAMP-PKA pathway and upon filamentation (Sudbery, 2011; Zheng et al., 2004). Although almost all hyphal-specific genes are dispensable for hyphal growth, Hgc1 is required for maintenance of filamentation in response to many cues, because together with Cdc28, it regulates processes required for filamentous growth (Zheng et al., 2004). The Hgc1-Cdc28 complex phosphorylates regulators of cell polarity, membrane trafficking, and cell separation to drive continuous, cell-cycle-independent hyphal development (Wang, 2016; Zheng et al., 2004) (Figure 1). One of its targets is the GTPase-activating protein Rga2, which negatively regulates the GTPase Cdc42 to suppress polarized growth (Zheng et al., 2007). Upon hyphal induction, Hgc1-Cdc28 phosphorylates and inactivates Rga2, allowing Cdc42 to localize to hyphal tips, where it facilitates hyphal growth (Zheng et al., 2007). In addition, both Clb2-Cdc28 and Hgc1-Cdc28 phosphorylate the polarisome scaffold protein Spa2, localizing the polarisome to the hyphal tip, where it assembles actin cables (Wang et al., 2016). Hgc1-Cdc28 also regulates polarized secretion, which is critical for hyphal development (Wang, 2016). For example, Hgc1-Cdc28 phosphorylates the exocyst subunit Exo84, promoting its recycling at the growing tip, which is necessary for efficient hyphal extension (Caballero-Lima and Sudbery, 2014). Lastly, a hallmark of filamentous growth is that cells do not separate following cytokinesis, which is influenced by Hgc1. Hgc1-Cdc28 contributes to the suppression of cell separation by phosphorylating the transcription factor Efg1, blocking expression of cell separation genes (Wang et al., 2009). Overall, through association with the master cell-cycle regulator Cdc28, Hgc1 plays a crucial role in filamentation (Figure 1).
CDK complexes are also involved in regulating transcriptional responses in filaments. The forkhead family transcription factor Fkh2 normally undergoes cell-cycle-dependent phosphorylation to induce expression of genes that regulate cell-cycle progression and cell proliferation (Bensen et al., 2002; Greig et al., 2015). However, almost immediately after hyphal induction, Ccn1/Cln3-Cdc28, the cell wall integrity kinase Cbk1, and its regulatory subunit Mob2 acutely phosphorylate Fkh2 in a cell-cycle-independent manner. This redirects Fkh2 to specifically enhance expression of hyphal-specific genes such as HGC1, causing misregulation of its normal target genes (Greig et al., 2015) (Figure 1). As such, deletion of FKH2 or mutation of the relevant phosphorylation sites causes abnormal hyphal formation, potentially because of the up-regulation of cyclin CLB4, which suppresses polarized growth (Bensen et al., 2002; Greig et al., 2015). Additional contributions of the CDK complexes to the morphogenetic transcriptional program remain to be explored but are likely substantial, considering the diverse Cdc28 clients and effects on the cell cycle and morphogenesis.
Another important family of proteins whose regulation by CDK complexes is central to both cell-cycle progression and morphogenesis are the septins. Septins are guanosine triphosphate (GTP)-binding proteins that form ring-like complexes at the site of cytokinesis and recruit proteins that regulate cell-cycle progression. Loss of the septin Cdc10 or Cdc11 leads to defects in cytokinesis and septin assembly and causes abnormally shaped filaments in hyphal-promoting conditions (Warenda and Konopka, 2002), whereas loss of the septin Sep7 causes abnormal cell separation during hyphal growth (González-Novo et al., 2008). Septins are tightly regulated by the opposing activities of CDKs and phosphatases that alter septin phosphorylation status (Han et al., 2019; Liu et al., 2016). Immediately upon hyphal induction, Ccn1-Cdc28 phosphorylates Cdc11, and this phosphorylation is subsequently maintained by Hgc1-Cdc28 throughout hyphal development (Sinha et al., 2007) (Figure 1). Mutating the phosphorylation sites abolishes Cdc11’s hyphal-specific phosphorylation and impairs maintenance of polarized growth (Sinha et al., 2007). Changes in septin ring dynamics during hyphal growth are also regulated by phosphorylation of Sep7 through Hgc1-Cdc28-mediated phosphorylation of the kinase Gin4, which phosphorylates Sep7 (Li et al., 2012). Furthermore, deletion of genes encoding either of the protein phosphatase 2A (PP2A) subunits, Pph21 and Tpd3, or the PP2A regulators, Cdc55 or Rts1, results in hyperphosphorylation of Sep7; this causes abnormal septin rings and defective cell division and largely abolishes hyphal growth in response to serum (Han et al., 2019; Liu et al., 2016). Overall, phosphoregulation of septins by CDK complexes is central to their dynamics during cell-cycle progression and morphogenesis.
Altogether, this demonstrates the association between CDK complexes and critical cell-cycle components in driving filamentation and exemplifies the tight link between cell-cycle regulation and morphogenesis (Figure 1). Identifying the complete repertoire of CDK targets with a role in morphogenesis could be achieved by investigating CDK targets in closely related species. For example, in the model yeast Saccharomyces cerevisiae, other proteins involved in polarized growth, such as Boi1 and its paralog Boi2 (McCusker et al., 2007), require CDK phosphorylation to localize to bud tips and promote apical growth. Further characterization of the regulation of Cdc28 activity by the CDK inhibitors Sol1 and Sic1 (Atir-Lande et al., 2005), the CDK-activating kinase Cak1 (Woolford et al., 2016), and the CDK-inhibiting kinase Swe1 (Wightman et al., 2004) will likely reveal additional connections among the cell cycle, morphogenesis, and proteostasis, as discussed in subsequent sections.
Cell-cycle checkpoints are required for morphogenesis in response to cell-cycle arrest
Many checkpoint pathways monitor the cell cycle to ensure critical events are completed correctly before progressing to the next stage. In C. albicans, genotoxic stresses that interrupt normal cell-cycle progression and activate checkpoints induce filamentation (Berman, 2006). This morphogenic change occurs in response to arrest at almost any stage of the cell cycle, including pharmacological treatments that cause DNA damage, perturbation of DNA synthesis, or disruption of mitosis (Berman, 2006). Treatments that cause DNA replication stress, as with the compounds hydroxyurea (HU) or aphidicolin (AC), or DNA damage, by exposure to UV radiation or methyl methane sulfonate (MMS), cause S-phase arrest, resulting in cell elongation followed by pseudohyphal growth upon resumption of the cell cycle (Bachewich et al., 2005; Shi et al., 2007). Similarly, mitotic arrest caused by the microtubule-disrupting agent nocodazole results in pseudohyphal growth (Bai et al., 2002). Filamentation also occurs when the cell cycle is disrupted by deletion or depletion of critical cell-cycle components (Berman, 2006). Complex regulatory networks involving checkpoints are responsible for the resulting filamentation, which can be specific to different arrest conditions.
Checkpoint proteins are critical for morphogenetic responses to DNA replication and DNA damage stress, because they are activated to halt cell-cycle progression and ensure correct and complete DNA replication before progressing to mitosis (Berman, 2006). Central to both DNA damage and DNA replication checkpoints is the protein kinase Rad53, whose phosphorylation and activation trigger diverse cellular responses to facilitate repair, whereas its dephosphorylation and deactivation allow the cell cycle to resume (Sun et al., 2011). In the absence of Rad53, cells are unable to filament in response to genotoxic stresses or depletion of the ribonucleotide-reductase subunit gene RNR2, which blocks DNA replication by depleting deoxyribonucleotide triphosphate (dNTP) pools (Loll-Krippleber et al., 2014; Shi et al., 2007). In addition, depletion of either the DNA repair protein Rad52 or the histone acetyl transferase Hat1 leads to an accumulation of spontaneous DNA damage that induces filamentous growth by triggering the DNA damage checkpoint (Andaluz et al., 2006; Tscherner et al., 2012); however, direct involvement of Rad53 has not yet been elucidated. Strikingly, mutating Rad53’s FHA1 domain, which is important for mediating protein interactions, uncouples Rad53 functions in regulating filamentous growth and mediating cell-cycle arrest (Shi et al., 2007). Overall, Rad53 is essential for morphogenesis induced by genotoxic stress, potentially by directly enabling filamentation (Loll-Krippleber et al., 2014; Shapiro et al., 2011; Shi et al., 2007).
Proteins that regulate Rad53 function or upstream checkpoint components that transduce DNA stress signals to Rad53 are also involved in filamentation in response to genotoxic stresses, although their roles can be specific to the arrest condition. Rad9, a component of the DNA damage checkpoint, is required for filamentous growth induced by DNA damage, but not DNA replication stress (Shi et al., 2007). Phosphatases that catalyze checkpoint deactivation and allow the cell cycle to resume upon DNA repair also have roles in morphogenesis. Deleting genes encoding either subunit of the PP2A Pph3/Psy2 complex or its regulator Tip41 enhances MMS-induced filamentous growth and delays recovery to yeast-form growth following removal of MMS because of impaired deactivation of Rad53 (Feng et al., 2016; Wang et al., 2012). In contrast, Pph3, Psy2, and Tip41 are dispensable for recovery from HU treatment, but the protein phosphatase 1 Glc7 and its regulator Shp1 may play a role (Feng et al., 2016). Overall, multiple conditions arrest the cell cycle during DNA synthesis, and complex networks of checkpoint proteins mediate the resulting switch to filamentous growth.
Perturbation of mitosis can also result in filamentous growth that depends on checkpoints. The spindle assembly checkpoints monitor mitotic nuclear division and delay progression until proper spindle formation and chromosome attachment occur. Different M-phase arrest conditions rely on distinct checkpoint components to enable filamentous growth. Depletion of the cell-cycle regulatory polo-like kinase Cdc5 causes mitotic arrest, resulting in elongated buds that later switch to hyphal growth (Bachewich et al., 2003). In addition, cytoplasmic dynein assists with nuclear migration during mitosis; in its absence, defective nuclear dynamics slow mitotic progression, leading to pseudohyphal growth (Finley et al., 2008). Filamentous growth caused by either of these conditions requires the spindle checkpoint factor Bub2 (Bachewich et al., 2005; Finley et al., 2008). In contrast, pseudohyphal growth induced by nocodazole requires the spindle assembly checkpoint factor Mad2, but not its upstream kinase Mps1 (Bai et al., 2002; Kamthan et al., 2014). Failure to exit mitosis by depleting cells of the kinase Cdc15 or the GTPase Tem1 that function in the mitotic exit network also results in filamentation, and this phenotype is terminal (Bates, 2018; Milne et al., 2014). Altogether, this shows that checkpoint proteins play an integral role in faithful completion of the cell cycle and in filamentation in response to genotoxic stress (Shapiro et al., 2011). Future research should expand on the investigations into whether cell-cycle checkpoint proteins can directly influence filamentous growth, independent of cell-cycle arrest, thereby adding a layer of complexity in the connections between cell-cycle and morphogenetic regulation.
The UPP influences proteostasis, the cell cycle, and filamentation
The UPP is a highly conserved pathway in all eukaryotes that is instrumental for maintaining cellular proteostasis and regulating cell-cycle progression. Proteasomal proteolysis is important for turnover of abnormal or short-lived proteins, degradation of denatured and misfolded proteins following environmental stress, and cell-cycle control. Perturbation of the UPP has devastating consequences on the cell, leading to aggregation of misfolded proteins, disruption of the cell cycle, and toxicity (Finley et al., 2012). In C. albicans, disruption of many critical components or regulators of the UPP has dramatic effects on morphogenesis (Hossain et al., 2020; Kornitzer, 2019).
Proteins are targeted to the UPP by the protein modifier ubiquitin. Ubiquitin is covalently linked to substrates through a cascade of enzymatic reactions driven by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s) (Finley et al., 2012; Weissman, 2001) (Figure 2). E3s form the largest group of proteins involved in ubiquitination and mediate the exquisite selectivity of ubiquitination by directing substrate specificity. The fate of ubiquitinated proteins is established by the nature of ubiquitin linkages; polyubiquitinated substrates are preferentially targeted to the proteasome for degradation, whereas monoubiquitinated substrates play diverse cellular roles, including DNA repair, chromatin dynamics, mRNA export, and trafficking of membrane proteins (Finley et al., 2012; Weissman, 2001) (Figure 2). In C. albicans, polyubiquitin is encoded by UBI4, comprising three consecutive, head-to-tail ubiquitin repeats, which is cleaved to generate ubiquitin monomers upon translation (Sepulveda et al., 1996) (Figure 2). Ubiquitination is important for cell-cycle progression and morphogenesis in C. albicans, because depletion of UBI4 induces the formation of hyphal and pseudohyphal growth with abnormal nuclear segregation patterns (Leach et al., 2011; Roig and Gozalbo, 2003). Furthermore, the putative E2 Rad6 represses morphogenesis; deletion of RAD6 induces filamentation mediated by the transcription factor Efg1, likely through cAMP-PKA signaling (Leng et al., 2000). A thorough characterization of all C. albicans ubiquitin-modifying enzymes, such as the E1 Uba1 and homologs of the 12 S. cerevisiae E2s, to the regulation of C. albicans protein degradation and morphogenesis remains to be achieved and is likely to reveal additional functional connections (Finley et al., 2012).
Figure 2. The ubiquitin-proteasome pathway influences proteostasis, the cell cycle, and filamentation.
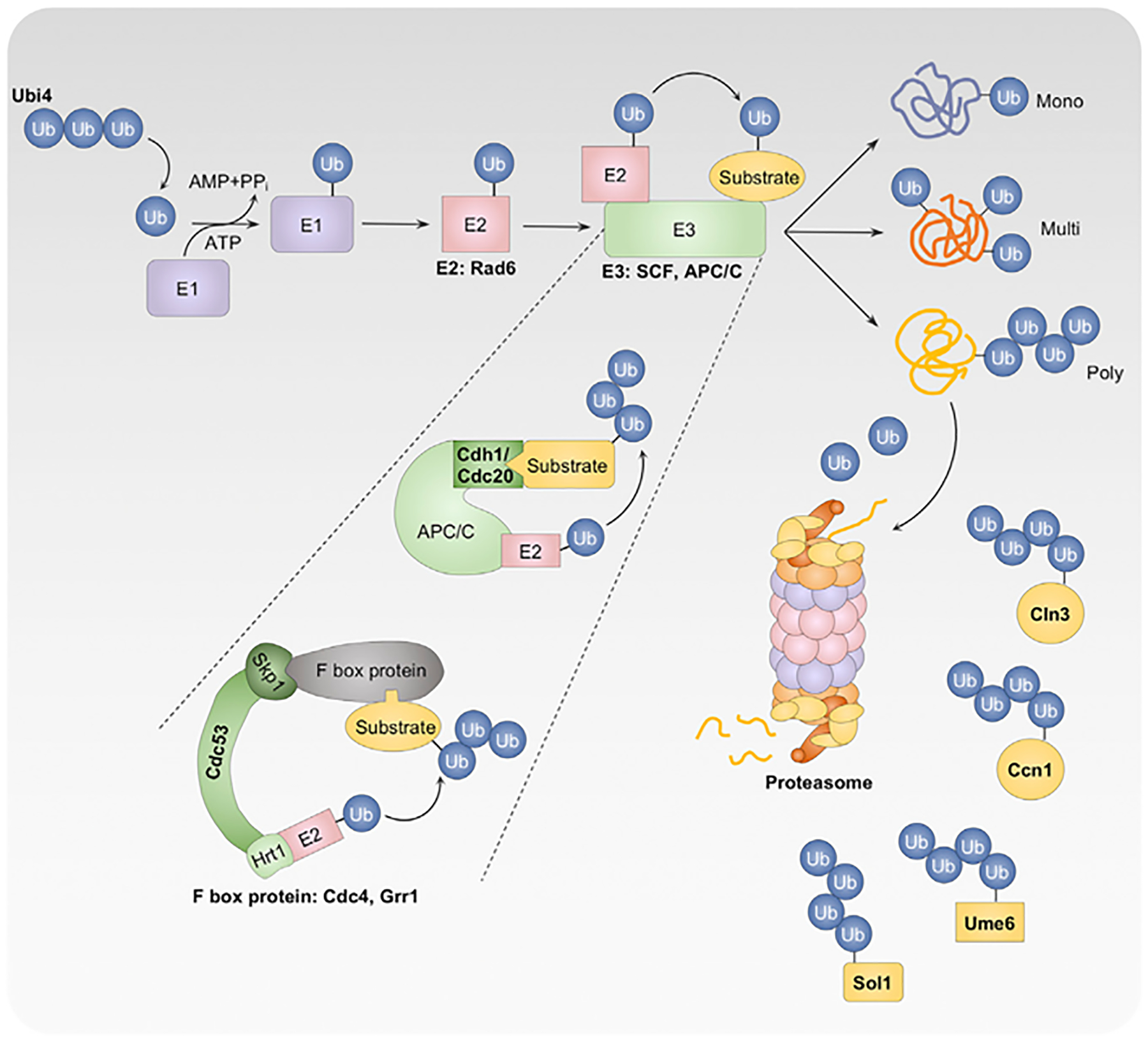
The polyubiquitin Ubi4 is cleaved to generate ubiquitin monomers. Ubiquitin is activated by a ubiquitin-activating enzyme (E1) in an ATP-dependent manner; transferred to a ubiquitin-conjugating enzyme (E2), such as the putative E2 Rad6; and subsequently covalently attached to substrate proteins by a ubiquitin ligase (E3). E3 is responsible for substrate selection and specificity. Addition of a single ubiquitin molecule to a substrate generates monoubiquitinated proteins, whereas multiple rounds of ubiquitination generate multi- or polyubiquitinated proteins. Polyubiquitinated proteins are preferentially targeted to the proteasome for degradation. The Skp1-Cullin-F-box (SCF) complex (consisting of the linker protein Skp1, scaffold protein Cdc53, and F-box protein) and the anaphase-promoting complex/cyclosome (APC/C) complex (including the coactivators Cdc20 and Cdh1) are important for proteasome-mediated turnover of proteins involved in the cell cycle and filamentation. UPP proteins and substrates with known roles in regulating C. albicans filamentation are indicated in bold.
In addition to its role in proteostasis, the UPP is crucial for degradation of cyclins and CDK inhibitors to ensure orderly progression through the cell cycle (Adams, 2004). Degradation of these key cell-cycle regulators is coordinated by two multiprotein E3 complexes: the Skp1-Cullin-F-box (SCF) complex and the anaphase-promoting complex/cyclosome (APC/C) complex. SCF complexes consist of the linker protein Skp1, the scaffold protein Cdc53/cullin, and a substrate recognition F-box protein, which provides substrate specificity (Figure 2). In S. cerevisiae, the cullin binds the E3 Hrt1, which is responsible for the recruitment and activation of E2 (Figure 2) (Finley et al., 2012). The SCF complex regulates cell-cycle progression by coordinating the temporal and spatial degradation of cyclins, CDKs, and transcription factors (Willems et al., 2004). Impairment of SCF complex activity through depletion of the essential cullin gene CDC53 or deletion of genes encoding the non-essential F-box proteins Grr1 or Cdc4 induces morphogenesis (Atir-Lande et al., 2005; Li et al., 2006; Sela et al., 2012; Trunk et al., 2009). Perturbation of SCF components leads to increased stability of SCF substrates, including the SCFGrr1 substrates, the G1 cyclins Ccn1 and Cln3, the SCFCdc4 substrates, the CDK inhibitor Sol1, and the positive transcriptional regulator of filamentation Ume6 (Atir-Lande et al., 2005; Li et al., 2006; Mendelsohn et al., 2017). As Ccn1 and Cln3 promote apical growth, their increased stability is expected to drive elongated growth in cells lacking GRR1 (Li et al., 2006). Sol1 represses Clb2-Cdc28; therefore, increased stability of Sol1 or its overexpression would inhibit apical to isotropic growth, resulting in highly elongated buds (Atir-Lande et al., 2005). Ume6 is a filament-specific transcription factor that regulates filament-specific gene expression in response to filament-inducing cues, and its overexpression is sufficient to drive hyphal formation (Banerjee et al., 2008; Carlisle et al., 2009). Altogether, the SCF complex is involved in regulating the stability of diverse proteins with roles in morphogenesis, and increased stability of SCF substrates drives morphogenesis upon perturbation of SCF activity (Figure 2).
The conserved APC/C is a 13-subunit E3 complex (Finley et al., 2012) that is required for mitotic progression through regulation of CDK activity, separation of sister chromatids, and disassembly of mitotic spindle (Simpson-Lavy et al., 2010). The activity of APC/C depends on its two coactivators, Cdc20 and Cdh1, which bind to APC/C at distinct stages of the cell cycle and mediate the specificity of substrates for ubiquitination and subsequent proteolysis (Manchado et al., 2010). In S. cerevisiae, Cdc20 regulates the degradation of proteins associated with the metaphase-anaphase transition and mitotic exit; toward the end of mitosis and during the G1 phase, Cdh1 controls degradation of mitotic regulators (Finley et al., 2012; Manchado et al., 2010; Simpson-Lavy et al., 2010). Although little is known about APC/C in C. albicans, initial characterization of Cdc20 and Cdh1 have illuminated connections with C. albicans morphogenesis. Depletion of C. albicans CDC20 leads to increased accumulation of Clb2 and Cdc5, blocking cells in early or late mitosis and inducing morphogenesis (Chou et al., 2011). Interestingly, deletion of CDH1 causes variable effects on morphology and the cell cycle, resulting in defects in mitotic exit; a mix of yeast, pseudohyphae, and elongated buds; and elevated levels of Cdc5 (Chou et al., 2011). Further exploration is required to determine whether perturbating the core APC/C subunits in C. albicans can induce morphogenesis by disrupting proteolysis.
Polyubiquitinated proteins targeted for the UPP are degraded by the proteasome. The proteasome is a highly conserved, essential, 33-subunit protein complex composed of a 20S core particle containing the active sites and flanked by two 19S regulatory particles involved in substrate recognition (Finley et al., 2012). Proteasome-mediated protein degradation not only prevents the toxic accumulation of damaged and misfolded proteins but also regulates protein quality control in the nucleus and the endoplasmic reticulum, as well as DNA repair, transcription, and cell-cycle progression (Finley et al., 2012). Proteasome substrates are often nuclear proteins, and proteasomes predominantly localize to the nucleus of proliferating yeast and mammalian cells (Enenkel, 2014). Proteasome inhibition interferes with cell-cycle progression and results in the accumulation of polyubiquitinated proteins, with impacts on C. albicans morphogenesis (Hossain et al., 2020) (Figure 2). Pharmacological inhibition of the proteasome with two structurally distinct inhibitors or genetic depletion of most proteasome subunits induces C. albicans filamentation in the absence of an inducing cue. Filaments formed in response to proteasome inhibition display aberrant cellular morphology compared with filaments induced by serum, with defective septum formation and nuclear segregation. The current paradigm is that proteasome inhibition causes an accumulation of ubiquitinated proteins that overwhelms the functional capacity of chaperones and induces filamentation (discussed in the following section) (Hossain et al., 2020). The proteasome is one of the newest additions in the list of proteostasis regulators that control C. albicans morphogenesis, and it establishes a new layer of connection for proteostasis, the cell cycle, and morphogenesis.
The UPP is undoubtedly crucial for the maintenance of cell-cycle progression and morphogenesis through regulated proteolysis. Although the UPP has been demonstrated to regulate morphogenesis through effects on the cell cycle and chaperones, protein degradation pathways also influence filamentation by altering the stability of regulators of morphogenesis. Hyphal initiation requires downregulation of the morphogenetic repressor Nrg1 (Braun et al., 2001; Murad et al., 2001). Nrg1 degradation is promoted by the kinase Sok1, which is repressed by the transcriptional repressor Cup9. In response to filament-inducing conditions, Cup9 degradation is mediated through the E3 Ubr1, relieving Sok1 repression, inducing Nrg1 degradation, and thereby enabling hyphal formation (Lu et al., 2014). Ubr1 is also important for degradation of Ume6 in the presence of O2, limiting levels of the positive regulator of morphogenesis and maintaining yeast-form growth (Lu et al., 2019). Similarly, in the presence of physiological CO2, the CDK Ssn3 phosphorylates Ume6 and targets it for degradation via SCFGrr1. However, the protein phosphatase Ptc2 dephosphorylates Ssn3 in the presence of high concentrations of CO2, reducing Ume6 phosphorylation and degradation and thereby promoting hyphal development (Lu et al., 2019). Altogether, this demonstrates that the C. albicans UPP mediates proteotoxic stress and modulates levels of cell-cycle and morphogenetic regulators, with profound effects on cell-cycle progression and morphogenesis. Future studies into the proteosome and the APC/C and SCF complexes are required to elucidate the kinetics of degradation of regulators of the cell cycle and morphogenesis, which can help determine the major driver of filamentation induced upon perturbation of the UPP.
Morphogenetic regulation by the heat shock protein Hsp90
Molecular chaperones are essential for facilitating protein folding and refolding of misfolded and aggregated proteins, which can be detrimental for cellular health and survival. Hsp90 is a highly conserved chaperone present from bacteria to mammals that functions in concert with cochaperones to regulate the stability and activation of many client proteins, both in normal conditions and in response to environmental stress (Taipale et al., 2010). Hsp90 interacts with approximately 10% of the yeast proteome, including many components of critical signal transduction pathways, enabling Hsp90 to influence diverse essential cellular processes (http://www.picard.ch/downloads/Hsp90interactors.pdf).
Maintaining proteostasis is essential not only for cellular viability and growth but also for proper regulation of fungal virulence. As such, C. albicans Hsp90 profoundly affects many virulence traits, including morphogenesis. Inhibition of C. albicans Hsp90 abrogates drug resistance, impairs tolerance to many stresses, sensitizes drug-resistant biofilms, and attenuates virulence in a mouse model of infection (O’Meara et al., 2017). Hsp90 governs temperature-dependent morphogenesis by inhibiting morphogenetic signaling through the cAMP-PKA pathway (Shapiro et al., 2009). Hsp90 and its cochaperone Sgt1 sequester Cyr1 in an inactive state, repressing the cAMP-PKA pathway under basal conditions (Shapiro et al., 2012a) (Figure 3). Compromise of Hsp90 function relieves this repression and induces C. albicans filamentation in the absence of an inducing cue (Shapiro et al., 2009) (Figure 3). Similarly, conditions that impair Hsp90 function stimulate signaling through the cAMP-PKA pathway and lead to an induction of morphogenesis. For example, growth at elevated temperatures results in global problems in protein folding that overwhelm the functional capacity of Hsp90, relieving Hsp90-mediated repression of morphogenesis (Shapiro et al., 2009). Most filament-inducing cues require a concomitant rise in temperature to 37°C to induce morphogenesis, highlighting the importance of relief of chaperone-mediated repression of morphogenetic signaling (Shapiro et al., 2011).
Figure 3. Hsp90 regulates C. albicans morphogenesis through its interactions with client proteins.
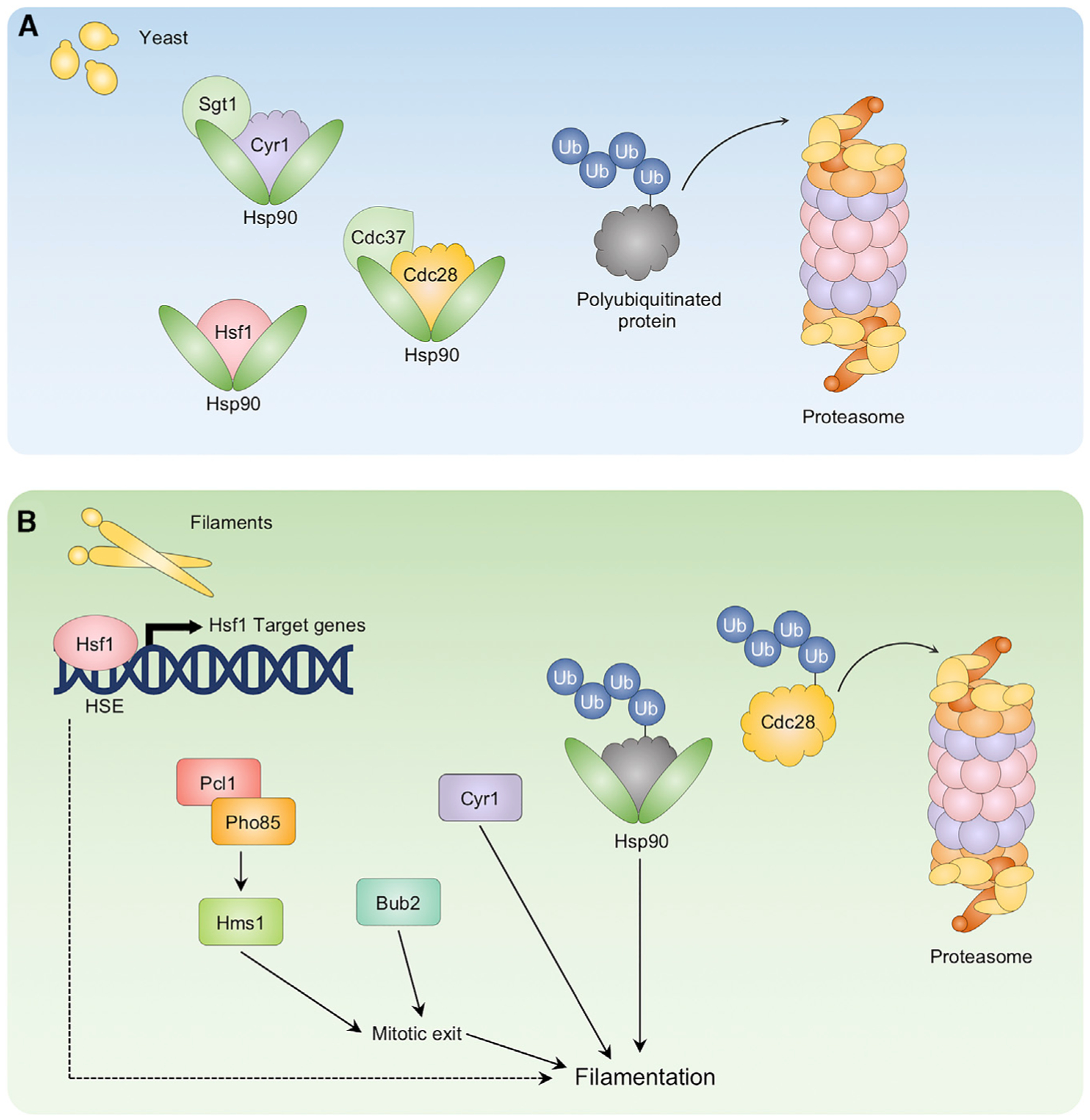
(A) Hsp90 and its cochaperone Sgt1 physically interact with Cyr1, keeping it in an inactive conformation, repressing morphogenesis, and maintaining the yeast form of growth. Hsp90 also binds Hsf1, repressing Hsf1 activity. Hsp90, along with its cochaperone Cdc37, maintains the stability of Cdc28, ensuring proper cell-cycle progression. The proteasome maintains protein homeostasis through regulated degradation of polyubiquitinated proteins.
(B) Conditions that compromise Hsp90’s function, such as elevated temperature or inhibition of Hsp90 or the proteasome, relieve repression of Hsf1 and the cAMP-PKA pathway, inducing the heat shock response and morphogenesis, respectively. Inhibition of Hsp90 also leads to Cdc28 destabilization and degradation by the proteasome, further affecting cell-cycle progression and influencing morphogenesis via Bub2, Pho85-Pcl1-Hms1 signaling, and the mitotic exit network. The role of Hsf1 in morphogenesis upon compromise of Hsp90 is unknown (indicated by dashed lines).
C. albicans Hsp90 has pleiotropic effects on diverse cellular processes, including cell-cycle progression. Hsp90 and its client proteins are key mediators of cell-cycle control, and Hsp90 regulates filamentation through multiple cell-cycle control pathways. Filaments induced by perturbation of Hsp90 function display defects in cell-cycle progression, with a delay in mitotic exit and defects in cytokinesis, and they share structural similarities with filaments induced by cell-cycle arrest, but not serum (Bachewich et al., 2005; Bensen et al., 2005; Hossain et al., 2020; Senn et al., 2012). This could be mediated through Hsp90’s role in regulating its client protein, Cdc28 (Senn et al., 2012). Hsp90 inhibition destabilizes Cdc28, and depletion of CDC28 halts cell-cycle progression and induces morphogenesis (Senn et al., 2012; Umeyama et al., 2006) (Figure 3). There is precedent for additional roles of Hsp90 in regulating cell-cycle progression in S. cerevisiae, in which several cell-cycle regulators have been implicated as Hsp90 interactors, including Cdc5, the DNA helicase Chl1 involved in mitotic sister chromatid cohesion and chromosome segregation, and the kinase Swe1 that regulates the G2/M transition (Goes and Martin, 2001; Khurana et al., 2018; Mollapour et al., 2010). In addition, as with filaments induced by depletion of Cdc5, filaments induced in response to Hsp90 inhibition depend on Bub2, suggesting that activation of the Bub2/Bfa1 checkpoint is important for induction of filaments with defective progression through or exit from mitosis (Bachewich et al., 2005; Senn et al., 2012) (Figure 3). The importance of the mitotic exit in filaments induced in response to Hsp90 inhibition is highlighted through the CDK Pho85 and the transcription factor Hms1. In S. cerevisiae, Pho85 and Hms1 influence mitotic exit through Cdc14, and Pho85 forms a complex with the cyclin Pcl1 and phosphorylates Hms1 (Keniry et al., 2004). In C. albicans, Pho85-Pcl1-Hms1 signaling is important for filamentation in response to Hsp90 inhibition and elevated temperature (Shapiro et al., 2012b) (Figure 3). Interestingly, Hms1 is dispensable for filamentation in response to other inducing cues, including serum and HU, indicating specificity in the requirement for mitotic exit in morphogenesis (Shapiro et al., 2012b). Work to identify additional C. albicans Hsp90 client proteins, particularly ones with roles in regulating the cell cycle, and the circuitry through which Hsp90 regulates morphogenesis is expected to reveal additional connections among C. albicans Hsp90, cell-cycle progression, and morphogenesis.
Hsf1 levels control morphogenesis via functional connections with Hsp90 and the cell cycle
Environmental stresses such as thermal insults induce an evolutionarily conserved heat shock response to upregulate molecular chaperones like Hsp90 and ameliorate proteotoxic stress (Veri et al., 2018b). In eukaryotes, induction of this heat shock response depends on the heat shock transcription factor Hsf1, which is conserved from yeast to humans (Veri et al., 2018b). C. albicans Hsf1 is dynamically regulated by environmental conditions and is activated in response to elevated temperature. Activated Hsf1 binds to heat shock elements (HSEs) in promoter regions and upregulates expression of genes involved in the heat shock response, including those encoding the chaperone proteins Hsp90, Hsp70, and Hsp104 (Leach et al., 2016; Nicholls et al., 2009). Increased abundance of chaperones, including Hsp90, facilitates protein repair, refolding, and degradation, thereby enabling cells to adapt to thermal insults (Leach et al., 2012a; Nicholls et al., 2009). Even in the absence of heat stress, Hsf1 is essential for cell viability, because it plays a central role in regulating proteostasis by controlling the expression of core cellular regulators such as Hsp90 (Leach et al., 2016).
In addition to heat shock transcriptional programs, Hsf1 controls expression of genes important for filamentation, pathogenesis, adhesion, and biofilm formation (Leach et al., 2016). Perturbation of HSF1 levels alters cellular morphology in the absence of filament-inducing cues (Veri et al., 2018a). Depletion of HSF1 impairs Hsp90 function independently of effects on HSP90 expression, likely due to the misregulation of Hsp90 co-chaperones and other proteostasis regulators (Veri et al., 2018a). Hsf1 has pleiotropic effects on morphogenesis, because depletion of HSF1 blocks filamentation in response to various solid hyphal-inducing media (Nair et al., 2017). Overexpression of HSF1 also induces filamentation, albeit through a mechanism independent of Hsp90 (Veri et al., 2018a). Instead, the abundance of Hsf1 expands its set of transcriptional targets, because many genes have HSEs in their promoter regions that are not normally bound when Hsf1 is limited. This leads to upregulated expression of positive regulators of filamentation, including BRG1 and UME6, and downregulated expression of negative regulators of filamentation, such as NRG1, driving morphogenesis (Veri et al., 2018a). Thus, Hsf1 governs morphogenesis through distinct mechanisms (Veri et al., 2018a). Hsf1 also plays an essential role in virulence (Nicholls et al., 2011), suggesting that Hsf1 could regulate virulence traits beyond thermotolerance and morphogenesis that remain to be explored (Veri et al., 2018b).
Misregulation of Hsf1 effects the cell cycle. Filaments induced by HSF1 depletion are often multinucleate, indicating defective cell-cycle progression (Veri et al., 2018a). Although connections between Hsf1 and cell cycle in C. albicans are poorly understood, in S. cerevisiae, Hsf1 function is required for spindle pole body duplication, likely through transcriptional activation of chaperone genes that assist in its assembly (Zarzov et al., 1997). In addition, cells harboring S. cerevisiae HSF1 but lacking its transcriptional activation domain undergo reversible arrest at G2/M when grown at elevated temperatures, but they can be rescued by the heat-inducible Hsp90 isoform HSP82 (Morano et al., 1999). In human cells, phosphorylation and degradation of HSF1 are required for progression through mitosis, demonstrating an important cell-cycle regulatory function that is independent from HSF1’s role as a transcriptional activator (Lee et al., 2008). Involvement of Hsf1 in C. albicans cell-cycle regulation could be a promising avenue for future research given its central role in governing proteostasis.
As critical mediators of proteostasis, the UPP, Hsp90, and Hsf1 have highly interconnected roles in regulating protein homeostasis, which influences C. albicans morphogenesis. Hsp90 and Hsf1 have many functional relationships, in addition to Hsf1 regulating HSP90 expression and modulating filamentation through alterations in Hsp90 function. Hsp90 negatively regulates Hsf1 activity through physical interaction (Leach et al., 2012b) (Figure 3) and modulates nucleosome occupancy in response to heat shock at Hsf1 DNA binding sites, thereby diminishing target gene expression (Leach et al., 2016). Upon temperature upshifts, increases in unfolded and misfolded proteins titrate Hsp90 away from repressing Hsf1, thereby inducing the heat shock response (Leach et al., 2012a). This temperature-dependent control of the Hsf1-Hsp90 autoregulatory circuit has widespread implications on proteostasis and fungal virulence (Leach and Cowen, 2014). As previously highlighted, proteasome inhibition also results in an accumulation of misfolded, ubiquitinated proteins that titrate Hsp90 away from its client proteins. This overwhelms Hsp90’s function, induces the heat shock response via Hsf1, and relieves Hsp90-mediated repression of cAMP-PKA signaling to induce filamentation (Hossain et al., 2020). In addition, Hsp90 inhibition blocks cycling of the chaperone, leading to the destabilization of its client proteins, which are targeted to the proteasome for degradation (Schulte et al., 1995; Whitesell et al., 1994) (Figure 3). Perturbation of Hsp90 function also increases levels of the proteasome to meet the global demand for proteolysis of misfolded Hsp90 clients (O’Meara et al., 2019). Altogether, this implicates proteostasis regulators as core hubs for orchestrating morphogenesis.
Filamentation induced by perturbation of cell-cycle or proteostasis signals through PKA
As a central hub for morphogenetic signaling, cAMP-PKA signaling is critical for morphogenesis induced by perturbation of the cell cycle and proteostasis. Intriguingly, although arresting the cell cycle at different stages or inhibiting different regulators of proteostasis induces similar polarized growth phenotypes, the underlying genetic requirements can differ (Figure 4). In response to HU treatment, the GTPase Ras1 is required to stimulate the adenyl cyclase Cyr1 and activate cAMP-PKA signaling, thereby triggering polarized growth (Bachewich et al., 2005; Chen et al., 2018). HU induces filamentation even in the absence of the downstream transcription factor Efg1 and the transcription factor Cph1 (Bachewich et al., 2003). In contrast, filamentation induced by depletion of CDC5 does not require Ras1 or the downstream transcription factor Efg1, but it does rely on Cyr1 (Bachewich et al., 2003, 2005). In addition, filaments induced by depletion of RAD52 require Cyr1 but are independent of Efg1 (Andaluz et al., 2006). Although CLN3 depletion activates cAMP-PKA signaling via Cyr1, loss of Ras1 or Efg1 has an intermediate effect on hyphal progression (Bachewich and Whiteway, 2005; Chen et al., 2018). Filamentation in response to CLN3 depletion depends on the hyphal-specific genes EFG1, HGC1, UME6, and FLO8, which is not the case for HU-induced pseudohyphae (Chen et al., 2018). Altogether, this demonstrates that filamentation induced by cell-cycle arrest signals through the cAMP-PKA pathway, through interactions with either Ras1 or Cyr1, but the downstream transcription factors remain unclear (Figure 4). Similarly, filamentation induced by both Hsp90 and proteasome inhibition requires many components of the cAMP-PKA pathway—including Ras1, Cyr1, and the PKA subunits Tpk1 and Tpk2—but not Efg1, highlighting that additional downstream regulators remain elusive (Hossain et al., 2020; Shapiro et al., 2009). In contrast, filaments induced by HSF1 depletion, which are also thought to be caused by altered Hsp90 function, are contingent on Efg1. Therefore, although filaments induced in response to Hsp90 inhibition and HSF1 depletion use some of the same genetic circuitry, including Ras1, depletion of HSF1 governs morphogenesis through additional mechanisms independent of Hsp90 (Veri et al., 2018a) (Figure 4). Overexpression of HSF1 induces the morphogenetic program independently of cAMP-PKA signaling through Ras1 but requires Efg1 (Veri et al., 2018a) (Figure 4).
Figure 4. Morphogenesis induced by perturbation of cell-cycle or proteostasis signals through the cAMP-PKA pathway.
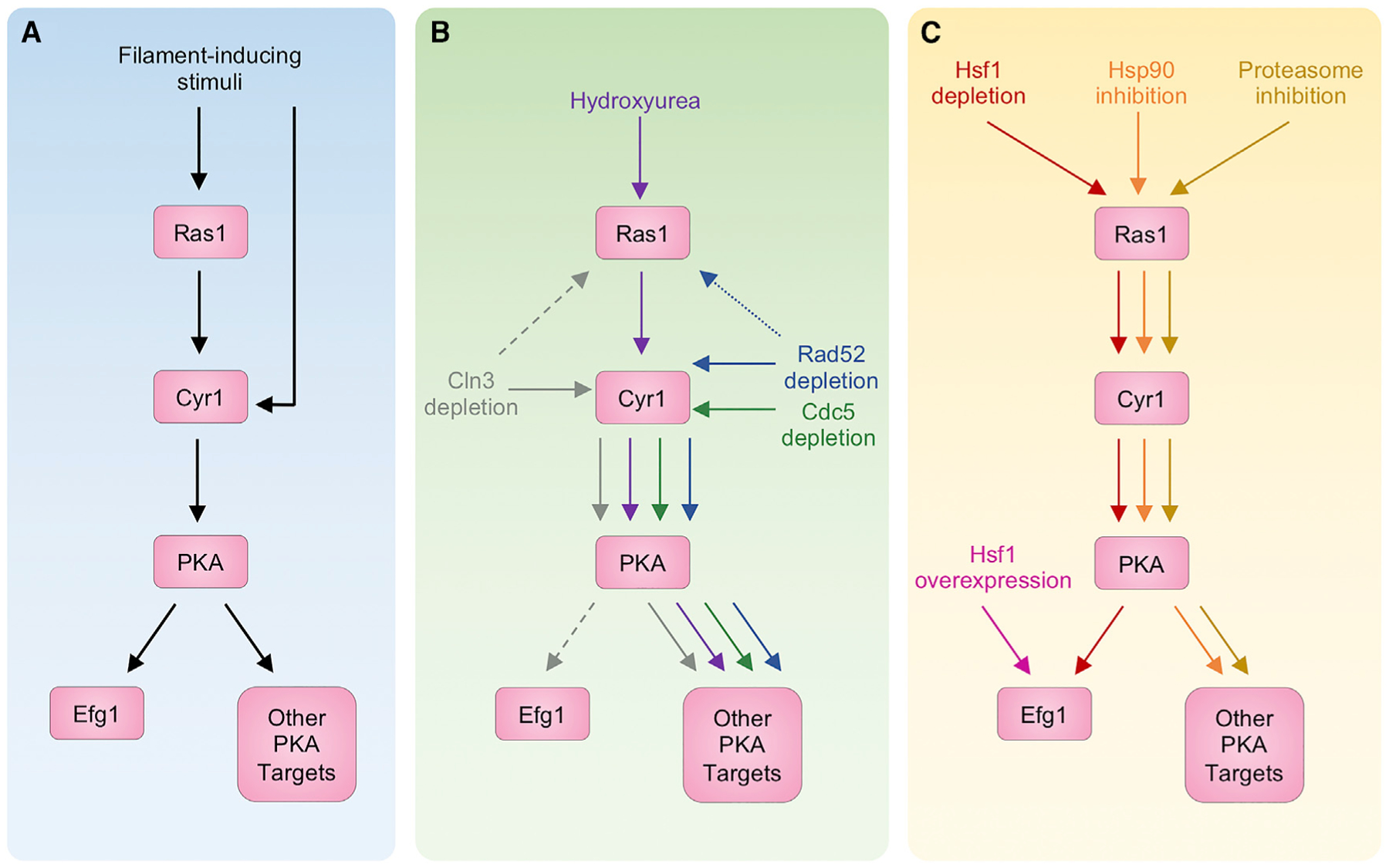
(A) Signaling through the cAMP-PKA pathway is required for filamentation in response to many cues.
(B) Disrupting cell-cycle progression by treating with the DNA synthesis inhibitor hydroxyurea (in purple), depleting the checkpoint protein Rad52 (in blue), or loss of the polo-like kinase Cdc5 (in green) induces filamentation in a manner that depends on the adenyl-cyclase Cyr1 but is independent of the PKA transcription factor Efg1, implicating PKA targets that remain to be identified. Filaments induced by hydroxyurea treatment, but not Cdc5 depletion, also require Ras1. The dependence of filaments induced by depletion of Rad52 on Ras1 is unknown (indicated with dotted lines). Hyphal growth in response to depletion of Cln3 requires signaling through Cyr1 but has only moderate dependence on Ras1 and Efg1 (indicated by dashed lines).
(C) Filamentation induced by inhibition of Hsp90 (in orange) or the proteasome (in yellow) or by depletion of Hsf1 (in red) requires signaling through Ras1, Cyr1, and the PKA subunits Tpk1 and Tpk2. Although filaments induced by Hsf1 depletion depend on Efg1, inhibition of Hsp90 or the proteasome induces filamentation through additional, unknown transcription factors downstream of PKA. Hsf1 overexpression (in pink) induces filamentation independently of cAMP-PKA signaling but requires Efg1.
Further comprehensive analysis is necessary to unravel the complexities of how perturbation of the cell cycle or proteostasis is sensed and transduced through different components of the cAMP-PKA pathway, with emphasis on determining the PKA-activated transcription factors that mediate this signaling and activate the morphogenetic program. In addition to their divergent requirements on the cAMP-PKA pathway, filaments induced in response to perturbation of the cell cycle or proteostasis differ from serum-induced filaments in their cellular morphology, septin formation, and cell-cycle progression (Hossain et al., 2020; Senn et al., 2012). Large-scale functional genomic analyses to characterize the genetic requirements for filamentation in response to serum and perturbation of the cell cycle and proteostasis will be instrumental in understanding the many complexities through which these filamentation programs differ. These analyses can overcome the limitations of comparing results among multiple groups of researchers that use different genetic backgrounds and variable growth conditions. In addition, there is a growing appreciation for the importance of studying genotype-phenotype relationships in diverse genetic backgrounds and in clinical isolates, because there is precedence for circuit diversification in regulation of biofilm and hyphal formation in C. albicans (Huang et al., 2019). Altogether, systematic genetic analyses of filamentation in response to inhibition of the cell cycle or proteostasis, compared with serum, have the potential to reveal the mechanisms through which the cell cycle and proteostasis are connected and the ways in which they diverge in their regulation of morphogenesis.
Concluding remarks
The mortality rates for systemic fungal infections are unacceptably high, demanding the development of novel antifungal treatments. Currently, there are only three major classes of antifungals available to treat invasive fungal infections—the azoles, the echinocandins, and the polyenes—and their use is hampered by a limited spectrum of activity, host toxicity, and the emergence of drug-resistant infections (Robbins et al., 2016). Although the close evolutionary relationship between fungi and humans often impedes antifungal drug discovery, innovative treatment strategies like targeting fungal virulence are emerging as promising alternative approaches (Vila et al., 2017).
Morphogenetic flexibility is one of the most critical C. albicans virulence traits. These reversible transitions among yeast, pseudohyphal, and hyphal forms enable the plasticity of C. albicans to thrive as both an asymptomatic commensal and a deadly pathogen in human hosts. The morphogenetic switch influences its detection and clearance by immune cells; its colonization, dissemination, and invasion of different organ sites; and the level of damage it causes to the host (Noble et al., 2017). Morphogenesis is also critical for the formation of drug-resistant biofilms on indwelling medical devices such as catheters and pacemakers (Shapiro et al., 2011). Given that morphogenesis is an essential virulence trait, C. albicans uses many complex and interconnected signaling pathways to maintain tight regulation of morphogenetic transitions in the host (Sudbery, 2011). Elucidating the mechanisms governing filamentation has the potential to cripple virulence of this deadly fungus and enable the development of novel antifungal treatments.
Regulation of C. albicans morphogenesis requires the concerted action of many cellular homeostasis pathways, mediated through critical cell-cycle and proteostasis regulators such as CDKs, heat shock response proteins, and the proteasome. Disruption of any of these regulators induces morphogenesis, often through effects on both protein homeostasis and the cell cycle. For example, inhibition of the proteasome leads to increased accumulation of misfolded proteins that overwhelms Hsp90’s functional capacity and disrupts the coordinated degradation of cyclins that are required for proper progression through the cell cycle, leading to morphogenesis (Hossain et al., 2020). Beyond the UPP, Hsp90, and Hsf1, the role of other proteostasis pathways such as autophagy and vacuolar degradation pathways, unfolded protein response, or translation in regulating morphogenesis has been underexplored. Although there is some evidence for the vacuole being important for filamentation in C. albicans and precedence for the vacuole being important for cell-cycle progression in S. cerevisiae, future work on these global proteostasis processes is expected to reveal additional mechanistic insights on the connections among proteostasis, morphogenesis, and the cell cycle (Bernardo et al., 2008; Jin and Weisman, 2015; Palmer et al., 2005; Poltermann et al., 2005; Rane et al., 2013, 2014).
In addition to their interconnected roles in regulating morphogenesis, many cell-cycle and proteostasis regulators are critical to tolerance of environmental stresses in the host during infection. Fungal Hsf1 is essential for thermal adaptation to temperature fluctuations during infection, including elevated temperatures experienced during febrile episodes, and blocking Hsf1 activation leads to thermosensitivity and reduced virulence in a murine model of infection (Nicholls et al., 2011). The C. albicans spindle assembly checkpoint protein Mad2 is essential for virulence in murine models, likely due to cell death caused by failure to arrest the cell cycle after DNA damage caused by reactive oxygen species from host immune cells (Bai et al., 2002). As critical regulators of morphogenesis, cell-cycle and proteostasis proteins may affect other C. albicans virulence traits, such as the formation of biofilms or the production of adhesins, secreted aspartyl proteases, and candidalysin. Lastly, because cellular homeostasis is also critical for viability and proliferation, many core regulators of cell-cycle progression and proteostasis, such as Cdc28 (Umeyama et al., 2006), Cdc5 (Bachewich et al., 2003), the proteasome (Hossain et al., 2020), Hsp90 (Shapiro et al., 2009), and Hsf1 (Nicholls et al., 2009), are essential for fungal growth. This can complicate assessing the role of these proteins in regulating filamentation or connections with other essential processes, because it is difficult to compromise essential pathways without confounding effects on the growth of the fungus. Nonetheless, this work provides many intriguing opportunities for antifungal drug development, because targeting cell-cycle and proteostasis regulators can disable the virulence of C. albicans and in some cases impair fungal growth.
The development of antivirulence therapies targeting morphogenesis will require the discovery of novel chemical matter that can disarm fungal virulence without causing toxicity to the human host. One promising approach is to identify regulators of the C. albicans cell cycle and protein homeostasis that are critical for virulence but are fungal specific or evolutionarily divergent between fungi and humans. To do this, future work should continue to dissect the genes required for filamentation in response to perturbation of the cell cycle and proteostasis and elucidate how signals are transduced to the cAMP-PKA pathway and downstream transcriptional regulators. Identification of additional regulators of the cell cycle and proteostasis and assessment of their contributions to fungal morphogenesis and virulence would also be illuminating. For example, a deep characterization of the targets of Cdc28 and its cyclin complexes could identify proteins through which CDKs orchestrate hyphal development and virulence that could be exploited. A powerful approach to targeting cell-cycle and proteostasis regulators for antivirulence treatments is to identify fungal-specific inhibitors of these processes. There are also opportunities to exploit targets that might be conserved in the host. For example, although Hsp90 is a promising target for antifungal therapies because of its central role in fungal drug resistance, stress response, and virulence, the available Hsp90 inhibitors cause toxicity in systemic models of fungal infection due to their inhibition of host Hsp90 (O’Meara et al., 2017). To address this, structure-guided drug design efforts have recently made significant progress in developing fungal-selective Hsp90 inhibitors (Whitesell et al., 2019), demonstrating that antifungals targeting these pathways hold great promise. It is exquisitely clear that innovative approaches to systematic analysis of regulators of the cell cycle and proteostasis and of chemical matter that can be exploited to inhibit these regulators can enable a novel class of therapeutics that cripple fungal virulence.
ACKNOWLEDGMENTS
We thank all members of the Cowen lab for helpful discussions. S.H. is supported by the Ontario Graduate Scholarship, and E.L. is supported by the Canada Graduate Scholarships-Master’s program by the Canadian Institutes of Health Research. L.E.C. is supported by the Canadian Institutes of Health Research Foundation Grant (FDN-154288) and a National Institutes of Health NIAID R01 (R01AI127375). L.E.C. is a Canada Research Chair (Tier 1) in Microbial Genomics & Infectious Disease and codirector of the CIFAR Fungal Kingdom: Threats & Opportunities program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DECLARATION OF INTERESTS
L.E.C. is a cofounder and shareholder in Bright Angel Therapeutics, a platform company for development of novel antifungal therapeutics. L.E.C. is also a consultant for Boragen, a small-molecule development company focused on leveraging the unique chemical properties of boron chemistry for crop protection and animal health.
REFERENCES
- Adams J (2004). The proteasome in cell-cycle regulation. In Proteasome Inhibitors in Cancer Therapy, Adams J, ed. (Humana Press; ), pp. 77–84. [Google Scholar]
- Andaluz E, Ciudad T, Gómez-Raja J, Calderone R, and Larriba G (2006). Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol. Microbiol 59, 1452–1472. [DOI] [PubMed] [Google Scholar]
- Atir-Lande A, Gildor T, and Kornitzer D (2005). Role for the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis. Mol. Biol. Cell 16, 2772–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C, and Whiteway M (2005). Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot. Cell 4, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C, Thomas DY, and Whiteway M (2003). Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 14, 2163–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C, Nantel A, and Whiteway M (2005). Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol. Microbiol 57, 942–959. [DOI] [PubMed] [Google Scholar]
- Bai C, Ramanan N, Wang YM, and Wang Y (2002). Spindle assembly checkpoint component CaMad2p is indispensable for Candida albicans survival and virulence in mice. Mol. Microbiol 45, 31–44. [DOI] [PubMed] [Google Scholar]
- Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, López-Ribot JL, and Kadosh D (2008). UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19, 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S (2018). Candida albicans Cdc15 is essential for mitotic exit and cytokinesis. Sci. Rep 8, 8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Filler SG, and Berman J (2002). A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Clemente-Blanco A, Finley KR, Correa-Bordes J, and Berman J (2005). The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol. Biol. Cell 16, 3387–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J (2006). Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol 9, 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo SM, Khalique Z, Kot J, Jones JK, and Lee SA (2008). Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genet. Biol 45, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongomin F, Gago S, Oladele RO, and Denning DW (2017). Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi (Basel) 3, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Kadosh D, and Johnson AD (2001). NRG1, a repressor of filamentous growth in C.albicans, is down-regulated during filament induction. EMBO J. 20, 4753–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Lima D, and Sudbery PE (2014). In Candida albicans, phosphorylation of Exo84 by Cdk1-Hgc1 is necessary for efficient hyphal extension. Mol. Biol. Cell 25, 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, López-Ribot JL, and Kadosh D (2009). Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. USA 106, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapa y Lazo B, Bates S, and Sudbery P (2005). The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot. Cell 4, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zeng G, and Wang Y (2018). G1 and S phase arrest in Candida albicans induces filamentous growth via distinct mechanisms. Mol. Microbiol 110, 191–203. [DOI] [PubMed] [Google Scholar]
- Chou H, Glory A, and Bachewich C (2011). Orthologues of the anaphase-promoting complex/cyclosome coactivators Cdc20p and Cdh1p are important for mitotic progression and morphogenesis in Candida albicans. Eukaryot. Cell 10, 696–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C (2014). Proteasome dynamics. Biochim. Biophys. Acta 1843, 39–46. [DOI] [PubMed] [Google Scholar]
- Feng J, Duan Y, Sun W, Qin Y, Zhuang Z, Zhu D, Sun X, and Jiang L (2016). CaTip41 regulates protein phosphatase 2A activity, CaRad53 deactivation and the recovery of DNA damage-induced filamentation to yeast form in Candida albicans. FEMS Yeast Res. 16, fow009. [DOI] [PubMed] [Google Scholar]
- Finley KR, Bouchonville KJ, Quick A, and Berman J (2008). Dynein-dependent nuclear dynamics affect morphogenesis in Candida albicans by means of the Bub2p spindle checkpoint. J. Cell Sci 121, 466–476. [DOI] [PubMed] [Google Scholar]
- Finley D, Ulrich HD, Sommer T, and Kaiser P (2012). The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192, 319–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goes FS, and Martin J (2001). Hsp90 chaperone complexes are required for the activity and stability of yeast protein kinases Mik1, Wee1 and Swe1. Eur. J. Biochem 268, 2281–2289. [DOI] [PubMed] [Google Scholar]
- González-Novo A, Correa-Bordes J, Labrador L, Sánchez M, Vázquez de Aldana CR, and Jiménez J (2008). Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol. Biol. Cell 19, 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig JA, Sudbery IM, Richardson JP, Naglik JR, Wang Y, and Sudbery PE (2015). Cell cycle-independent phospho-regulation of Fkh2 during hyphal growth regulates Candida albicans pathogenesis. PLoS Pathog. 11, e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Pan C, Wang Y, Wang N, Wang Y, and Sang J (2019). The PP2A regulatory subunits, Cdc55 and Rts1, play distinct roles in Candida albicans’ growth, morphogenesis, and virulence. Fungal Genet. Biol 131, 103240. [DOI] [PubMed] [Google Scholar]
- Hossain S, Veri AO, and Cowen LE (2020). The proteasome governs fungal morphogenesis via functional connections with Hsp90 and cAMP-protein kinase A signaling. mBio 11, e00290–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MY, Woolford CA, May G, McManus CJ, and Mitchell AP (2019). Circuit diversification in a biofilm regulatory network. PLoS Pathog. 15, e1007787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, and Weisman LS (2015). The vacuole/lysosome is required for cell-cycle progression. ELife 4, e08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamthan M, Nalla VK, Ruhela D, Kamthan A, Maiti P, and Datta A (2014). Characterization of a putative spindle assembly checkpoint kinase Mps1, suggests its involvement in cell division, morphogenesis and oxidative stress tolerance in Candida albicans. PLoS ONE 9, e101517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry ME, Kemp HA, Rivers DM, and Sprague GF Jr. (2004). The identification of Pcl1-interacting proteins that genetically interact with Cla4 may indicate a link between G1 progression and mitotic exit. Genetics 166, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana N, Bakshi S, Tabassum W, Bhattacharyya MK, and Bhattacharyya S (2018). Hsp90 Is essential for Chl1-mediated chromosome segregation and sister chromatid cohesion. mSphere 3, e00225–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer D (2019). Regulation of Candida albicans hyphal morphogenesis by endogenous signals. J. Fungi (Basel) 5, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg BJ, and Arendrup MC (2015). Invasive Candidiasis. N. Engl. J. Med 373, 1445–1456. [DOI] [PubMed] [Google Scholar]
- Leach MD, and Cowen LE (2014). To sense or die: mechanisms of temperature sensing in fungal pathogens. Curr. Fungal Infect. Rep 8, 185–191. [Google Scholar]
- Leach MD, Stead DA, Argo E, MacCallum DM, and Brown AJP (2011). Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol. Microbiol 79, 1574–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MD, Tyc KM, Brown AJP, and Klipp E (2012a). Modelling the regulation of thermal adaptation in Candida albicans, a major fungal pathogen of humans. PLoS ONE 7, e32467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MD, Budge S, Walker L, Munro C, Cowen LE, and Brown AJP (2012b). Hsp90 orchestrates transcriptional regulation by Hsf1 and cell wall remodelling by MAPK signalling during thermal adaptation in a pathogenic yeast. PLoS Pathog. 8, e1003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MD, Farrer RA, Tan K, Miao Z, Walker LA, Cuomo CA, Wheeler RT, Brown AJP, Wong KH, and Cowen LE (2016). Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans. Nat. Commun 7, 11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-J, Kim E-H, Lee JS, Jeoung D, Bae S, Kwon SH, and Lee Y-S (2008). HSF1 as a mitotic regulator: phosphorylation of HSF1 by Plk1 is essential for mitotic progression. Cancer Res. 68, 7550–7560. [DOI] [PubMed] [Google Scholar]
- Leng P, Sudbery PE, and Brown AJ (2000). Rad6p represses yeast-hypha morphogenesis in the human fungal pathogen Candida albicans. Mol. Microbiol 35, 1264–1275. [DOI] [PubMed] [Google Scholar]
- Lew DJ, and Reed SI (1993). Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol 120, 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WJ, Wang YM, Zheng XD, Shi QM, Zhang TT, Bai C, Li D, Sang JL, and Wang Y (2006). The F-box protein Grr1 regulates the stability of Ccn1, Cln3 and Hof1 and cell morphogenesis in Candida albicans. Mol. Microbiol 62, 212–226. [DOI] [PubMed] [Google Scholar]
- Li C-R, Au Yong JY, Wang Y-M, and Wang Y (2012). CDK regulates septin organization through cell-cycle-dependent phosphorylation of the Nim1-related kinase Gin4. J. Cell Sci 125, 2533–2543. [DOI] [PubMed] [Google Scholar]
- Liu Q, Han Q, Wang N, Yao G, Zeng G, Wang Y, Huang Z, Sang J, and Wang Y (2016). Tpd3-Pph21 phosphatase plays a direct role in Sep7 dephosphorylation in Candida albicans. Mol. Microbiol 101, 109–121. [DOI] [PubMed] [Google Scholar]
- Loeb JDJ, Sepulveda-Becerra M, Hazan I, and Liu H (1999). A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol. Cell. Biol 19, 4019–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll-Krippleber R, d’Enfert C, Feri A, Diogo D, Perin A, Marcet-Houben M, Bougnoux M-E, and Legrand M (2014). A study of the DNA damage checkpoint in Candida albicans: uncoupling of the functions of Rad53 in DNA repair, cell cycle regulation and genotoxic stress-induced polarized growth. Mol. Microbiol 91, 452–471. [DOI] [PubMed] [Google Scholar]
- Lu Y, Su C, Unoje O, and Liu H (2014). Quorum sensing controls hyphal initiation in Candida albicans through Ubr1-mediated protein degradation. Proc. Natl. Acad. Sci. USA 111, 1975–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Su C, Ray S, Yuan Y, and Liu H (2019). CO2 signaling through the Ptc2-Ssn3 axis governs sustained hyphal development of Candida albicans by reducing Ume6 phosphorylation and degradation. mBio 10, e02320–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Eguren M, and Malumbres M (2010). The anaphase-promoting complex/cyclosome (APC/C): cell-cycle-dependent and -independent functions. Biochem. Soc. Trans 38, 65–71. [DOI] [PubMed] [Google Scholar]
- McCusker D, Denison C, Anderson S, Egelhofer TA, Yates JR 3rd, Gygi SP, and Kellogg DR (2007). Cdk1 coordinates cell-surface growth with the cell cycle. Nat. Cell Biol 9, 506–515. [DOI] [PubMed] [Google Scholar]
- Mendelsohn S, Pinsky M, Weissman Z, and Kornitzer D (2017). Regulation of the Candida albicans hypha-inducing transcription factor Ume6 by the CDK1 cyclins Cln3 and Hgc1. mSphere 2, e00248–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne SW, Cheetham J, Lloyd D, Shaw S, Moore K, Paszkiewicz KH, Aves SJ, and Bates S (2014). Role of Candida albicans Tem1 in mitotic exit and cytokinesis. Fungal Genet. Biol 69, 84–95. [DOI] [PubMed] [Google Scholar]
- Mollapour M, Tsutsumi S, Donnelly AC, Beebe K, Tokita MJ, Lee M-J, Lee S, Morra G, Bourboulia D, Scroggins BT, et al. (2010). Swe1Wee1-dependent tyrosine phosphorylation of Hsp90 regulates distinct facets of chaperone function. Mol. Cell 37, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano KA, Santoro N, Koch KA, and Thiele DJ (1999). A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol. Cell. Biol 19, 402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, Schnell N, Talibi D, Marechal D, Tekaia F, et al. (2001). NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20, 4742–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R, Shariq M, Dhamgaye S, Mukhopadhyay CK, Shaikh S, and Prasad R (2017). Non-heat shock responsive roles of HSF1 in Candida albicans are essential under iron deprivation and drug defense. Biochim. Biophys. Acta Mol. Cell Res 1864, 345–354. [DOI] [PubMed] [Google Scholar]
- Nicholls S, Leach MD, Priest CL, and Brown AJP (2009). Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol. Microbiol 74, 844–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls S, MacCallum DM, Kaffarnik FAR, Selway L, Peck SC, and Brown AJP (2011). Activation of the heat shock transcription factor Hsf1 is essential for the full virulence of the fungal pathogen Candida albicans. Fungal Genet. Biol 48, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, and Johnson AD (2010). Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet 42, 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Gianetti BA, and Witchley JN (2017). Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol 15, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, and Cowen LE (2015). Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat. Commun 6, 6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Robbins N, and Cowen LE (2017). The Hsp90 chaperone network modulates Candida Virulence traits. Trends Microbiol. 25, 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, O’Meara MJ, Polvi EJ, Pourhaghighi MR, Liston SD, Lin Z-Y, Veri AO, Emili A, Gingras A-C, and Cowen LE (2019). Global proteomic analyses define an environmentally contingent Hsp90 interactome and reveal chaperone-dependent regulation of stress granule proteins and the R2TP complex in a fungal pathogen. PLoS Biol. 17, e3000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer GE, Kelly MN, and Sturtevant JE (2005). The Candida albicans vacuole is required for differentiation and efficient macrophage killing. Eukaryot. Cell 4, 1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín J, Bardetti P, Castanheira S, de la Torre A, and Tenorio-Gómez M (2016). Virulence-specific cell cycle and morphogenesis connections in pathogenic fungi. Semin. Cell Dev. Biol 57, 93–99. [DOI] [PubMed] [Google Scholar]
- Poltermann S, Nguyen M, Günther J, Wendland J, Härtl A, Künkel W, Zipfel PF, and Eck R (2005). The putative vacuolar ATPase subunit Vma7p of Candida albicans is involved in vacuole acidification, hyphal development and virulence. Microbiology (Reading) 151, 1645–1655. [DOI] [PubMed] [Google Scholar]
- Polvi EJ, Li X, O’Meara TR, Leach MD, and Cowen LE (2015). Opportunistic yeast pathogens: reservoirs, virulence mechanisms, and therapeutic strategies. Cell. Mol. Life Sci 72, 2261–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane HS, Bernardo SM, Raines SM, Binder JL, Parra KJ, and Lee SA (2013). Candida albicans VMA3 is necessary for V-ATPase assembly and function and contributes to secretion and filamentation. Eukaryot. Cell 12, 1369–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane HS, Bernardo SM, Hayek SR, Binder JL, Parra KJ, and Lee SA (2014). The contribution of Candida albicans vacuolar ATPase subunit V1B, encoded by VMA2, to stress response, autophagy, and virulence is independent of environmental pH. Eukaryot. Cell 13, 1207–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins N, Wright GD, and Cowen LE (2016). Antifungal drugs: the current armamentarium and development of new agents. Microbiol. Spectr 4, FUNK-0002–2016. [DOI] [PubMed] [Google Scholar]
- Roig P, and Gozalbo D (2003). Depletion of polyubiquitin encoded by the UBI4 gene confers pleiotropic phenotype to Candida albicans cells. Fungal Genet. Biol 39, 70–81. [DOI] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Monteagudo C, and Lopez-Ribot JL (2003). Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2, 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte TW, Blagosklonny MV, Ingui C, and Neckers L (1995). Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J. Biol. Chem 270, 24585–24588. [DOI] [PubMed] [Google Scholar]
- Sela N, Atir-Lande A, and Kornitzer D (2012). Neddylation and CAND1 independently stimulate SCF ubiquitin ligase activity in Candida albicans. Eukaryot. Cell 11, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn H, Shapiro RS, and Cowen LE (2012). Cdc28 provides a molecular link between Hsp90, morphogenesis, and cell cycle progression in Candida albicans. Mol. Biol. Cell 23, 268–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda P, Cervera AM, Lopez-Ribot JL, Chaffin WL, Martinez JP, and Gozalbo D (1996). Cloning and characterization of a cDNA coding for Candida albicans polyubiquitin. J. Med. Vet. Mycol 34, 315–322. [PubMed] [Google Scholar]
- Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, Heitman J, and Cowen LE (2009). Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol 19, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Robbins N, and Cowen LE (2011). Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev 75, 213–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Zaas AK, Betancourt-Quiroz M, Perfect JR, and Cowen LE (2012a). The Hsp90 co-chaperone Sgt1 governs Candida albicans morphogenesis and drug resistance. PLoS ONE 7, e44734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Sellam A, Tebbji F, Whiteway M, Nantel A, and Cowen LE (2012b). Pho85, Pcl1, and Hms1 signaling governs Candida albicans morphogenesis induced by high temperature or Hsp90 compromise. Curr. Biol 22, 461–470. [DOI] [PubMed] [Google Scholar]
- Shi Q-M, Wang Y-M, Zheng X-D, Lee RTH, and Wang Y (2007). Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol. Biol. Cell 18, 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Lavy KJ, Oren YS, Feine O, Sajman J, Listovsky T, and Brandeis M (2010). Fifteen years of APC/cyclosome: a short and impressive biography. Biochem. Soc. Trans 38, 78–82. [DOI] [PubMed] [Google Scholar]
- Sinha I, Wang Y-M, Philp R, Li C-R, Yap WH, and Wang Y (2007). Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev. Cell 13, 421–432. [DOI] [PubMed] [Google Scholar]
- Sudbery PE (2011). Growth of Candida albicans hyphae. Nat. Rev. Microbiol 9, 737–748. [DOI] [PubMed] [Google Scholar]
- Sudbery P, Gow N, and Berman J (2004). The distinct morphogenic states of Candida albicans. Trends Microbiol. 12, 317–324. [DOI] [PubMed] [Google Scholar]
- Sun LL, Li WJ, Wang HT, Chen J, Deng P, Wang Y, and Sang JL (2011). Protein phosphatase Pph3 and its regulatory subunit Psy2 regulate Rad53 dephosphorylation and cell morphogenesis during recovery from DNA damage in Candida albicans. Eukaryot. Cell 10, 1565–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, and Lindquist S (2010). HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol 11, 515–528. [DOI] [PubMed] [Google Scholar]
- Trunk K, Gendron P, Nantel A, Lemieux S, Roemer T, and Raymond M (2009). Depletion of the cullin Cdc53p induces morphogenetic changes in Candida albicans. Eukaryot. Cell 8, 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherner M, Stappler E, Hnisz D, and Kuchler K (2012). The histone acetyltransferase Hat1 facilitates DNA damage repair and morphogenesis in Candida albicans. Mol. Microbiol 86, 1197–1214. [DOI] [PubMed] [Google Scholar]
- Umeyama T, Kaneko A, Niimi M, and Uehara Y (2006). Repression of CDC28 reduces the expression of the morphology-related transcription factors, Efg1p, Nrg1p, Rbf1p, Rim101p, Fkh2p and Tec1p and induces cell elongation in Candida albicans. Yeast 23, 537–552. [DOI] [PubMed] [Google Scholar]
- Veri AO, Miao Z, Shapiro RS, Tebbji F, O’Meara TR, Kim SH, Colazo J, Tan K, Vyas VK, Whiteway M, et al. (2018a). Tuning Hsf1 levels drives distinct fungal morphogenetic programs with depletion impairing Hsp90 function and overexpression expanding the target space. PLoS Genet. 14, e1007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veri AO, Robbins N, and Cowen LE (2018b). Regulation of the heat shock transcription factor Hsf1 in fungi: implications for temperature-dependent virulence traits. FEMS Yeast Res. 18, foy041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila T, Romo JA, Pierce CG, McHardy SF, Saville SP, and Lopez-Ribot JL (2017). Targeting Candida albicans filamentation for antifungal drug development. Virulence 8, 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y (2016). Hgc1-Cdc28-how much does a single protein kinase do in the regulation of hyphal development in Candida albicans? J. Microbiol 54, 170–177. [DOI] [PubMed] [Google Scholar]
- Wang A, Raniga PP, Lane S, Lu Y, and Liu H (2009). Hyphal chain formation in Candida albicans: Cdc28-Hgc1 phosphorylation of Efg1 represses cell separation genes. Mol. Cell. Biol 29, 4406–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Gao J, Li W, Wong AH-H, Hu K, Chen K, Wang Y, and Sang J (2012). Pph3 dephosphorylation of Rad53 is required for cell recovery from MMS-induced DNA damage in Candida albicans. PLoS ONE 7, e37246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Huang Z-X, Au Yong JY, Zou H, Zeng G, Gao J, Wang Y, Wong AH-H, and Wang Y (2016). CDK phosphorylates the polarisome scaffold Spa2 to maintain its localization at the site of cell growth. Mol. Microbiol 101, 250–264. [DOI] [PubMed] [Google Scholar]
- Warenda AJ, and Konopka JB (2002). Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 13, 2732–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM (2001). Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol 2, 169–178. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, De Costa B, Myers CE, and Neckers LM (1994). Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. USA 91, 8324–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Robbins N, Huang DS, McLellan CA, Shekhar-Guturja T, LeBlanc EV, Nation CS, Hui R, Hutchinson A, Collins C, et al. (2019). Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus. Nat. Commun 10, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R, Bates S, Amornrrattanapan P, and Sudbery P (2004). In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J. Cell Biol 164, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Schwab M, and Tyers M (2004). A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695, 133–170. [DOI] [PubMed] [Google Scholar]
- Woolford CA, Lagree K, Xu W, Aleynikov T, Adhikari H, Sanchez H, Cullen PJ, Lanni F, Andes DR, and Mitchell AP (2016). Bypass of Candida albicans filamentation/biofilm regulators through diminished expression of protein kinase Cak1. PLoS Genet. 12, e1006487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzov P, Boucherie H, and Mann C (1997). A yeast heat shock transcription factor (Hsf1) mutant is defective in both Hsc82/Hsp82 synthesis and spindle pole body duplication. J. Cell Sci 110, 1879–1891. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wang Y, and Wang Y (2004). Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23, 1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X-D, Lee RTH, Wang Y-M, Lin Q-S, and Wang Y (2007). Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 26, 3760–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]


