Abstract
Bats can harbor zoonotic pathogens causing emerging infectious diseases, but their status as hosts for bacteria is limited. We aimed to investigate the distribution, prevalence and genetic diversity of Borrelia in bats and bat ticks in Hubei Province, China, which will give us a better understanding of the risk of Borrelia infection posed by bats and their ticks. During 2018–2020, 403 bats were captured from caves in Hubei Province, China, 2 bats were PCR-positive for Borrelia. Sequence analysis of rrs, flaB and glpQ genes of positive samples showed 99.55%-100% similarity to Candidatus Borrelia fainii, a novel human-pathogenic relapsing fever Borrelia species recently reported in Zambia, Africa and Eastern China, which was clustered together with relapsing fever Borrelia species traditionally reported only in the New World. Multilocus sequence typing (MLST) and pairwise genetic distances further confirmed the Borrelia species in the bats from Central China as Candidatus Borrelia fainii. No Borrelia DNA was detected in ticks collected from bats. The detection of this human-pathogenic relapsing fever Borrelia in bats suggests a wide distribution of this novel relapsing fever Borrelia species in China, which may pose a threat to public health in China.
Author summary
Bats have a wide range of species and distribution, these mammals have been proposed as the natural reservoirs of pathogens for many years. Previous research focus mainly on viral agents in bats, while the infection of Borrelia spirochetes in bats is much neglected. Borrelia species causing Lyme disease and relapsing fever have been found in a wide range of ticks. Relapsing fever has long been recognized as major cause of disease and death in Africa, but as more and more cases reported in different continents, relapsing fever has become an emergent disease. There is growing evidence that bats can play a role in carrying Borrelia species. In this study, we captured different species of bats from caves during 2018–2020 in Central China, tested liver tissue for Borrelia by PCR, and applied multilocus sequence typing (MLST) to identify this bacterium. Our study found human pathogenic relapsing fever Candidatus Borrelia fainii in Central China, suggesting bat borne Borrelia spirochetes may pose a threat to public health in China.
Introduction
Relapsing fever is a zoonosis caused by the relapsing fever group spirochetes of the genus Borrelia, and its clinical symptoms include recurrent febrile episodes with headache, myalgia, chills, and nausea [1]. To date, 27 relapsing fever Borrelia species spirochetes have been identified [2], but additional species have been proposed. Typically, relapsing fever borreliae were thought to be transmitted by soft ticks, with the exception of the human louse-borne B. recurrentis as well as the hard tick-borne B. lonestari and B. miyamotoi [3–5]. Historically, the relapsing fever borreliae were classified by the “one vector one species” concept, and in each region, a specific relationship existed between soft ticks and Borrelia spp. [6]. According to the epidemic areas and genetic lineage of the causative agent, the relapsing fever borreliae were rather arbitrarily divided into two subgroups: the Old World borreliae (the Palearctic and Afrotropic ecozone) and the New World borreliae (Nearctic ecozone) [7].
Bats have attracted much attention from their close relationship with emerging viral infectious diseases, including Nipah, Hendra, SARS, MERS and the recent worldwide pandemic COVID-19 [8]. It is worth noting that previous research focused mainly on viral agents in bats, while the infection and prevalence of bacteria in bats are much neglected [9]. Knowing whether bats can act as carriers of pathogenic bacteria is a common interest for public health [10–12]. Ticks, as one of the parasites of bats, act as the most important arthropod vectors for transmitting microbial pathogens to animals and humans. Once bitten by these ticks, the risk of infection will increase. Therefore, the role of both bats and their ticks in carrying or transmitting bacteria deserves more study. A recent study found bats and bat soft ticks from a cave in Zambia, Africa showed a high infection rate with Candidatus Borrelia fainii, which was a novel human-pathogenic relapsing fever Borrelia species clustered together with the New World relapsing fever borreliae. It proposed that bats and bat soft ticks contributed to the environmental cycle of this Borrelia species as hosts and vectors, respectively [13]. Our group also found that one Myotis bat from Shandong Province in Eastern China was infected with Candidatus Borrelia fainii, as the first report of this novel relapsing fever Borrelia species in Asia [14]. Since only a single bat was found to be infected with the Borrelia species, the role of bats in the transmission of Borrelia cannot be determined in China. In this study, we aimed to investigate the distribution, prevalence and genetic diversity of Borrelia in bats and bat ticks from Hubei Province, China, which will give us a better understanding of the risk of Borrelia infection posed by bats and their ticks.
Material and methods
Ethical statement
The collection of bats for microbiological studies was approved by the Ethics Committee of the Medical School, Wuhan University (WHU2020-YF0023), and all efforts were made to minimize discomfort to the animals.
Bat sampling
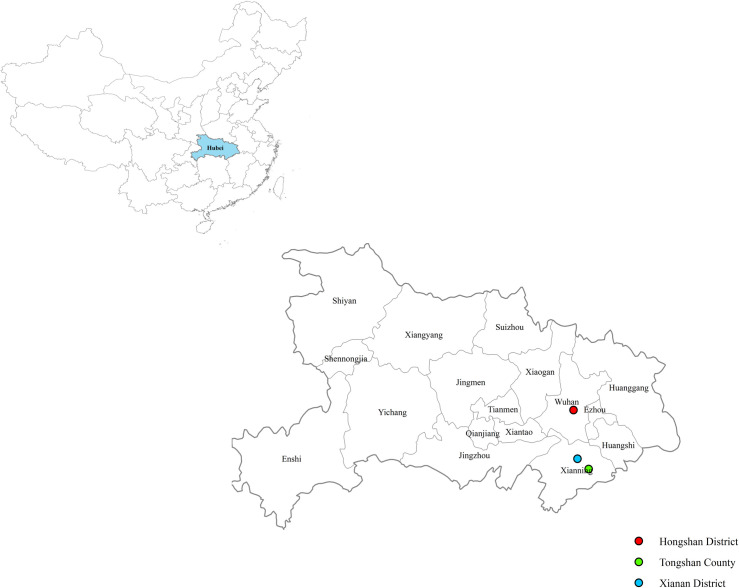
From May 2018 to August 2020, bats were collected from caves in Xianning City and Wuhan City of Hubei Province in Central China (Fig 1). Bats were captured using mist nets settled near the entrance of the cave at sunset when bats left for night feeding, and bats were collected early the next morning. Bats were euthanized by an overdose of chloral hydrate administered via intraperitoneal injection, and tissue samples were collected and stored at -80°C. Bat species were identified with morphology, and then confirmed by amplifying and sequencing the cytochrome B (cytB) gene as described previously [15].
Fig 1. Map showing the location where bats were collected in Hubei Province, China.
The map constructed using ArcGIS 10.6 software.
Molecular detection of relapsing fever borreliae in bats
DNA was extracted from the liver tissue of each bat with QIAGEN DNA Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Bats were initially screened with the universal 16S rRNA (rrs) primers of bacteria [16], and two bats were found positive for relapsing fever Borrelia. To further characterize the relapsing fever Borrelia from the bats, Borrelia flagellin (flaB) [17] and the glycerophosphodiester phosphodiesterase (glpQ) [18] genes were amplified (S1 Table) with PCR.
The PCR reaction was conducted in a 15 μL mixture containing 7.5 μL of 2X Taq Master Mix (Vazyme Biotech, China), 0.5 μL of 10 μM of each forward and reverse primer (Sangon Biotech, Shanghai, China), 4.5 μL of nuclease-free water, and 2 μL liver DNA of each sample. Nuclease-free water was used as a negative control. To prevent contamination, no positive control was used in the study. PCR was performed under the following conditions: 1 denaturing cycle at 95°C for 5 min followed by 40 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 90 s, and an additional final cycle at 72°C for 10 min. Each PCR assay included a negative control (distilled water instead of DNA template).PCR products were analyzed with 1.2% agarose gel electrophoresis, and bands of expected size (1,494 bp for rrs, 753 bp for flaB, 599 bp for glpQ fragment 1, and 453 bp for glpQ fragment 2) were excised from gels, and purified with a Gel Extraction Kit (TSINGKE Biological Technology, China). The purified amplicons were inserted into the pMD19-T vector (TaKaRa, Shiga, Japan) for cloning, and at least three positive clones were selected for sequencing with universal primers M13-47 and M13-48. Chromatograms were checked with Chromas 2.5.1 (Technelysium, Tewantin, QLD) to ensure the accuracy of sequencing. The 2 glpQ fragments were assembled into an 880 bp sequence with Lasergene 7.1 (DNASTAR, Madison, WI, USA). The obtained sequences were compared to those in public databases using the Nucleotide Basic Local Alignment Search Tool (BLASTn) on the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Molecular detection of relapsing fever borreliae in bat ticks
Ticks were collected from bats, and tick species were identified by PCR targeting tick mitochondrial 16S rDNA as previously described [19] (S1 Table). DNA was extracted from pools of ticks with AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA). Examination of the Borrelia DNA in the ticks was performed by PCR and the sequencing of amplicons as described above.
Phylogenetic analysis
Phylogenetic analysis was conducted using MEGA 7.0. ClustalW software to align the sequences. Phylogenetic trees were constructed based on neighbor-joining method with the Kimura 2-parameter model in MEGA 7.0, bootstrap values were calculated with 1, 000 replicates.
Multilocus sequence typing of relapsing fever borreliae
For relapsing fever Borrelia-positive samples, MLST was attempted by PCR amplification of 8 housekeeping genes (clpA, clpX, nifS, pepX, pyrG, recG, rplB and uvrA) with degenerate primers kindly provided by Dr. Gabriele Margos, who is the curator of the Borrelia MLST database (https://pubmlst.org/borrelia/) (S1 Table). The amplicons were cloned for sequencing as described above.
Sequences were analyzed using the sequence query function of the Borrelia MLST database, and each gene was compared with its existing alleles. If the sequence was new to the database, new allele numbers were assigned after being submitted to the Borrelia MLST database, and based on the allelic profile of the 8 housekeeping genes, the Borrelia species would be assigned an existing or new (for those with new combinations of alleles or novel alleles) ST number.
Sequences of the 8 housekeeping genes were trimmed as described in the Borrelia MLST database, and concatenated in the order of clpA, clpX, nifS, pepX, pyrG, recG, rplB and uvrA. Concatenated sequences were imported into MEGA 7.0. After being aligned with ClustalW, a phylogenetic tree was constructed by using the neighbor-joining method with the Kimura 2-parameter model in MEGA 7.0.
Pairwise genetic distances were calculated by using the Kimura-2 model, and relapsing fever Borrelia species were identified using the threshold (98.3% similarity, genetic distance 0.017) [20]. Reference sequences of Borrelia species included in the analyses were obtained from the Borrelia MLST database as well as the GenBank.
Results
Bat and tick sampling
From May 2018 to August 2020, a total of 403 bats were sampled from caves in Xianning City and Wuhan City, Hubei Province, China. By amplifying and sequencing the cytB gene, bats were identified into five species of two families (Table 1). Fourteen ticks were collected from bats sampled in July 2020, and they were identified as 8 Carios vespertilionis and 6 Ixodes simplex. The cytB gene of representative bat species and the 16S rDNA sequences of bat ticks were deposited in GenBank with accession No. MH888178, MH888180, MW085077-MW085079 and MW132810-MW132811.
Table 1. Summary of bat sampling information.
| Sampling date | Sampling area | Bat family | Bat species | Bats No. |
|---|---|---|---|---|
| May 2018 | Xianan District, Xianning (29°78′N, 114°31′E) | Vespertilionidae | Myotis davidii | 42 |
| Myotis adversus | 15 | |||
| Myotis altarium | 2 | |||
| Miniipterus schreibersii | 9 | |||
| July 2020 | Tongshan County, Xianning (29°61′N, 114°48′E) | Vespertilionidae | Myotis davidii | 124 |
| Myotis adversus | 38 | |||
| Rhinolophidae | Rhinolophus pusillus | 154 | ||
| August 2020 | Hongshan District, Wuhan (30°50′N, 114°34′E) | Vespertilionidae | Myotis davidii | 16 |
| Myotis adversus | 1 | |||
| Myotis altarium | 1 | |||
| Rhinolophidae | Rhinolophus pusillus | 1 | ||
| Total | 403 |
Molecular detection of relapsing fever borreliae
Two out of 403 bats were positive for relapsing fever Borrelia by PCR amplification of rrs, flaB and glpQ. The 2 positive bats were Rh. pusillus (bat ID: XN788) and My. davidii (bat ID: XN888) collected from Xianning City in July 2020. Borrelia DNA was also detected in the spleen, lung, kidney and blood samples of Borrelia-positive bats. Tick was not found on these two infected bats, and no Borrelia DNA was detected in ticks collected from bats.
Based on rrs, flaB and glpQ genes, relapsing fever Borrelia detected in bats from this study were closely related to Borrelia sp. Qtaro and Borrelia sp. SD065, which were designated as Candidatus Borrelia fainii, and were reported in Zambia and China, respectively (Table 2). Neighbor-joining phylogenetic trees constructed based on rrs, flaB and glpQ genes showed that Borrelia detected in bats of this study clustered with a group of human pathogenic New World relapsing fever borreliae (Fig 2). The rrs, flaB and glpQ genes of relapsing fever Borrelia of this study were deposited in GenBank with accession No. MT913156-MT913157 (rrs), MT919991-MT919992 (flaB) and MT975516, MT919993 (glpQ).
Table 2. Best matches to Borrelia gene sequences found in GenBank.
| Borrelia-positive bat ID number | Bat species | Borrelia gene | Closest match in GenBank | Accession number of closest match | Nucleotide identify | Accession number of this study |
|---|---|---|---|---|---|---|
| XN788 | Rhinolophus pusillus | rrs | Borrelia sp. Qtaro | LC382043 | 99.93% (1493/1494) | MT913156 |
| flaB | Borrelia sp. SD065 | MG832413 | 99.86% (750/751) | MT919991 | ||
| glpQ | Borrelia sp. SD065 | MG921625 | 100%(880/880) | MT975516 | ||
| XN888 | Myotis davidii | rrs | Borrelia sp. Qtaro | LC382043 | 99.93% (1493/1494) | MT913157 |
| flaB | Borrelia sp. SD065 | MG832413 | 100% (751/751) | MT919992 | ||
| glpQ | Borrelia sp. SD065 | MG921625 | 99.55% (876/880) | MT919993 |
Fig 2.
Phylogenetic analysis of (A) rrs, (B) flaB and (C) glpQ gene of relapsing fever Borrelia species. The sequences M60967 from (A) and L42876 from (B) are outgroup. Phylogenetic tree was drawn using the neighbor-joining method with the Kimura 2-parameter model in MEGA 7.0. Phylogenetic tree representing the relationships among Borrelia identified in this study (50% boot cutoff). Bootstrap values are indicated at the nodes. Scale bar indicates the degree of divergence represented by a given length of branch. Boldface indicates the taxonomic position of Borrelia. Borrelia species identified in bats from this study were shown in red.
MLST analysis
For Borrelia-positive Rh. pusillus (bat ID: XN788), sequence query showed that the 8 housekeeping genes were exactly the same as ST927, which was previously reported by our team in a Myotis ricketti bat from Shandong Province, China [14]. For Borrelia-positive My. davidii (bat ID: XN888), the 8 loci of MLST were all novel alleles, and the 8 loci were submitted to the Borrelia MLST database and novel allele numbers were assigned: clpA (303), clpX (263), nifS (238), pepX (267), pyrG (278), recG (294), rplB (256), and uvrA (270), and sequence typing was assigned as ST938. Phylogenetic analysis based on the concatenated 8 housekeeping genes (4,776 bp, in the order of clpA-clpX-nifS-pepX-pyrG-recG-rplB-uvrA) revealed that the Borrelia species found in bats from this study clustered with a group of human pathogenic relapsing fever Borrelia spirochetes, and were most related to Candidatus Borrelia fainii, which was recently identified in a febrile patient, bats and soft ticks from Zambia [13], as well as in a bat from Shandong Province, China [14] (Fig 3). Phylogenetic trees based on each of the 8 loci of relapsing fever Borrelia were shown in S1–S8 Figs. Genetic distance analysis of the concatenated 8 loci (4,776 bp) revealed a value of 0.015 compared to Candidatus Borrelia fainii strain Qtar (S2 Table). Herein, the Borrelia species detected in bats from this study was identified as Candidatus Borrelia fainii.
Fig 3. Phylogenetic analysis of borreliae based on concatenated sequences of the 8 loci in the order of clpA, clpX, nifS, pepX, pyrG, recG, rplB and uvrA.
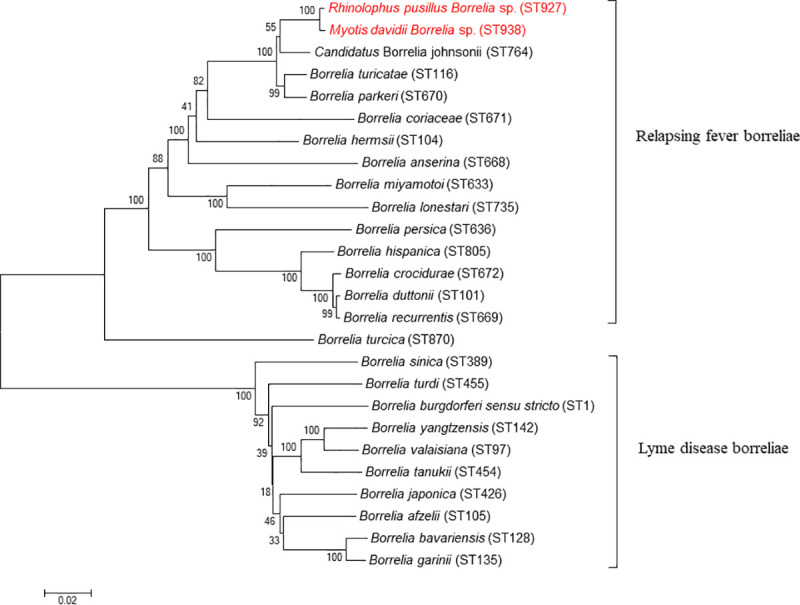
Phylogenetic tree was drawn using the neighbor-joining method with the Kimura 2-parameter model in MEGA 7.0. Phylogenetic tree representing the relationships among Borrelia identified in this study (50% boot cutoff). Bootstrap values are indicated at the nodes. Scale bar indicates the degree of divergence represented by a given length of branch. Boldface indicates the taxonomic position of Borrelia. Borrelia sequences identified in bats from this study were shown in red.
Discussion
In this study, we found that two bats captured from Xianning City, Hubei Province of China were positive for Candidatus Borrelia fainii, which was recently isolated from a patient diagnosed with relapsing fever in Zambia, and clustered together with the relapsing fever borreliae [13]. Candidatus Borrelia fainii is closely related to a group of New World relapsing fever borreliae, including B. turicatae, B. parkeri and Candidatus Borrelia johnsonii, which were endemic in USA. Borrelia turicatae and B. parkeri cause tick-borne relapsing fever in humans[21,22], and they are predominantly prevalent in the southwestern of USA [23]. Candidatus Borrelia johnsonii was firstly reported in bat soft ticks distributed in the USA [24]. A recent study found that Candidatus Borrelia johnsonii was associated with tick-borne diseases in humans [20].
Previous reports of Borrelia species in bats were only individual cases [25,26]. A novel Old World relapsing fever Borrelia species (CPB1) was found responsible for fatal borreliosis in a Pipistrelle bat from the UK [25]. Subsequently, Borrelia species CPB1 was detected in soft ticks from bats in France [27], indicating the association of relapsing fever Borrelia spirochetes and bat soft ticks. A recent study found that bats and bat soft ticks collected from a cave in Zambia showed a high infection rate for Candidatus Borrelia fainii, and proposed that bats and bat soft ticks contributed to the environmental cycle of Candidatus Borrelia fainii as hosts and vectors, respectively [13]. However, our previous study found that only one out of 145 bats collected from Shandong Province, China was infected with Candidatus Borrelia fainii [14]. In this study, only 2 out of 403 bats were found positive for Candidatus Borrelia fainii. The Borrelia-positive bat species found in China in this study include Myotis ricketti in Shandong Province [14], Rhinolophus pusillus and Myotis davidii in Hubei Province of this study. Myotis ricketti is distributed in China, Vietnam and Laos, Rhinolophus pusillus can be found in Southern China, India, Nepal, Myanmar, Vietnam, Thailand, Malaysia and Indonesia, while Myotis davidii is endemic to China (http://www.bio.bris.ac.uk/research/bats/China bats/). Besides, the liver and spleen of the two Borrelia-positive bats appeared to be enlarged. Borrelia DNA was also detected in spleen, lung, and kidney tissue as well as blood samples of Borrelia-positive bats, indicating multiorgan infection resulted from spirochetemia. However none of the ticks collected from these bats was positive for Borrelia by PCR, possibly due to the small sample size. Given the facts that this relapsing fever Borrelia species infection could be fatal to bats and the infection rate of this Borrelia species in bats was low, Borrelia infection in bats might be incidental events, just as the cases in humans. Further studies are needed to investigate the reservoir and vector of this novel relapsing fever Borrelia species in China.
Besides Candidatus Borrelia fainii, there were also several reports of the relapsing fever Borrelia found in Africa that phylogenetically clustered together with relapsing fever borreliae reported in the New World. A new human pathogenic Borrelia species was identified in Ornithodoros ticks from Tanzania, and it grouped together with the relapsing fever borreliae reported in the New World rather than the relapsing fever-inducing spirochetes that were known to be endemic in East Africa [11,28]. Another two studies described the discovery of a novel relapsing fever Borrelia, Candidatus Borrelia kalaharica, in travelers returning from the Kalahari Desert, South Africa [29,30].
The presence of Candidatus Borrelia fainii in Zambia, as well as the discovery of Borrelia species (B. lonestari and B. miyamotoi) that were related to relapsing fever borreliae but were transmitted by hard ticks rather than soft ticks challenged previous taxonomies based largely on microbe-vector specificity and geographic distribution. With more and more studies, we might have a better understanding as to the classification of relapsing fever borreliae.
Conclusively, our studies found that the human pathogenic relapsing fever Candidatus Borrelia fainii in a new location in China and further study is needed to determine its true distribution in China. However, with the emergence of this human-pathogenic relapsing fever Borrelia species in China, it’s important to find the reservoir and vector of it.
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Phylogenetic trees of S1–S8 Figs were drawn using the neighbor-joining method with the Kimura 2-parameter model with an alignment of the eight allelic sequences derived from the Borrelia MLST database (https://pubmlst.org/borrelia/) as well as from the GenBank. Corresponding GenBank number was shown in the brackets, and the sequences with * represents that the sequences were only submitted to the GenBank. Sequences amplified from this study were shown in red.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
Many people helped to conduct this study. The authors thank Yuan Jiang for constructing the map. The authors also extend appreciation to the researchers and students at the Wuhan University for their kind support.
Data Availability
The cytB gene of representative bat species and the 16S rDNA sequences of bat ticks were deposited in GenBank with accession No. MH888178, MH888180, MW085077-MW085079 and MW132810-MW132811, respectively. The Borrelia sequences in bats of this study were deposited in GenBank with accession numbers: MT913156-MT913157 (rrs), MT919991-MT919992 (flaB) and MT975516, MT919993 (glpQ). The relapsing fever Borrelia of this study was deposited in the Borrelia MLST database (https://pubmlst.org/borrelia/) as ST938 (https://pubmlst.org/bigsdb?page=profileInfo&db=pubmlst_borrelia_seqdef&scheme_id=1&profile_id=938).
Funding Statement
This work was supported by the National Natural Science Funds of China (grant numbers: 81971939)(XJY)(http://www.nsfc.gov.cn/) and the China Postdoctoral Science Foundation Funded Project (grant numbers: 2019M662720)(HJH)(http://jj.chinapostdoctor.org.cn/website/index.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cutler SJ. Possibilities for relapsing fever reemergence. Emerg Infect Dis. 2006;12(3):369–74. Epub 2006/05/18. 10.3201/eid1203.050899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour AG and Schwan TG. Borrelia 2018. Bergey’s Manual of Systematics of Archaea and Bacteria, John Wiley & Sons, Inc. [Google Scholar]
- 3.Cutler SJ. Relapsing fever borreliae: A Global Review. Clin Lab Med. 2015;35(4):847–65. Epub 2015/11/26. 10.1016/j.cll.2015.07.001 . [DOI] [PubMed] [Google Scholar]
- 4.Michalik J, Wodecka B, Liberska J, Dabert M, Postawa T, Piksa K, et al. Diversity of Borrelia burgdorferi sensu lato species in Ixodes ticks (Acari: Ixodidae) associated with cave-dwelling bats from Poland and Romania. Ticks Tick Borne Dis. 2020;11(1):101300. Epub 2019/10/22. 10.1016/j.ttbdis.2019.101300 . [DOI] [PubMed] [Google Scholar]
- 5.Stanek G, Wormser GP, Gray J, Strle F. Lyme borreliosis. Lancet (London, England). 2012;379(9814):461–73. Epub 2011/09/10. 10.1016/s0140-6736(11)60103-7 . [DOI] [PubMed] [Google Scholar]
- 6.Felsenfeld O. Borreliae, human relapsing fever, and parasite-vector-host relationships. Bacteriol Rev. 1965;29(1):46–74. Epub 1965/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour AG. Phylogeny of a relapsing fever Borrelia species transmitted by the hard tick Ixodes scapularis. Infect Genet Evol. 2014;27:551–8. Epub 2014/05/13. 10.1016/j.meegid.2014.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han HJ, Wen HL, Zhou CM, Chen FF, Luo LM, Liu JW, et al. Bats as reservoirs of severe emerging infectious diseases. Virus Res. 2015;205:1–6. Epub 2015/05/23. 10.1016/j.virusres.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mühldorfer K. Bats and bacterial pathogens: a review. Zoonoses Public Health. 2013;60(1):93–103. Epub 2012/08/07. 10.1111/j.1863-2378.2012.01536.x . [DOI] [PubMed] [Google Scholar]
- 10.Cutler SJ, Ruzic-Sabljic E, Potkonjak A. Emerging borreliae—Expanding beyond Lyme borreliosis. Mol Cell Probes. 2017;31:22–7. Epub 2016/08/16. 10.1016/j.mcp.2016.08.003 . [DOI] [PubMed] [Google Scholar]
- 11.Mitani H, Talbert A, Fukunaga M. New World relapsing fever Borrelia found in Ornithodoros porcinus ticks in central Tanzania. Microbiol Immunol. 2004;48(7):501–5. Epub 2004/07/24. 10.1111/j.1348-0421.2004.tb03545.x . [DOI] [PubMed] [Google Scholar]
- 12.Trape JF, Diatta G, Arnathau C, Bitam I, Sarih M, Belghyti D, et al. The epidemiology and geographic distribution of relapsing fever borreliosis in West and North Africa, with a review of the Ornithodoros erraticus complex (Acari: Ixodida). PLoS One. 2013;8(11):e78473. Epub 2013/11/14. 10.1371/journal.pone.0078473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu Y, Nakao R, Hang’ombe BM, Sato K, Kajihara M, Kanchela S, et al. Human borreliosis caused by a New World relapsing fever Borrelia-like organism in the Old World. Clin Infect Dis. 2019;69(1):107–12. Epub 2018/11/14. 10.1093/cid/ciy850 . [DOI] [PubMed] [Google Scholar]
- 14.Han HJ, Liu JW, Wen HL, Li ZM, Lei SC, Qin XR, et al. Pathogenic New World relapsing fever Borrelia in a Myotis bat, Eastern China, 2015. Emerg Infect Dis. 2020;26(12):3083–5. Epub 2020/11/22. 10.3201/eid2612.191450 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii A, Ueno K, Orba Y, Sasaki M, Moonga L, Hang’ombe BM, et al. A nairovirus isolated from African bats causes haemorrhagic gastroenteritis and severe hepatic disease in mice. Nat Commun. 2014;5:5651. Epub 2014/12/03. 10.1038/ncomms6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans NJ, Brown JM, Demirkan I, Singh P, Getty B, Timofte D, et al. Association of unique, isolated treponemes with bovine digital dermatitis lesions. J Clin Microbiol. 2009;47(3):689–96. Epub 2009/01/16. 10.1128/jcm.01914-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Assous MV, Wilamowski A, Bercovier H, Marva E. Molecular characterization of tickborne relapsing fever Borrelia, Israel. Emerg Infect Dis. 2006;12(11):1740–3. Epub 2007/02/08. 10.3201/eid1211.060715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toledo A, Anda P, Escudero R, Larsson C, Bergstrom S, Benach JL. Phylogenetic analysis of a virulent Borrelia species isolated from patients with relapsing fever. J Clin Microbiol. 2010;48(7):2484–9. Epub 2010/05/14. 10.1128/jcm.00541-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takano A, Fujita H, Kadosaka T, Takahashi M, Yamauchi T, Ishiguro F, et al. Construction of a DNA database for ticks collected in Japan: application of molecular identification based on the mitochondrial 16S rDNA gene. Med. Entomol. Zool. 2014;65(1):13–21. 10.7601/mez.65.13 [DOI] [Google Scholar]
- 20.Kingry LC, Anacker M, Pritt B, Bjork J, Respicio-Kingry L, Liu G, et al. Surveillance for and discovery of Borrelia species in US patients suspected of tickborne illness. Clin Infect Dis. 2018;66(12):1864–71. Epub 2017/12/23. 10.1093/cid/cix1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen AM, Pietralczyk E, Lopez JE, Brooks C, Schriefer ME, Wozniak E, et al. Diagnosis and management of Borrelia turicatae infection in febrile soldier, Texas, USA. Emerg Infect Dis. 2017;23(5):883–4. Epub 2017/04/19. 10.3201/eid2305.162069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dworkin MS, Schwan TG, Anderson DE Jr. Tick-borne relapsing fever in North America. The medical clinics of North America. 2002;86(2):417–33, viii-ix. Epub 2002/05/02. 10.1016/s0025-7125(03)00095-6 . [DOI] [PubMed] [Google Scholar]
- 23.Schwan TG, Raffel SJ, Schrumpf ME , Policastro PF, Rawlings JA, Lane RS, et al. Phylogenetic analysis of the spirochetes Borrelia parkeri and Borrelia turicatae and the potential for tick-borne relapsing fever in Florida. J Clin Microbiol. 2005;43(8):3851–9. Epub 2005/08/06. 10.1128/JCM.43.8.3851-3859.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwan TG, Raffel SJ, Schrumpf ME, Gill JS, Piesman J. Characterization of a novel relapsing fever spirochete in the midgut, coxal fluid, and salivary glands of the bat tick Carios kelleyi. Vector Borne Zoonotic Dis (Larchmont, NY). 2009;9(6):643–7. Epub 2009/03/14. 10.1089/vbz.2008.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans NJ, Bown K, Timofte D, Simpson VR, Birtles RJ. Fatal borreliosis in bat caused by relapsing fever spirochete, United Kingdom. Emerg Infect Dis. 2009;15(8):1331–3. Epub 2009/09/16. 10.3201/eid1508.090475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marinkelle CJ, Grose ES. Species of Borrelia from a Colombian bat (Natalus tumidirostris). Nature. 1968;218(5140):487. Epub 1968/05/04. 10.1038/218487a0 . [DOI] [PubMed] [Google Scholar]
- 27.Socolovschi C, Kernif T, Raoult D, Parola P. Borrelia, Rickettsia, and Ehrlichia species in bat ticks, France, 2010. Emerg Infect Dis. 2012;18(12):1966–75. Epub 2012/11/23. 10.3201/eid1812.111237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kisinza WN, McCall PJ, Mitani H, Talbert A, Fukunaga M. A newly identified tick-borne Borrelia species and relapsing fever in Tanzania. Lancet (London, England). 2003;362(9392):1283–4. Epub 2003/10/25. 10.1016/s0140-6736(03)14609-0 . [DOI] [PubMed] [Google Scholar]
- 29.Fingerle V, Pritsch M, Wächtler M, Margos G, Ruske S, Jung J, et al. "Candidatus Borrelia kalaharica" detected from a febrile traveller returning to Germany from Vacation in Southern Africa. PLoS Negl Trop Dis. 2016;10(3):e0004559. Epub 2016/04/01. 10.1371/journal.pntd.0004559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stete K, Rieg S, Margos G, Häcker G, Wagner D, Kern WV, et al. Case report and genetic sequence analysis of Candidatus Borrelia kalaharica, Southern Africa. Emerg Infect Dis. 2018;24(9):1659–64. Epub 2018/08/21. 10.3201/eid2409.171381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Phylogenetic trees of S1–S8 Figs were drawn using the neighbor-joining method with the Kimura 2-parameter model with an alignment of the eight allelic sequences derived from the Borrelia MLST database (https://pubmlst.org/borrelia/) as well as from the GenBank. Corresponding GenBank number was shown in the brackets, and the sequences with * represents that the sequences were only submitted to the GenBank. Sequences amplified from this study were shown in red.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
The cytB gene of representative bat species and the 16S rDNA sequences of bat ticks were deposited in GenBank with accession No. MH888178, MH888180, MW085077-MW085079 and MW132810-MW132811, respectively. The Borrelia sequences in bats of this study were deposited in GenBank with accession numbers: MT913156-MT913157 (rrs), MT919991-MT919992 (flaB) and MT975516, MT919993 (glpQ). The relapsing fever Borrelia of this study was deposited in the Borrelia MLST database (https://pubmlst.org/borrelia/) as ST938 (https://pubmlst.org/bigsdb?page=profileInfo&db=pubmlst_borrelia_seqdef&scheme_id=1&profile_id=938).