Vaccine protects against B1.1.7 variant
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B1.1.7 (VOC 202012/01) variant that emerged in late 2020 in the United Kingdom has many changes in the spike protein gene. Three of these are associated with enhanced infectivity and transmissibility, and there are concerns that B.1.1.7 might compromise the effectiveness of the vaccine. Muik et al. compared the neutralization efficacy of sera from 40 subjects immunized with the BioNTech-Pfizer mRNA vaccine BNT162b2 against a pseudovirus bearing the Wuhan reference strain or the lineage B.1.1.7 spike protein (see the Perspective by Altmann et al.). Serum was derived from 40 subjects in two age groups 21 days after the booster shot. The vaccine remained effective against B.1.1.7 with a slight but significant decrease in neutralization that was more apparent in participants under 55 years of age. Thus, the vaccine provides a significant “cushion” of protection against this variant.
Science, this issue p. 1152; see also p. 1103
Despite the many genetic changes in the B.1.1.7 (VOC 202012/01) 2020 UK variant of SARS-CoV-2, the BioNTech-Pfizer mRNA vaccine remains protective.
Abstract
Recently, a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineage called B.1.1.7 (variant of concern: VOC 202012/01), which is reported to spread more efficiently and faster than other strains, emerged in the United Kingdom. This variant has an unusually large number of mutations, with 10 amino acid changes in the spike (S) protein, raising concerns that its recognition by neutralizing antibodies may be affected. In this study, we tested SARS-CoV-2-S pseudoviruses bearing either the Wuhan reference strain or the B.1.1.7 lineage spike protein with sera of 40 participants who were vaccinated in a previously reported trial with the messenger RNA–based COVID-19 vaccine BNT162b2. The immune sera had slightly reduced but overall largely preserved neutralizing titers against the B.1.1.7 lineage pseudovirus. These data indicate that the B.1.1.7 lineage will not escape BNT162b2-mediated protection.
In a phase 3 trial conducted in the United States, Argentina, Brazil, South Africa, Germany, and Turkey, the BioNTech-Pfizer mRNA vaccine BNT162b2 was 95% effective in preventing COVID-19 through the data cutoff date of 14 November 2020 (1). The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineage B.1.1.7 (variant of concern: VOC 202012/01) was discovered to have emerged in the United Kingdom in September 2020 (2), and it subsequently increased in prevalence, showed enhanced transmissibility, and spread to other countries and continents (3). B.1.1.7 has a series of mutations in its spike (S) protein: ΔH69/V70, ΔY144, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H (H, His; V, Val; Y, Tyr; N, Asn; A, Ala; D, Asp; G, Gly; P, Pro; T, Thr; I, Ile; S, Ser). One of these mutations, N501Y, was of particular concern because it is located in the receptor binding site. The spike with this mutation binds more tightly to its cellular receptor, ACE-2 (4), and virus with this mutation has an increased host range that includes mice (5). BNT162b2-immune sera neutralized SARS-CoV-2 (USA/WA-1/2020 background strain) with an introduced N501Y mutation as efficiently as they neutralized SARS-CoV-2 without the mutation (6). Further, 19 pseudoviruses, each bearing a SARS-CoV-2 S with a different mutation found in circulating virus strains, were also neutralized as efficiently as nonmutant SARS-CoV-2 S–bearing pseudoviruses by BNT162b2-immune sera (7). However, it was still unclear whether a virus with the full set of mutations in the lineage B.1.1.7 spike, each of which may potentially interfere with antibody binding, would be neutralized efficiently by BNT162b2-immune sera.
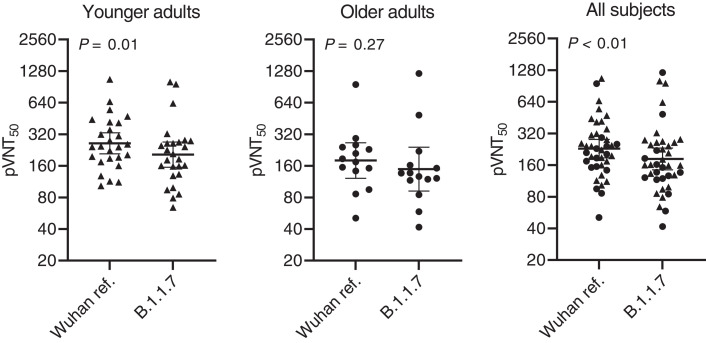
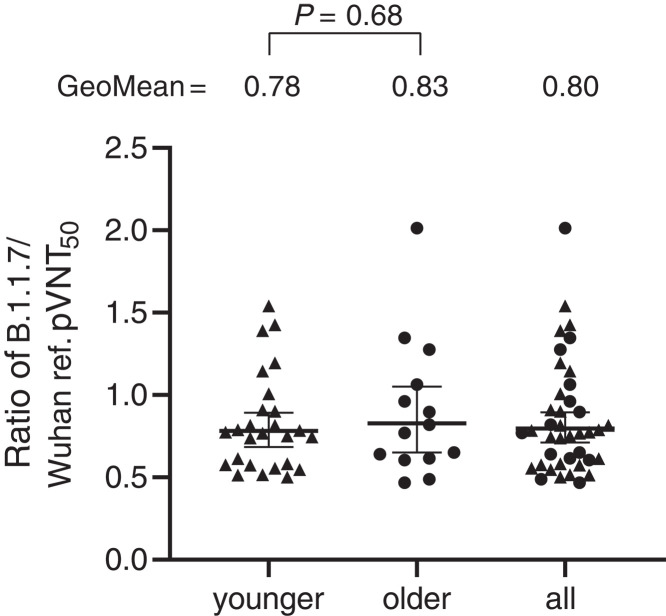
To answer this question, we generated vesicular stomatitis virus (VSV)–SARS-CoV-2-S pseudoviruses bearing the Wuhan reference strain or the lineage B.1.1.7 spike protein (fig. S1). An unbiased set of sera of 40 participants in the previously reported German phase 1/2 trial (7)—drawn from 26 younger (aged 23 to 55 years) and 14 older adults (aged 57 to 73 years) at 7 or 21 days after the booster immunization with 30 μg of BNT162b2 (fig. S2)—was tested for neutralization of SARS-CoV-2 Wuhan and lineage B.1.1.7 spike–pseudotyped VSV by a 50% neutralization assay [50% pseudovirus neutralization titer (pVNT50)]. The 50% neutralization geometric mean titers (GMTs) of the sera against the SARS-CoV-2 lineage B.1.1.7 spike–pseudotyped VSV for the younger adult group and the full analysis set were slightly but statistically significantly reduced compared with the GMTs against the Wuhan reference spike–pseudotyped VSV (Fig. 1 and table S1). GMTs were not significantly different for the older adult group. The calculated geometric mean ratio with 95% confidence interval (CI) of the B.1.1.7 pseudotype and the Wuhan pseudotype GMTs was 0.78 (95% CI: 0.68 to 0.89) for the younger group and 0.83 (95% CI: 0.65 to 1.1) for the older adults [0.80 (95% CI: 0.71 to 0.89) in aggregate] (Fig. 2). No statistical difference in the ratio was observed between the younger and the older vaccinated participants.
Fig. 1. 50% pseudovirus neutralization titers (pVNT50) of 40 sera from BNT162b2 vaccine recipients against VSV-SARS-CoV-2-S pseudovirus bearing the Wuhan reference strain or lineage B.1.1.7 spike protein.
Sera from n = 26 younger adults (aged 23 to 55 years; indicated by triangles) and n = 14 older adults (aged 57 to 73 years; indicated by circles) drawn at either day 29 or day 43 (7 or 21 days after vaccine dose two) were tested. Statistical significance of the difference between the neutralization of the VSV-SARS-CoV-2-S pseudovirus bearing the Wuhan or lineage B.1.1.7 spike protein was calculated by a Wilcoxon matched-pairs signed rank test. Two-tailed P values are reported. GMTs and 95% CIs are indicated.
Fig. 2. pVNT50 ratio of SARS-CoV-2 lineage B.1.1.7 to Wuhan reference strain spike–pseudotyped VSV.
Triangles represent sera from younger adults (aged 23 to 55 years), and circles represent sera from older adults (aged 57 to 73 years). Sera were drawn on either day 29 or day 43 (7 or 21 days after vaccine dose two). Geometric means of the pVNT50 ratios of SARS-CoV-2 lineage B.1.1.7 to Wuhan spike–pseudotyped VSV and 95% CIs are indicated. The difference in distribution of titer ratios between younger and older adults was tested for statistical significance with a two-tailed Mann-Whitney U test.
On the basis of experience from studying antibody correlates of disease protection for influenza virus vaccines, a 20% reduced titer does not indicate a biologically relevant change in neutralization activity (8, 9). The largely preserved neutralization of pseudoviruses bearing the B.1.1.7 spike by BNT162b2-immune sera makes it unlikely that the U.K. variant virus will escape BNT162b2-mediated protection.
A potential limitation of the work may be the use of a nonreplicating pseudovirus system. However, previous reports have shown good concordance between pseudotype neutralization and SARS-CoV-2 neutralization assays (10, 11). Still, concordance may vary between different SARS-CoV-2 strains and remains to be demonstrated for the SARS-CoV-2 B.1.1.7 lineage. Additional experiments will be needed to confirm efficient neutralization of B.1.1.7 lineage clinical isolates. This study has evaluated sera elicited by the recommended regimen of two doses administered 21 days apart and does not provide insight into neutralization if the recommended dosing regimen is not followed. The ongoing evolution of SARS-CoV-2 necessitates continuous monitoring of the biological relevance of changes for maintained protection by the currently authorized vaccines. Unlike the protocol for influenza vaccines, the degree of reduction in neutralization that might indicate a need for a strain change has not yet been established for COVID-19 vaccines. A previous study demonstrated that BNT162b2 elicits both a polyepitopic CD8+ T cell response to the encoded spike protein and virus-neutralizing antibodies (7). Given the multiple potential mediators of protection elicited by BNT162b2, it is possible that vaccine efficacy could be preserved in the longer term, even with substantial losses of neutralization by vaccine-elicited sera. This view is further supported by the rapid onset of disease protection ~12 days after the first dose of BNT162b2, at a time when neutralizing antibody titers are still very low (1). Without an established correlate of protection, clinical effectiveness data will be needed to provide definitive assessment of vaccine-mediated protection against viral variants.
Although sustained neutralization of the current B.1.1.7 variant is reassuring, preparation for potential COVID-19 vaccine strain change is prudent. Adaptation of the vaccine to a new virus strain would be facilitated by the flexibility of mRNA-based vaccine technology.
Acknowledgments
We thank the BioNTech German clinical trial (NCT04380701, EudraCT: 2020-001038-36) participants, from whom the postimmunization human sera were obtained. We thank the many colleagues at BioNTech and Pfizer who developed and produced the BNT162b2 vaccine candidate. We thank S. Jägle and N. Beckmann for logistical support. Funding: This work was supported by BioNTech and Pfizer. Author contributions: U.Ş., Ö.T., A.M., and P.R.D. conceived and conceptualized the work. K.A.S. and A.M. planned and supervised experiments. A.M., A.-K.W., J.M., B.S., H.C., W.C., and R.S. performed experiments. A.M., D.M., H.C., and K.A.S. analyzed data. U.Ş., Ö.T., A.M., P.R.D., and K.A.S. interpreted data and wrote the manuscript. All authors supported the review of the manuscript. Competing interests: U.Ş. and Ö.T. are management board members and employees at BioNTech SE. A.M., A.-K.W., J.M., B.S., and D.M. are employees at BioNTech SE. U.Ş., Ö.T., and A.M. are inventors on patents and patent applications related to RNA technology and COVID-19 vaccine. U.Ş., Ö.T., A.M., J.M., and B.S. have securities from BioNTech SE. K.A.S., W.C., H.C., R.S., and P.R.D. are employees at Pfizer and may have securities from Pfizer. Data and materials availability: A table of the neutralization titers is provided in table S1. Materials are available from the authors under a material transfer agreement with BioNTech. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
Supplementary Materials
science.sciencemag.org/content/371/6534/1152/suppl/DC1
Materials and Methods
Figs. S1 to S3
Tables S1 and S2
Reference (12)
MDAR Reproducibility Checklist
References and Notes
- 1.Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Pérez Marc G., Moreira E. D., Zerbini C., Bailey R., Swanson K. A., Roychoudhury S., Koury K., Li P., Kalina W. V., Cooper D., Frenck R. W. Jr.., Hammitt L. L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Jansen K. U., Gruber W. C.; C4591001 Clinical Trial Group , Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). 10.1056/NEJMoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. Rambaut, N. Loman, O. Pybus, W. Barclay, J. Barrett, A. Carabelli, T. Connor, T. Peacock, D. L. Robertson, E. Volz; COVID-19 Genomics Consortium UK (CoG-UK), Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. virological.org (2020); https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
- 3.E. Volz, S. Mishra, M. Chand, J. C. Barrett, R. Johnson, L. Geidelberg, W. R. Hinsley, D. J. Laydon, G. Dabrera, Á. O’Toole, R. Amato, M. Ragonnet-Cronin, I. Harrison, B. Jackson, C. V. Ariani, O. Boyd, N. J. Loman, J. T. McCrone, S. Gonçalves, D. Jorgensen, R. Myers, V. Hill, D. K. Jackson, K. Gaythorpe, N. Groves, J. Sillitoe, D. P. Kwiatkowski, The COVID-19 Genomics UK (COG-UK) consortium, S. Flaxman, O. Ratmann, S. Bhatt, S. Hopkins, A. Gandy, A. Rambaut, N. M. Ferguson, Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv 2020.12.30.20249034 [Preprint]. 4 January 2021. 10.1101/2020.12.30.20249034. 10.1101/2020.12.30.20249034 [DOI]
- 4.Starr T. N., Greaney A. J., Hilton S. K., Ellis D., Crawford K. H. D., Dingens A. S., Navarro M. J., Bowen J. E., Tortorici M. A., Walls A. C., King N. P., Veesler D., Bloom J. D., Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 182, 1295–1310.e20 (2020). 10.1016/j.cell.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu H., Chen Q., Yang G., He L., Fan H., Deng Y. Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Y., Li X. F., Li J., Zhang N. N., Yang X., Chen S., Guo Y., Zhao G., Wang X., Luo D. Y., Wang H., Yang X., Li Y., Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C. F., Zhou Y., Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369, 1603–1607 (2020). 10.1126/science.abc4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.X. Xie, J. Zou, C. R. Fontes-Garfias, H. Xia, K. A. Swanson, M. Cutler, D. Cooper, V. D. Menachery, S. Weaver, P. R. Dormitzer, P.-Y. Shi, Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. bioRxiv 2021.01.07.425740 [Preprint]. 7 January 2021. 10.1101/2021.01.07.425740 [DOI]
- 7.U. Sahin, A. Muik, I. Vogler, E. Derhovanessian, L. M. Kranz, M. Vormehr, J. Quandt, N. Bidmon, A. Ulges, A. Baum, K. Pascal, D. Maurus, S. Brachtendorf, V. Lörks, J. Sikorski, P. Koch, R. Hilker, D. Becker, A.-K. Eller, J. Grützner, M. Tonigold, C. Boesler, C. Rosenbaum, L. Heesen, M.-C. Kühnle, A. Poran, J. Z. Dong, U. Luxemburger, A. Kemmer-Brück, D. Langer, M. Bexon, S. Bolte, T. Palanche, A. Schultz, S. Baumann, A. J. Mahiny, G. Boros, J. Reinholz, G. T. Szabó, K. Karikó, P.-Y. Shi, C. Fontes-Garfias, J. L. Perez, M. Cutler, D. Cooper, C. A. Kyratsous, P. R. Dormitzer, K. U. Jansen, Ö. Türeci, BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. medRxiv 2020.12.09.20245175 [Preprint]. 11 December 2020. 10.1101/2020.12.09.20245175. [DOI]
- 8.Smith D. J., Lapedes A. S., de Jong J. C., Bestebroer T. M., Rimmelzwaan G. F., Osterhaus A. D., Fouchier R. A., Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 (2004). 10.1126/science.1097211 [DOI] [PubMed] [Google Scholar]
- 9.Black S., Nicolay U., Vesikari T., Knuf M., Del Giudice G., Della Cioppa G., Tsai T., Clemens R., Rappuoli R., Hemagglutination inhibition antibody titers as a correlate of protection for inactivated influenza vaccines in children. Pediatr. Infect. Dis. J. 30, 1081–1085 (2011). 10.1097/INF.0b013e3182367662 [DOI] [PubMed] [Google Scholar]
- 10.Case J. B., Rothlauf P. W., Chen R. E., Liu Z., Zhao H., Kim A. S., Bloyet L.-M., Zeng Q., Tahan S., Droit L., Ilagan M. X. G., Tartell M. A., Amarasinghe G., Henderson J. P., Miersch S., Ustav M., Sidhu S., Virgin H. W., Wang D., Ding S., Corti D., Theel E. S., Fremont D. H., Diamond M. S., Whelan S. P. J., Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe 28, 475–485.e5 (2020). 10.1016/j.chom.2020.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel A. B., Kanevsky I., Che Y., Swanson K. A., Muik A., Vormehr M., Kranz L. M., Walzer K. C., Hein S., Güler A., Loschko J., Maddur M. S., Ota-Setlik A., Tompkins K., Cole J., Lui B. G., Ziegenhals T., Plaschke A., Eisel D., Dany S. C., Fesser S., Erbar S., Bates F., Schneider D., Jesionek B., Sänger B., Wallisch A.-K., Feuchter Y., Junginger H., Krumm S. A., Heinen A. P., Adams-Quack P., Schlereth J., Schille S., Kröner C., de la Caridad Güimil Garcia R., Hiller T., Fischer L., Sellers R. S., Choudhary S., Gonzalez O., Vascotto F., Gutman M. R., Fontenot J. A., Hall-Ursone S., Brasky K., Griffor M. C., Han S., Su A. A. H., Lees J. A., Nedoma N. L., Mashalidis E. H., Sahasrabudhe P. V., Tan C. Y., Pavliakova D., Singh G., Fontes-Garfias C., Pride M., Scully I. L., Ciolino T., Obregon J., Gazi M., Carrion R. Jr., Alfson K. J., Kalina W. V., Kaushal D., Shi P.-Y., Klamp T., Rosenbaum C., Kuhn A. N., Türeci Ö., Dormitzer P. R., Jansen K. U., Sahin U., BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 10.1038/s41586-021-03275-y, (2021). 10.1038/s41586-021-03275-y [DOI] [PubMed] [Google Scholar]
- 12.Berger Rentsch M., Zimmer G., A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLOS ONE 6, e25858 (2011). 10.1371/journal.pone.0025858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
science.sciencemag.org/content/371/6534/1152/suppl/DC1
Materials and Methods
Figs. S1 to S3
Tables S1 and S2
Reference (12)
MDAR Reproducibility Checklist