Abstract
Brown and beige fat are specialized adipose tissues that dissipate energy for thermogenesis by UCP1 (Uncoupling Protein-1)-dependent and independent pathways. Until recently, thermogenic adipocytes were considered a homogeneous population. However, recent studies have indicated that there are multiple subtypes or subpopulations that are distinct in developmental origin, substrate use, and transcriptome. Despite advances in single-cell genomics, unbiased decomposition of adipose tissues into cellular subtypes has been challenging because of the fragile nature of lipid-filled adipocytes. The protocol presented was developed to circumvent these obstacles by effective isolation of single nuclei from adipose tissue for downstream applications, including RNA sequencing. Cellular heterogeneity can then be analyzed by RNA sequencing and bioinformatic analyses.
Introduction
Studies have shown that brown adipose tissue (BAT) has a remarkable capacity to dissipate energy. Two types of thermogenic adipocytes with distinct developmental features exist in both rodents and humans: beige adipocytes and classical brown adipocytes. While classical brown adipocytes are located mostly in interscapular BAT depots, beige adipocytes sporadically emerge in white adipose tissue (WAT) in response to certain physiological cues, such as chronic cold exposure, a process referred to as "browning" or "beiging". Through the use of advanced imaging, it is now clear that adult humans have substantial depots of UCP1+ BAT, especially in the supraclavicular region1 , 2 , 3 , 4 . The amount of adult human BAT inversely correlates with adiposity and can be increased by external cues, such as chronic cold exposure5 , 6 or β3-adrenergic receptor agonist7. BAT-mediated energy expenditure may offer a viable approach to combat obesity.
Until recently, thermogenic adipocytes have been considered a homogeneous population. However, studies have revealed the existence of multiple subtypes or subpopulations that are distinct in developmental origin, substrate usage, and transcriptome8 , 9 , 10 . For instance, a type of beige adipocyte that preferentially uses glucose for thermogenesis, the g-beige adipocyte, was recently described10. The incomplete understanding of cell types in brown and beige adipose tissue and the lack of specific markers constitute a critical barrier to studying their biological functions.
Traditional methods for isolating subpopulations of cells are based on expression of only a few known marker genes. Recent advances in single-cell genomics enables the use of global gene expression data of single cells to provide an unbiased estimate of the number of subpopulations in a tissue. The ultimate goal of this protocol is to determine all adipose tissue subtypes under various thermogenic stimuli at a single-cell resolution. In contrast to other tissues and cell types, determining cellular subtypes of adipose tissue is challenging due to the fragility of lipid-filled adipocytes. This paper introduces a robust protocol to isolate single nuclei from adipose tissue for downstream application to snRNA sequencing. Importantly, recent literature comparing well-matched single-nuclei RNA sequencing (snRNA-seq) and single-cell RNA sequencing (scRNA-seq) datasets revealed that snRNA-seq is comparable to scRNA-seq in cell type detection, and superior in cellular coverage for a complex tissue like the brain11. This protocol combines a density gradient centrifugation method optimized for adipose tissues by Rosen et al.12 with a nuclei "cleanup" step with a MoFlo XDP High Speed Sorter. As seen in the representative results, an analysis of 7,500 single nuclei from mouse interscapular brown adipose tissue identified multiple cell types within seemingly homogeneous brown adipocytes. Overall, this simple and robust protocol can be applied to study tissue-level organization of adipocytes and adipose-resident cells, identification of subtype-specific marker genes, and development phenotyping of adipose-selective knockout/transgenic mice.
Protocol
Animal care and experimentation were performed according to procedures approved by the Institutional Animal Care and Use Committee at the Albert Einstein College of Medicine.
1. Preparation of tissue digestion and lysis buffers
- Prepare tissue digestion buffer.
- Prepare ~1 mL of digestion buffer for every gram of adipose tissue.
- Weigh out 1.5 U/mL collagenase D and 2.4 U/mL dispase II and add phosphate buffered saline (PBS).
- Prepare nuclei preparation buffer (NPB).
- Prepare 10 mM HEPES, 1.5 mM magnesium chloride, 10 mM potassium chloride, 250 mM sucrose, and 0.1% NP-40 in nuclease-free water. Mix well. Sucrose may take more time to dissolve than the other components.
- Prepare 0.5% bovine serum albumin (BSA) solution.
- Prepare 0.5% BSA in PBS containing EDTA. It is recommended to use sterile-filtered cell culture grade PBS (see Table of Materials).
Materials
| Name | Company | Catalog Number | Comments |
|---|---|---|---|
| autoMACS Rinsing Solution | Miltenyi Biotec | 130-091-222 | PBS with EDTA; sterile-filtered |
| BSA | Sigma | A1595 | |
| CaCl2 | Sigma | 21115 | |
| Cell filter 100 μm | Corning | 431752 | |
| Cell filter 40μm | Corning | 431750 | |
| CellTrics (30 μm) | Sysmex | 04-004-2326 | |
| Collagenase D | Roche | 11088866001 | |
| Countess II FL Automated Cell Counter | Invitrogen | AMQAF1000 | |
| DAPI | Sigma | D9542 | |
| Dispase II | Roche | 4942078001 | |
| HEPES | Sigma | H4034 | |
| KCl | Fisher | P217-3 | |
| MACS SmartStrainers (30 μm) | Miltenyi Biotec | 130-098-458 | Stackable filters |
| MgCl2 | Sigma | M1028 | |
| MoFloXDP Cell Sorter | Beckman Coulter | ML99030 | |
| NP-40 | Sigma | 74385 | |
| Protector RNase Inhibitor | Roche | 3335402001 | |
| Sucrose | Fisher | S5-3 |
2. Enzymatic digestion of adipose tissue
Dissect mice to extract adipose tissue according to Aune et al.13 . Collect isolated tissue in a dish containing PBS. Transfer the tissue to a paper towel to pat dry. Then place the tissue in a clean dish.
Add calcium chloride to the digestion buffer to a final concentration of 10 mM. Add a small volume of digestion buffer to the dish and thoroughly mince the adipose tissue. The volume of digestion buffer required will be based on the amount of tissue isolated. Usually ~1 mL or less is used.
Add about half of the amount of prepared digestion buffer (e.g., ~10 mL for five mice) to the minced tissue. Using a serological pipette, transfer the sample to a 50 mL conical centrifuge tube. Add any remaining amount of buffer to wash the dish and transfer to the 50 mL tube containing the sample. Pipette up and down several times. Then incubate with shaking at 200-210 rpm at 37 °C for 12-15 min.
3. Adipocyte isolation
After incubation, add 0.5% BSA at a 1:1 ratio to the sample. Mix well by pipetting up and down several times. Centrifuge the sample at 300 x g for 5 min at room temperature (RT) to spin down the stromal vascular fraction (SVF). The adipocyte fraction will remain in the top layer of the sample.
Place a 40 μm cell filter on top of a 50 mL conical centrifuge tube and filter the sample, collecting the top layer and leaving behind the SVF at the bottom of the tube. Transfer the top layer (adipocyte fraction) to a new tube by flipping the filter upside down over the tube and using 0.5% BSA to reverse wash the filter and collect the sample in the tube.
Bring the total volume of 0.5% BSA to 10-15 mL based on sample size and pipette up and down with a serological pipette or briefly shake up and down to mix the sample. Centrifuge the sample again at 300 x g for 5 min at RT.
4. Nuclei isolation
Use a wide-bore pipette tip and carefully transfer the top layer of the sample to a 100 μm cell filter placed on top of a new prechilled tube on ice, leaving behind any residual SVF.
Rinse/wash the filter with a sufficient amount of NPB and discard the filter. From this step forward have the sample on ice as often as possible and work quickly to avoid leaky and/or unhealthy nuclei. It is helpful to prechill microcentrifuge tubes to be used in future steps on ice.
Place the sample on ice for no more than 2 min with intermittent gentle inverting of the tube to mix. The incubation time on ice should be optimized for sample size and type of adipocytes. Centrifuge at 1,000 x g at 4 °C for 10 min.
-
Remove the supernatant and resuspend the pellet in 1 mL of NPB. Transfer the sample to a clean microcentrifuge tube. While transferring, avoid touching the walls of the tube, which contain debris and lipid. Add 0.6 U/μL RNase inhibitor to the sample and mix. Centrifuge again at 1,000 x g at 4 °C for 10 min.
NOTE: Centrifugation speed and time should be decreased at this step for smaller samples.
Remove the supernatant and resuspend in 1 mL of Nuclei Wash Buffer (2% BSA/PBS + 0.6 U/μL RNase inhibitor).
Double filter into a clean tube with stackable 30 μm filters. Wash filter with a small volume (~300 μL) of Nuclei Wash Buffer. Alternatively, for smaller samples, use a 30 μm filter that fits into a microcentrifuge tube and filter directly into it.
- Confirm successful isolation of healthy nuclei.
- Stain a small aliquot of the sample with trypan and use an automated cell counter to assess average dead size and percent dead cells. Average dead size for a sample containing mostly nuclei should range between 7-10 μm. Nuclei size is usually around 8 μm.
- Stain a small aliquot of the sample with DAPI and examine under a microscope with a 20x objective or higher. The nuclear membrane should be intact and the nuclei should be round.
5. FACS cleanup and nuclei concentration step
Proceed immediately to the FACS step to clean up nuclei and remove any excess debris. The collection buffer should contain Nuclei Wash Buffer. During FACS the buffer is diluted. Preparing a higher concentration of BSA and RNase inhibitor to account for dilution during sorting is helpful. The FACS collection block should be on cooling mode.
After sorting the nuclei, centrifuge at 500 x g for 5 min at 4 °C to concentrate the sample. Reconstitute in an appropriate volume of Nuclei Wash Buffer to achieve a concentration of 500-1,500 nuclei/μL. Do not attempt to remove all the supernatant. Doing so will result in loss of nuclei.
Use a hemocytometer and a DAPI stained aliquot for a final count and to visualize the nuclei. Then proceed immediately with snRNA-seq according to the manufacturer's protocol.
Representative Results
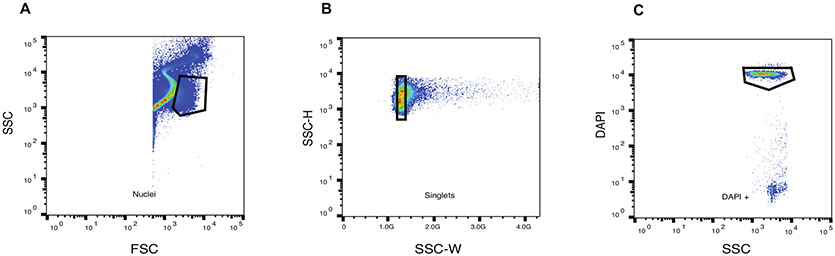
Unsorted adipocyte nuclei contain debris and doublets that create noise and high background in downstream single-cell RNA sequencing. The representative FACS gate strategy is shown in Figure 1. The nuclei were first selected based on forward scatter (FSC) and side scatter (SSC) (A), then, only singlets were selected based on the combination of width and heights of SSC (B). Finally, only DAPI-positive events were selected and sorted into collection buffer (C). This workflow provides highly-purified single nuclei with minimized nuclear aggregates and cellular debris (Figure 2).
Figure 1: Representative flow cytometry analysis of nuclei by MoFlo XDP High Speed Sorter.
(A) Size and granularity. (B) Detection of nuclear aggregates and multiplets. (C) Final sorting gate selecting only DAPI-positive events.
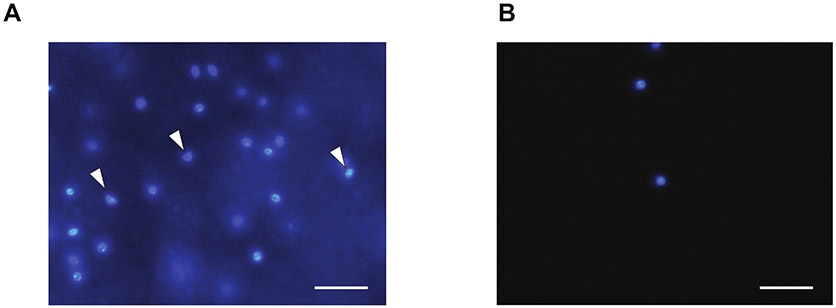
Figure 2: Representative image of nuclei isolated from brown adipocytes.
(A) Before and (B) after MoFLo XDP sorting. White arrows in A indicate nuclei without an intact membrane. Scale bar = 50 μm.
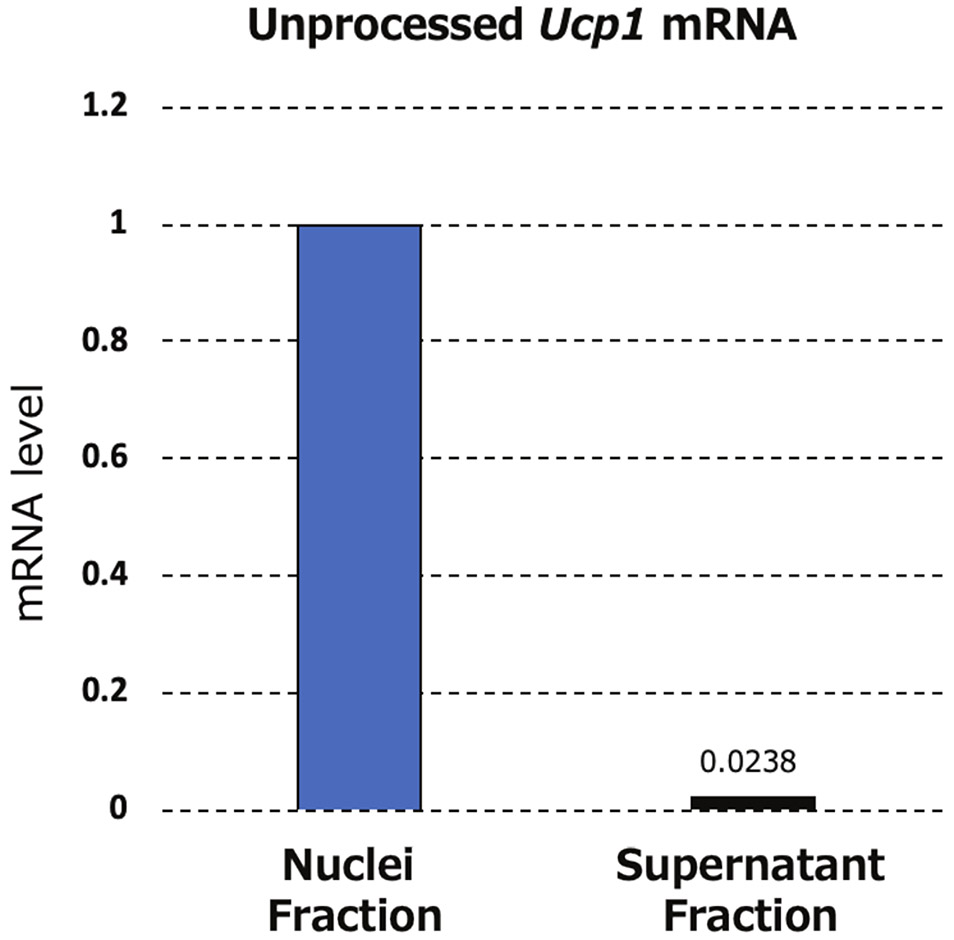
To confirm the integrity of the sorted nuclei, a small aliquot of the sample was inspected by microscope after DAPI staining. The nuclear membrane should be intact and the nuclei should be round (Figure 2B). To confirm the success of the purification, it is also recommended to collect supernatant (i.e., collection buffer) after centrifugation and run real-time qRT-PCR of a marker gene. Use of a primer set (primer sequences in Table 1) designed to amplify nascent, intron-containing UCP1, confirmed that the supernatant of sorted nuclei did not contain a detectable level of nascent Ucp1 mRNA (Figure 3). This step can be done for other marker genes highly abundant in brown, beige, or white adipocytes, such as Cidea, Pgc1-a, Adipoq, or Fabp4.
Table 1:
Primer sequences for real-time qPCR analysis.
| Gene | Species | Forward primer | Reverse primer |
|---|---|---|---|
| Ucp1 (Intron-containing) | mouse | GAT CTT CTC AGC CGG AGT TTC | CCT TCC TAA TAG CAC CCA TTC C |
Figure 3:
Representative qRT-PCR result for isolated nuclei and the supernatant.
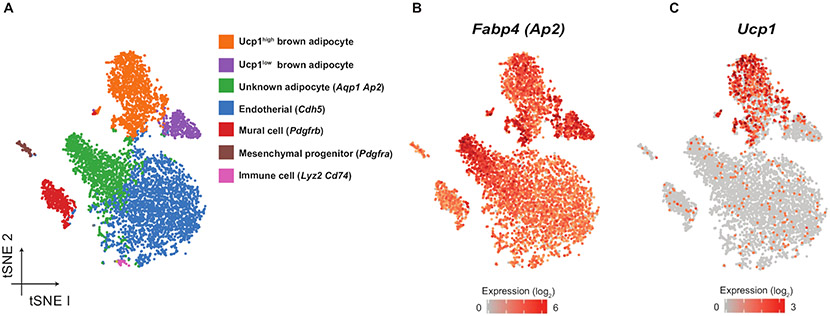
Nuclei isolated and confirmed through this protocol can be subjected to virtually any single-cell level gene expression platform. Use of the Chromium platform14 (10x Genomics) determined a transcriptome of 7,500 nuclei from interscapular brown adipose tissue from 8-week-old male C57BL/6 mice (Figure 4). t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction and K-means clustering revealed seven cell types, including brown adipocytes, endothelial cells, mural cells marked by Pdgfrb, adipocyte progenitors, and immune cells. All clusters expressed Fabp4, a pan-adipocyte gene, to varying degrees (Figure 4B). A subset of clusters expressed a high level of Ucp1 mRNA, whereas other clusters sporadically expressed Ucp1 (Figure 4A,C). This is consistent with a previous study showing that interscapular brown adipose tissue contains at least two distinct populations with high and low Ucp1 expression and thermogenic activity8 , 9 .
Figure 4: Representative 10x chromium snRNA-seq results for brown adipocytes isolated from 8-week-old male C57BL/6 mice under RT.
(A) t-SNE dimensionality reduction and K-Means clustering for 7,500 nuclei. k = 9. Canonical markers used for cluster annotations are shown in parentheses. (B) mRNA expression of Fabp4, an adipocyte-selective marker, in A. White = no expression, red = high expression. (C) mRNA expression of Ucp1, thermogenic marker in A. These data are from a single experiment. snRNA-seq data used for these figures was deposited in the Gene Expression Omnibus (GEO) under superseries accession number GSE144720.
Discussion
A straightforward and robust method to isolate single nuclei and study adipose tissue heterogeneity is presented. Compared to whole tissue RNA sequencing, this workflow offers an unbiased view of cellular heterogeneity and population-specific markers. This is significant and innovative for the advancement of adipocyte biology, molecular metabolism, and obesity research.
This protocol is particularly optimized for downstream application of snRNA-seq. The "cleanup" step to achieve isolation of healthy nuclei with the MoFlo XDP High Speed Sorter completely removes debris and aggregates from the crude suspension while maintaining nuclear membrane integrity. Therefore, this increases capture efficiency in the droplet formation and recovers thousands of single-nuclei transcriptomes from just one run without loss of the number of detected genes. In addition to droplet-based single-cell platforms, which provide a higher number of surveyed single nuclei or cells, sorter-based microplate and Fluidigm C1 platforms15 can also be used to obtain greater resolution and coverage of the transcriptome16. Because this workflow offers high-quality nuclei, assays for transposase-accessible chromatin using sequencing (i.e., ATAC-sequencing) can also be performed with minimum modifications to the existing scATAC-seq protocol17.
One limitation of this protocol is the loss of information from the removal of nonnuclear compartments, such as the cytosol and cellular membrane. For instance, emerging multimodal single-cell analyses18 , 19 that simultaneously measure cell membrane protein expression and the transcriptome cannot be applied to this protocol.
The future goal is to combine this protocol with nuclei multiplexing using barcoded antibodies20. Sorted nuclei from different adipose tissues under different experimental conditions will be barcoded and pooled using an antinuclear pore complex antibody and snRNA-seq will be performed. By sequencing these barcodes alongside the nuclear transcriptome, each nuclei can be assigned to its original sample, cross-sample multiplets can be clearly identified, and droplet-based systems can be "super-loaded" for significant cost reduction per nuclei.
Acknowledgments
We would like to thank David Reynolds from the Albert Einstein Genomics core and Jinghang Zhang from the Flow Cytometry Core for technical support. We acknowledge support from the National Institutes of Health (NIH) (DK110426) and Pilot and Feasibility Grants from the Einstein-Mount Sinai Diabetes Research Center (DK020541), and New York Obesity Research Center (DK026687) (all to K.S.). We also would like to thank Albert Einstein Cancer Center (CA013330) for core support.
Footnotes
Disclosures
The authors declare that they have no competing financial interests.
References
- 1.Cypess AM et al. Identification and importance of brown adipose tissue in adult humans. The New England Journal of Medicine. 360 (15), 1509–1517 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Marken Lichtenbelt WD et al. Cold-activated brown adipose tissue in healthy men. The New England Journal of Medicine. 360 (15), 1500–1508 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Virtanen KA et al. Functional brown adipose tissue in healthy adults. The New England Journal of Medicine. 360 (15), 1518–1525 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Nedergaard J, Bengtsson T, Cannon B Unexpected evidence for active brown adipose tissue in adult humans. American Journal of Physiology-Endocrinology and Metabolism. 293 (2), E444–E452 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Saito M et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 58 (7), 1526–1531 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoneshiro T et al. Recruited brown adipose tissue as an antiobesity agent in humans. The Journal of Clinical Investigation. 123 (8), 3404–3408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cypess AM et al. Activation of human brown adipose tissue by a β3-adrenergic receptor agonist. Cell Metabolism. 21 (1), 33–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song A et al. Low- and high-thermogenic brown adipocyte subpopulations coexist in murine adipose tissue. The Journal of Clinical Investigation. 130 (1), 247–257 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cinti S et al. CL316,243 and cold stress induce heterogeneous expression of UCP1 mRNA and protein in rodent brown adipocytes. The Journal of Histochemistry and Cytochemistry: Official Journal of the Histochemistry Society. 50 (1), 21–31 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Chen Y et al. Thermal stress induces glycolytic beige fat formation via a myogenic state. Nature. 565 (7738), 180–185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakken TE et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PloS One. 13 (12), e0209648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roh HC et al. Simultaneous Transcriptional and Epigenomic Profiling from Specific Cell Types within Heterogeneous Tissues In Vivo. Cell Reports. 18 (4), 1048–1061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aune UL, Ruiz L, Kajimura S Isolation and differentiation of stromal vascular cells to beige/brite cells. Journal of Visualized Experiments. (73), e50191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nature Communications. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollen AA et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nature Biotechnology. 32 (10), 1053–1058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi T et al. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nature Communications. 9 (1), 619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satpathy AT et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nature Biotechnology. 37 (8), 925–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nature Methods. 14 (9), 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson VM et al. Multiplexed quantification of proteins and transcripts in single cells. Nature Biotechnology. 35 (10), 936–939 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Gaublomme JT et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nature Communications. 10 (1), 2907 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]