ABSTRACT
Ranbp2 (also known as Nup358) is a member of the nucleoporin family, which constitutes the nuclear pore complex. Ranbp2 localizes at the nuclear membrane and was recently reported at the axon initial segment (AIS). However, we show that the anti-Ranbp2 antibody used in previous studies is not specific for Ranbp2. We mapped the antibody binding site to the amino acid sequence KPLQG, which is present in both Ranbp2 and neurofascin (Nfasc), a well-known AIS protein. After silencing neurofascin expression in neurons, the AIS was not stained by the antibody. Surprisingly, an exogenously expressed N-terminal fragment of Ranbp2 localizes at the AIS. We show that this fragment interacts with stable microtubules. Finally, using CRISPR/Cas9 in primary cultured neurons, we inserted an HA-epitope tag at N-terminal, C-terminal or internal sites of the endogenously expressed Ranbp2. No matter the location of the HA-epitope, endogenous Ranbp2 was found at the nuclear membrane but not the AIS. These results show that endogenously expressed Ranbp2 is not found at AISs.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Axon, Cytoskeleton, Antibody, Ranbp2, Nup358
Summary: Previous reports of axon initial segment Ranbp2 are shown to be based on NF186-cross-reacting antibodies and the intrinsic ability of an N-terminal fragment of Ranbp2 to bind to stable microtubules.
INTRODUCTION
The axon initial segment (AIS) is essential for the proper generation of axonal action potentials and the regulation of neuronal polarity (Leterrier, 2018). These properties and the maintenance of the AIS require the scaffolding and cytoskeletal proteins AnkyrinG (AnkG, also known as Ank3) and β4 spectrin (Liu et al., 2020; Zhou et al., 1998). In addition, the AIS is enriched with Na+ and K+ channels, the cell adhesion molecule neurofascin 186 (NF186) and microtubule-associated proteins, such as Trim46 and Ndel1 (Kuijpers et al., 2016; van Beuningen et al., 2015). Recently, we identified new AIS-associated proteins using differential proteomics and proximity biotinylation (Hamdan et al., 2020; Torii et al., 2020). Among the proteins identified as candidates, we found Ranbp2 (also known as Nup358), a member of the nucleoporin family that was previously reported to be a component of the nuclear pore complex (Goldberg, 2017). Immunostaining for Ranbp2 showed AIS localization. In a separate study, Khalaf et al. (2019) also reported Ranbp2 at the AIS and suggested that its AIS localization requires AnkG. Nuclear Ranbp2 is thought to be linked to the microtubule cytoskeleton, in which it may contribute to cell cycle control, nuclear transport and neuronal cytoarchitecture (Goldberg, 2017).
Given the potential link of Ranbp2 to the microtubule-based cytoskeleton, and the number of microtubule-associated proteins that regulate AIS function (Leterrier, 2018), we sought to determine the function of Ranbp2 at the AIS. However, we found that the anti-Ranbp2 antibody used in both previous studies, and that labels AIS (Hamdan et al., 2020; Khalaf et al., 2019), is not specific to Ranbp2. Instead, it cross-reacts with neurofascin (Nfasc). Epitope tagging of endogenous Ranbp2 at its N- or C-termini, or at an internal site, only showed Ranbp2 at the nuclear membrane; tagged endogenous Ranbp2 was not detected at the AIS. Nevertheless, a small fragment of Ranbp2, when exogenously expressed in neurons, can localize at the AIS. We show that this fragment binds stable microtubules that are enriched at the AIS. Our results show Ranbp2 is not an AIS protein and illustrate how multiple different types of control experiments must be used to identify and characterize new AIS proteins.
RESULTS
The anti-Ranbp2 antibody (A301-796A) detects Ranbp2 and neurofascin
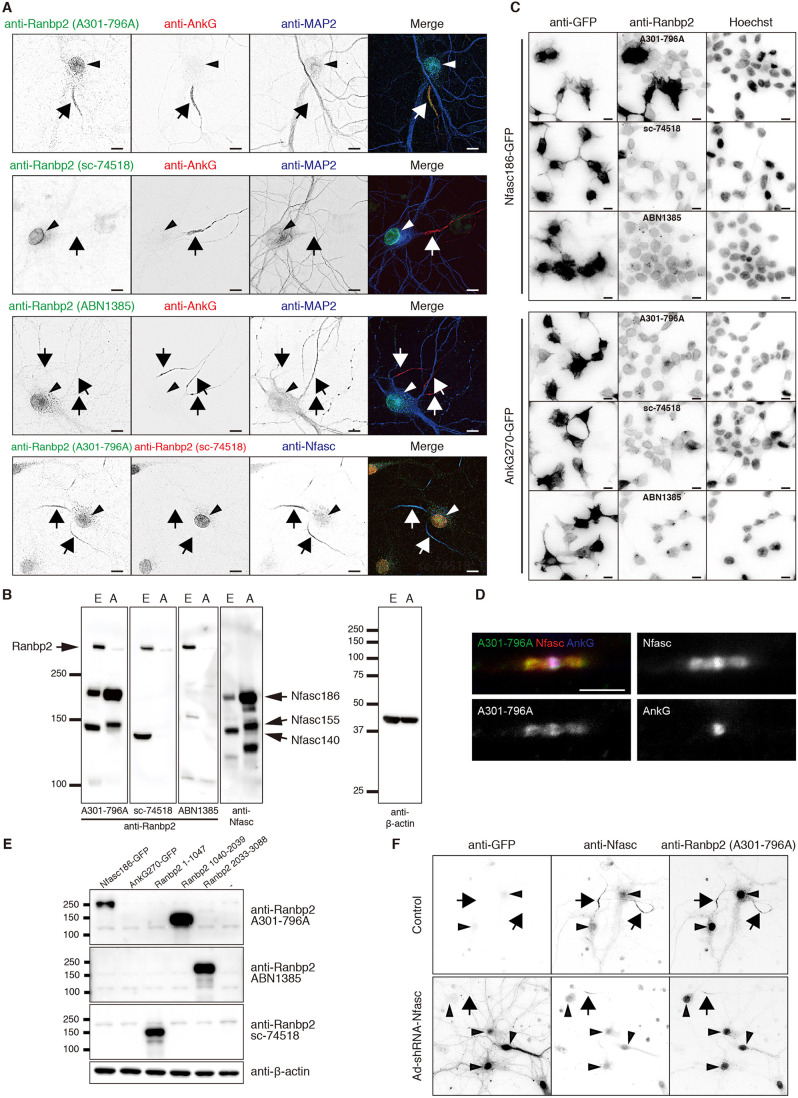
To determine whether Ranbp2 is located at the AIS, we compared three different anti-Ranbp2 antibodies (A301-796A, sc-74518 and ABN1385). We immunostained cultured hippocampal neurons at 10 days in vitro (DIV). Consistent with previous reports (Hamdan et al., 2020; Khalaf et al., 2019), the anti-Ranbp2 antibody (A301-796A) labeled nuclei, but also colocalized with AnkG at the AIS (Fig. 1A). The other two antibodies stained nuclei, but not AIS (Fig. 1A). Immunoblots of total brain homogenate from embryonic or adult rats using all three anti-Ranbp2 antibodies revealed 358 kDa Ranbp2. However, the anti-Ranbp2 antibody A301-796A also strongly labeled ∼190 and ∼150 kDa proteins (Fig. 1B). These molecular weights are similar to neurofascin splice variants, including a 140 kDa variant (NF140) expressed during early development (Zhang et al., 2015), a 155 kDa variant (NF155) found at paranodal junctions of myelinated axons (Tait et al., 2000), and a 186 kDa variant (NF186) found at the AIS and nodes of Ranvier (Davis et al., 1996). Similar to Khalaf et al. (2019), we found that immunostaining of myelinated axons using antibody A301-796A labeled nodes of Ranvier and paranodal junctions (Fig. 1D), the same locations at which NF186 and NF155 are clustered in high density.
Fig. 1.
The anti-Ranbp2 A301-796A antibody cross-reacts with neurofascin. (A) Primary cultured hippocampal neurons were fixed at DIV 10 and stained with combinations of anti-Ranbp2 antibodies (A301-796A, sc-74518 and ABN1385), anti-AnkG and anti-MAP2 antibodies. The arrows indicate the AIS and the arrowheads indicate the nuclei. (B) Immunoblotting of total brain homogenates from embryonic and adult rats. Blots were probed with anti-Ranbp2 antibodies (A301-796A, sc-74518, and ABN1385), anti-neurofascin (Nfasc) or anti-β-actin antibodies. E, embryonic brain; A, adult brain. Arrows indicate Ranbp2 or Nfasc splice variants. (C) Immunostaining of HEK293T cells transfected with GFP-tagged AnkyrinG 270 (AnkG270-GFP) or GFP-tagged NF186 (Nfasc186-GFP). Cells were stained with anti-Ranbp2 (A301-796A), anti-Ranbp2 (sc-74518), anti-Ranbp2 (ABN1385) and anti-GFP antibodies. Nuclei were counterstained with Hoechst. (D) Immunostaining of mouse sciatic nerve nodes of Ranvier using antibody A301-796A (green) and antibodies against AnkG (blue) or Nfasc (red). Scale bar: 5 μm. (E) Immunoblotting of HEK293T cells transfected with Nfasc186-GFP, AnkG270-GFP or Ranbp2 (amino acids 1-1047), Ranbp2 (amino acids 1040-2039) and Ranbp2 (amino acids 2033-3088). Blots were probed with anti-Ranbp2 antibodies (A301-796A, sc-74518 and ABN1385) and anti-β-actin antibodies. (F) Immunostaining of primary cultured neurons infected with adenovirus expressing shRNA against neurofascin (Ad-shRNA-Nfasc) at 0 DIV. Neurons were fixed at 14 DIV and stained with anti-GFP, anti-Nfasc and antibody A301-796A. Arrows indicate axon initial segments, and arrowheads indicate nuclei. All experiments were repeated at least two times. Scale bars: 10 µm.
To determine whether anti-Ranbp2 antibodies cross-react with NF186 or AnkG, the only other protein known to be present at the AIS, nodes and paranodes (Chang et al., 2014), we expressed GFP-tagged NF186 or AnkG in HEK293T cells and immunostained these cells with each anti-Ranbp2 antibody. The cells expressing NF186 were strongly labeled with anti-Ranbp2 antibody A301-796A, but not with the others (Fig. 1C). Furthermore, none of these antibodies labeled AnkG transfected cells. Immunoblotting of the transfected cells also showed that only the A301-796A antibody detected NF186, and none of the antibodies detected AnkG (Fig. 1E). We also found that the three anti-Ranbp2 antibodies used here recognize different regions of Ranbp2: A301-796A detects amino acids 1040-2039, ABN1385 detects amino acids 2033-3088, and sc-74518 detects amino acids 1-1047 (Fig. 1E).
To further confirm that the AIS immunoreactivity seen using the A301-796A antibodies reflects labeling of AIS NF186, we silenced expression of NF186 using a well-characterized NF186 shRNA (Hedstrom et al., 2007). We infected neurons at 0 DIV and immunostained the neurons at 14 DIV. The infection of neurons resulted in the efficient silencing of NF186. More importantly, AIS were not stained with the anti-Ranbp2 antibody A301-796A (Fig. 1F). Together, these results demonstrate that previous reports of Ranbp2 at the AIS actually detected NF186.
Antibody A301-796A recognizes the sequence KPLQG in both Ranbp2 and NF186
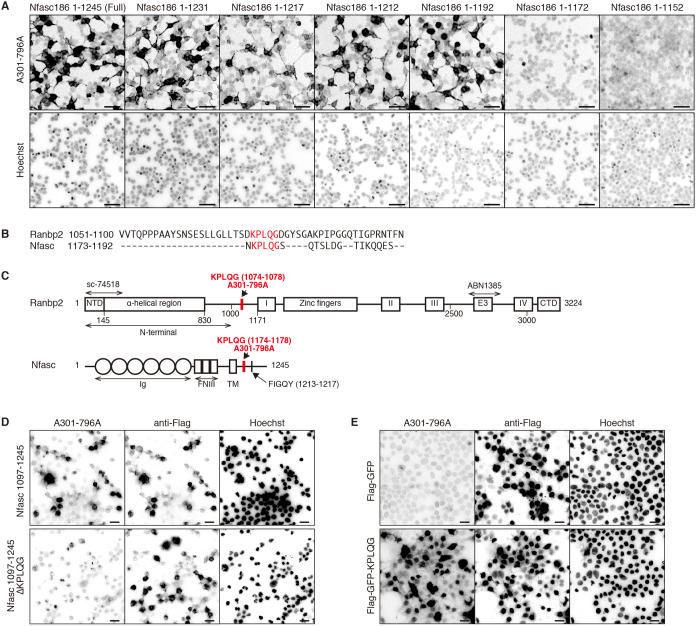
To determine the amino acid sequence recognized by the antibody A301-796A, we immunostained HEK293T cells transfected with full-length or truncated NF186. We found antibody A301-796A recognizes NF186 between amino acids 1173 to 1192 (Fig. 2A), a region common to all neurofascin splice variants. The antibody A301-796A was produced against the region of Ranbp2 between amino acids 1050 to 1100. A sequence alignment between amino acids 1173-1192 of NF186, and amino acids 1051-1100 of Ranbp2, revealed that the sequence KPLQG is common to NF186 and Ranbp2 (Fig. 2B,C). To confirm the necessity of these five amino acids for antibody A301-796A binding to NF186, we expressed the Flag-tagged C-terminal region of NF186, with or without the KPLQG sequence (Nfasc 1097-1245 or Nfasc 1097-1245 ΔKPLQG, respectively), in HEK293T cells. The antibody A301-796A showed strong reactivity against cells expressing Nfasc 1097-1245 but not against cells expressing Nfasc 1097-1245 ΔKPLQG (Fig. 2D). Thus, the KPLQG sequence is necessary for A301-796A binding. Expression of Flag-tagged GFP that was also tagged with the KPLQG peptide sequence at the both the N- and C-termini (Flag-GFP-KPLQG) in HEK293T cells was also sufficient to permit A301-796A binding (Fig. 2E). Thus, the amino acids KPLQG are both necessary and sufficient for binding by the antibody A301-796A.
Fig. 2.
Antibody A301-796A recognizes the peptide sequence KPLQG. (A) Immunostaining of HEK293T cells transfected with full-length or NF186 (Nfasc186) C-terminal truncation constructs. Cells were stained with the A301-796A antibody, whereas nuclei were labeled with Hoechst. (B) ClustalW sequence alignment between Ranbp2, including amino acids 1051-1100 and neurofascin, including amino acids 1173-1192. The KPLQG peptide sequences are highlighted in red. (C) Schematic protein structure of Ranbp2 and neurofascin. The KPLQG peptide sequences are highlighted in red. The region of Ranbp2 used to generate each of the antibodies used in this study are indicated above the protein structure. I, II, III and IV, Ran binding domains; CTD, C-terminal domain; E3, E3 ligase domain; FNIII, fibronectin type III domain; Ig, immunoglobulin domain; NTD, N-terminal domain; TM, transmembrane domain. FIGQY is the AnkG binding motif. (D) Immunostaining of HEK293T cells transfected with Flag-tagged neurofascin, including amino acids 1097-1245 (Nfasc 1097-1245) or Flag-tagged neurofascin 1097-1245 lacking the KPLQG peptide sequence (Nfasc 1097-1245 ΔKPLQG). Cells were stained with anti-Flag and A301-796A antibodies. Nuclei were labeled with Hoechst. (E) Immunostaining of HEK293T cells transfected with Flag-tagged GFP (Flag-GFP) or Flag-tagged GFP with the KPLQG peptide sequence (Flag-GFP-KPLQG). Cells were stained with anti-Flag and A301-796A antibodies. Nuclei were labeled with Hoechst. All experiments were repeated at least two times. Scale bars: 50 µm (A); 20 µm (D,E).
An exogenously expressed N-terminal fragment of Ranbp2 localizes at the AIS
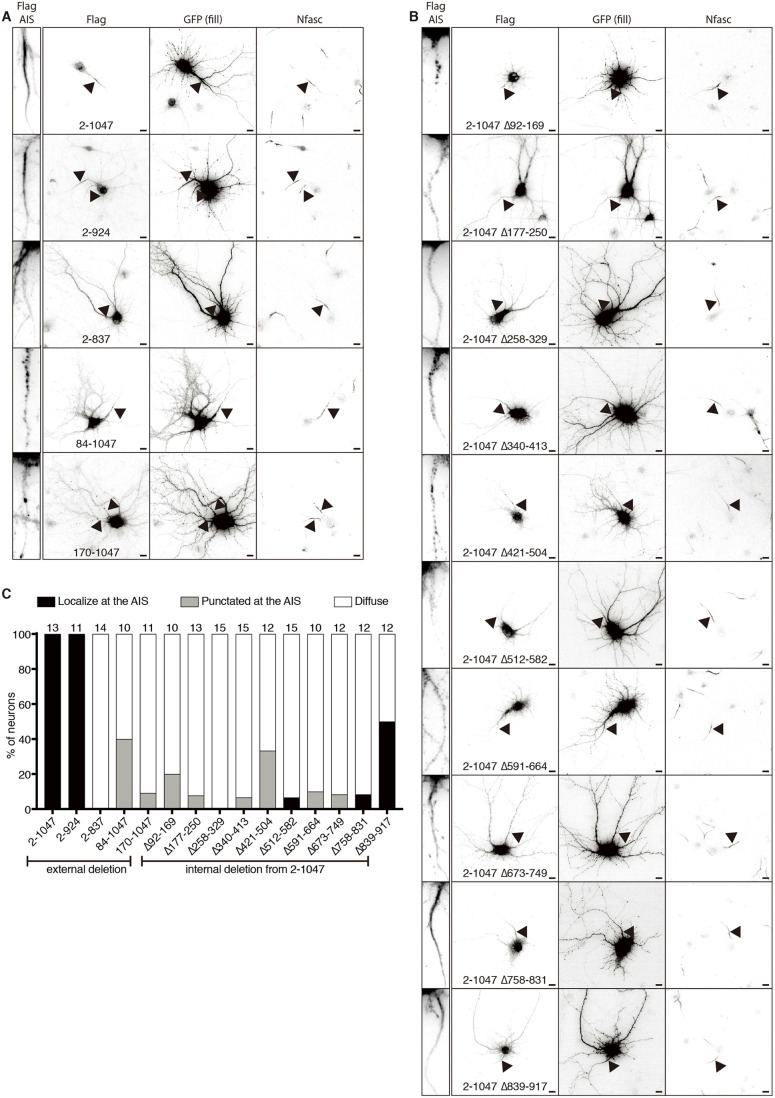
The results presented thus far suggest that immunostaining of the AIS using antibody A301-796A reflects cross-reactivity between Ranbp2 and NF186. However, Khalaf et al. (2019) showed that an exogenously overexpressed N-terminal fragment of Ranbp2 (amino acids 1-900) can also be clustered at the AIS. Because of this surprising result, we generated constructs expressing a Flag-tagged N-terminal fragment of Ranbp2 that is followed by GFP and self-cleaving P2A peptide sequence to visualize neuronal morphology. We immunostained neurons using antibodies against Flag, GFP and neurofascin to label the AIS (Fig. 3A,B). Like Khalaf et al. (2019), we found that the exogenously expressed N-terminal fragment of Ranbp2 (amino acids 2-1047) localized at the AIS (Fig. 3A, top panels). To further understand how the N-terminal fragment of Ranbp2 localizes at the AIS, we generated truncations of this N-terminal fragment. Among these, a construct including amino acids 2-924 still localized at the AIS in 100% of GFP+ neurons (Fig. 3A,C). However, further deletion disrupted its localization at the AIS (Fig. 3A,C). Internal deletions of the N-terminal fragment of Ranbp2 (amino acids 2-1047) also failed to be enriched at the AIS except for the N-terminal fragment of Ranbp2 with an internal deletion of amino acids 839-917 that localized at the AIS in 50% of GFP+ neurons (Fig. 3B,C). These results suggest that the entire structure of the N-terminal fragment of Ranbp2 is important for its localization at the AIS.
Fig. 3.
Localization of the N-terminal fragment of Ranbp2 in mature neurons. (A,B) Immunostaining of primary cultured neurons transfected with the indicated Ranbp2 constructs followed by GFP and self-cleaving P2A peptide sequence. Neurons were transfected at DIV 13, fixed at DIV 14 and stained with anti-Flag, anti-GFP and anti-neurofascin (Nfasc) antibodies. The arrowheads indicate the AIS. Left insets show anti-Flag staining at the AIS. Scale bars: 10 µm. (C) Quantification of the localization pattern of the exogenously expressed N-terminal fragments of Ranbp2 in neurons. The number of neurons counted is shown above each bar. Experiments in A and B were repeated at least two times, whereas the experiment in C was performed once with measurements taken from technical replicates.
Exogenously expressed N-terminal fragment of Ranbp2 colocalizes with stable microtubules
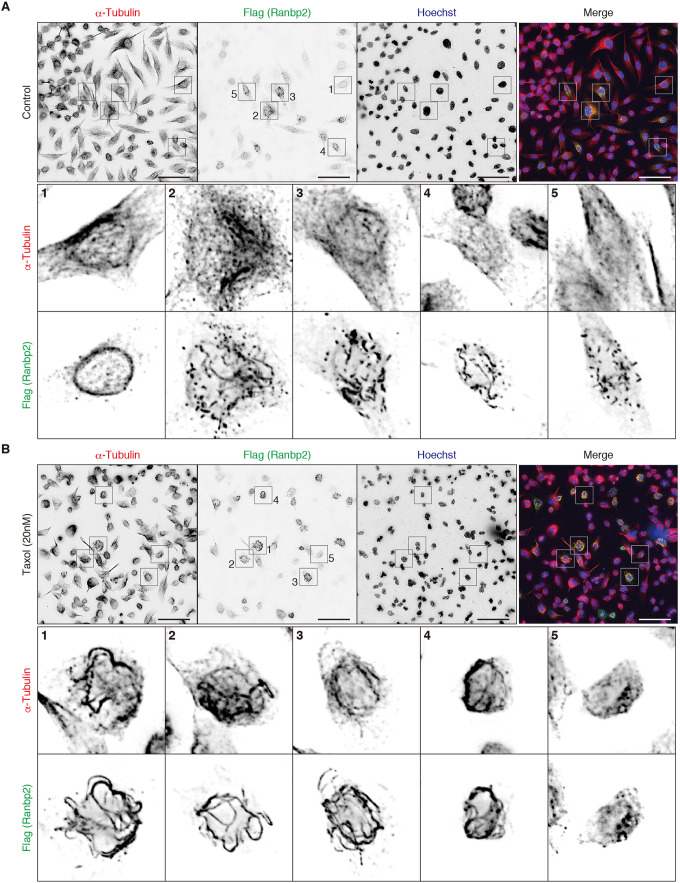
How can the exogenously expressed N-terminal fragment of Ranbp2, but not endogenous Ranbp2, be clustered at the AIS? The AIS is highly enriched with microtubules and has a unique microtubule organization (Palay et al., 1968; van Beuningen et al., 2015). As Ranbp2 is linked to microtubules, we considered whether interaction with microtubules might explain the difference. To test this, we expressed the Flag-tagged N-terminal fragment of Ranbp2 (amino acids 2-1047) in HeLa cells and compared its localization to alpha-tubulin. The Ranpb2 fragment localized at the nuclear membrane in cells that weakly expressed the Ranbp2 fragment (Fig. 4A1). However, in cells that strongly expressed the N-terminal fragment of Ranbp2, we found that it formed filaments (Fig. 4A2,A3) or comets (Fig. 4A4,A5) in the cytosol. However, these structures were not strongly associated with tubulins. After HeLa cells were treated with Taxol to stabilize microtubules, they formed thick bundles. We found that the exogenously expressed N-terminal fragment of Ranbp2 colocalized with the stabilized microtubules (Fig. 4B1-B4). Intriguingly, in Taxol-treated cells that weakly expressed the N-terminal fragment, the Flag-tagged Ranbp2 did not localize at the nuclear membrane but still associated with bundles of microtubules (Fig. 4B5). Together, these results suggest that the AIS localization of the N-terminal fragment of Ranbp2 is an artifact due to its intrinsic affinity for stable microtubules (Joseph and Dasso, 2008).
Fig. 4.
Exogenously expressed N-terminal fragment of Ranbp2 colocalizes with stable microtubules in Taxol-treated cells. (A,B) HeLa cells were transiently transfected with a Flag-tagged N-terminal fragment of Ranbp2, including amino acids 2-1047. Cells were stained with anti-Flag and anti-alpha-tubulin antibodies. Nuclei were labeled with Hoechst. Cells in B were treated with 20 nM Taxol. Higher magnification of insets 1 to 5 are shown in the bottom panels. All experiments were repeated at least two times. Scale bars: 50 µm.
Endogenously expressed Ranbp2 does not accumulate at the AIS
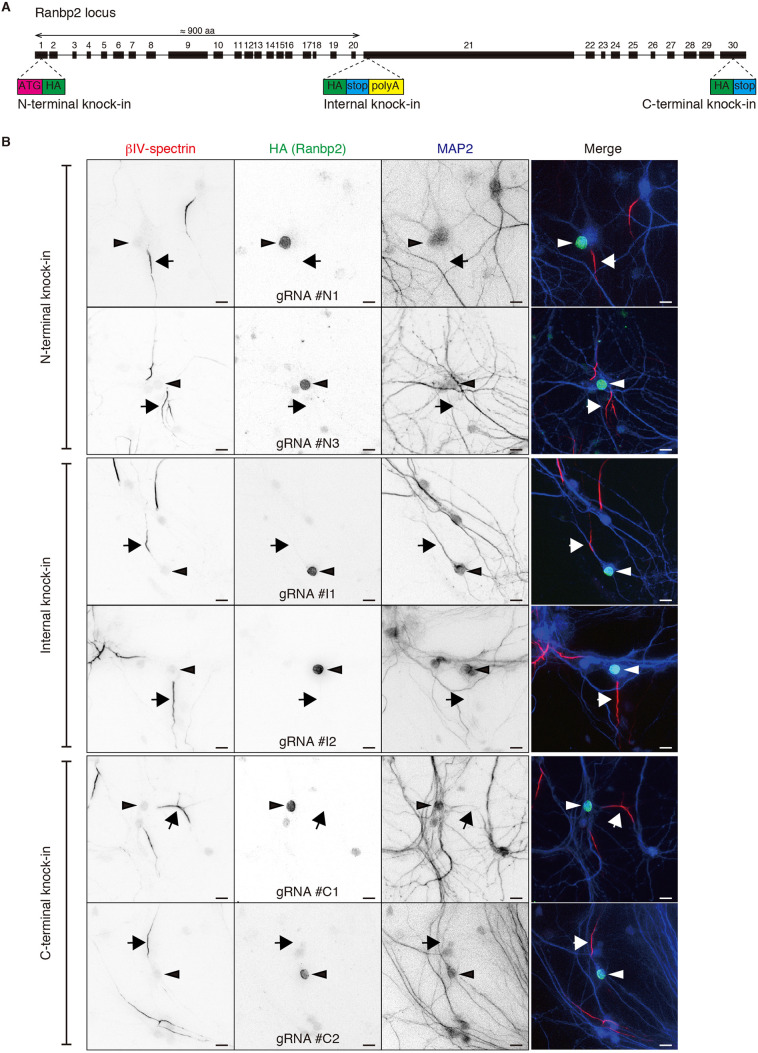
To determine the localization of endogenously expressed Ranbp2 without the use of anti-Ranbp2 antibodies, we used CRISPR/Cas9 to perform homology-independent insertion of a hemagglutinin (HA)-epitope tag into Ranbp2 in neurons (Gao et al., 2019; Suzuki and Izpisua Belmonte, 2018; Willems et al., 2020). We generated six independent single guide RNAs (sgRNAs) and three independent donors for different insertion sites (Fig. 5A). For N-terminal knock-in, an HA-epitope tag followed by a Kozak sequence was inserted after the initiation codon (ATG) of Ranbp2 at exon 1. For C-terminal knock-in, an HA-epitope tag was inserted before the stop codon of Ranbp2 at exon 30. For an internal knock-in, an HA-epitope tag followed by a stop codon and polyadenylation sequence was inserted at exon 21 of Ranbp2 to stop translation at the insertion site and to express the N-terminal fragment of Ranbp2 (amino acids around 1-900) that also localizes at the AIS after exogenous overexpression. Immunostaining of the HA-epitope tag showed clear localization of Ranbp2 at the nuclear membrane but not at the AIS of neurons using all six pairs of sgRNAs and donors (Fig. 5B). Thus, endogenous Ranbp2 does not localize to the AIS.
Fig. 5.
Endogenously expressed Ranbp2 does not accumulate at the AIS. (A) Schematic depicting insertion sites for labeling of endogenous Ranbp2 with an HA-epitope tag. (B) Immunolabeling of tagged Ranbp2 in cultured hippocampal neurons. Neurons were infected with AAV at DIV 1, fixed at DIV 13 and stained with anti-HA (green), anti-β4 spectrin to label AIS (red), and anti-MAP2 antibodies (blue). Arrows indicate AIS, whereas arrowheads indicate the nucleus. All experiments were repeated at least two times. Scale bars: 10 µm.
DISCUSSION
Two previous studies reported that Ranbp2 is localized at the AIS of neurons (Khalaf et al., 2019; Hamdan et al., 2020). In both previous reports, AIS Ranbp2 was shown by immunostaining using the anti-Ranbp2 antibody A301-796A. Khalaf et al. (2019) also showed that an N-terminal fragment of Ranbp2 expressed in neurons was recruited to the AIS. However, our results show that Ranbp2 is not found at the AIS and we explain why the previous studies were incorrect. We show that: (1) the antibody A301-796A strongly reacts with both NF186 and Ranbp2 at the common amino acid sequence KPLQG; (2) silencing of NF186 in neurons blocks AIS labeling using antibody A301-796A; (3) other antibodies that recognize Ranbp2 do not label AIS; (4) tagging of endogenous Ranbp2 in neurons using CRISPR/Cas9 shows only labeling of the nuclear membrane; and (5) the N-terminal fragment of Ranbp2 strongly associates with stabilized microtubules. The results presented here should serve as a cautionary tale and illustrate how multiple independent control experiments are necessary to validate antibody labeling and even overexpression studies (Buffington et al., 2012; Chang et al., 2010; Rhodes and Trimmer, 2006).
One of the most surprising results was the apparent inconsistency between the simple explanation of antibody cross-reactivity and the observation that when overexpressed in neurons, the N-terminal fragment of Ranbp2 could be found at the AIS. Unfortunately, searching for unique AIS localization motifs like those found in Na+ channels and NF186 was unsuccessful. Instead, our truncation and deletion studies suggest that the entire N-terminal fragment of Ranbp2 is important for its AIS localization. Although we did not find a unique AIS localization motif, we did find that stabilization of microtubules in HeLa cells was sufficient to cause exogenously expressed Ranbp2 to associate with microtubules. This observation is consistent with previous reports of an N-terminal fragment of Ranbp2 remodeling and stabilizing microtubules (Joseph and Dasso, 2008). Together, these observations help resolve the apparent inconsistency. AISs are known to have a unique microtubule organization with fascicles of microtubules crosslinked by the protein Trim46 (Palay et al., 1968; van Beuningen et al., 2015). In addition, the microtubule-associated protein Map6 (also known as STOP or stable-tubule only polypeptide) is enriched at the AIS at which it interacts with AnkG (Hamdan et al., 2020; Tortosa et al., 2017).
Why did Ranbp2 appear in two separate proteomics experiments designed to identify new AIS proteins? In the first, we generated a fusion protein between the microtubule-associated protein Ndel1 and BioID to perform proximity biotinylation at the AIS (Hamdan et al., 2020). We found that Ranbp2 co-purified with biotinylated proteins, suggesting close proximity to AIS Ndel1-BioID. However, the experiments reported here show that endogenous Ranbp2 is localized at the nuclear envelope, and neither the labeling of the Ndel1-BioID fusion protein nor the biotinylated proteins were found at the nuclear envelope. In the second experiment, we performed differential mass spectrometry between control and AnkG-deficient brains (Torii et al., 2020). Here, we found Ranbp2 was reduced after loss of AnkG, suggesting the amount of Ranbp2 in the neuron depends on AnkG. The experiments reported here suggest that the Ranbp2 identified in our previous proteomics experiments is an artifact and further emphasizes the need to perform careful and thorough controls when validating proteomic data sets.
MATERIALS AND METHODS
Animals
Timed pregnant ICR mice and SD rats were ordered from Charles River. All animal work and procedures were approved by the Institutional Animal Care and Use Committee at the Baylor College of Medicine, and were performed in accordance with the National Institutes of Health (NIH) guide for the humane care and use of animals.
Cell culture
HEK293T cells were a gift from the laboratory of Dr Ben Arenkiel (Baylor College of Medicine) and HeLa cells (provided by the Tissue Culture Core of Baylor College of Medicine) were purchased and grown on cell culture dishes at 37°C in Dulbecco's modified Eagle's Medium containing 10% heat-inactivated fetal bovine serum (GE Healthcare), 50 units/ml penicillin and 50 μg/ml streptomycin. Primary neuronal cultures were prepared as described previously (Torii et al., 2020).
Antibodies
The following mouse monoclonal antibodies were used: Ranbp2 (amino acids 1-300 at the N-terminus of Ranbp2 of human origin; Santa Cruz Biotechnology, Cat# sc-74518, RRID:AB_2176784, 1:20), Flag (MBL, M185, 1:10,000), alpha-tubulin (Developmental Studies Hybridoma Bank, clone 12G10; RRID: AB_2315509, 1:500), and beta-actin (Millipore Sigma-Aldrich, Cat# A1978, 1:10,000). Mouse monoclonal AnkG antibody (N106/65; RRID: AB_10675130, 1:200) was purchased from the University of California, Davis/NIH NeuroMab Facility. The rabbit polyclonal antibodies were sourced as follows: Ranbp2 (Bethyl Cat# A301-796A, RRID: AB_1211503, 1:200), Ranbp2 [generated against internal repeat IR1+2 domain (IR) of Ranbp2 of human origin; Millipore Sigma, Cat# ABN1385, 1:100] and GFP (Thermo Fisher Scientific, Cat# A-11122, RRID: AB_221569, 1:1000). The rabbit anti-βIV Spectrin SD antibody has been described previously (Yoshimura et al., 2017; 1:1000). We purchased the following chicken antibodies: pan-neurofascin (R&D Systems, Cat# AF3235; RRID: AB_10890736, 1:500) and MAP2 (EnCor, Cat# CACP-MAP2; RRID: AB_2138173, 1:5000). The rat monoclonal HA antibody (Cat# 11867423001, RRID: AB_390918, 1:500) was purchased from Roche. Hoechst 33258 (H3569, 1:100,000) was purchased from Thermo Fisher Scientific. Secondary antibodies were purchased from Thermo Fisher Scientific and Jackson ImmunoResearch Laboratories.
Plasmids
AnkG270-GFP and NF186-GFP were gifts from Dr Vann Bennett (Duke University, Durham, NC, USA). The generation of the truncation mutants of neurofascin 186 has been described previously (Torii et al., 2020). pUCmini-iCAP-PHP.S was obtained from Addgene (plasmid # 103006). The pHelper plasmid was purchased from Agilent Technologies. To generate truncated Ranbp2 constructs, including GFP and self-cleaving P2A peptide sequence, GFP was PCR amplified from the pEGFPC1 vector adding self-cleaving P2A peptide sequence, and then inserted into the CSII-CMV-MCS vector (a generous gift from Dr Hiroyuki Miyoshi, RIKEN Bioresource Center, Tsukuba, Japan). Truncated Ranbp2 fragments were next PCR amplified from the rat genome and inserted into the above vector. The Flag-tagged truncated Ranbp2 construct (amino acids 2-1047, 1040-2039 and 2033-3088) was generated by PCR amplification of the fragment from the rat genome and insertion into the p3XFLAG-CMV-7.1 vector. The Flag-tagged C-terminal region of NF186 (Nfasc 1097-1245) and the one lacking KPLQG (Nfasc 1097-1245 ΔKPLQG) were made by PCR amplification of the fragments followed by insertion into the p3XFLAG-CMV-7.1 vector. Flag-tagged GFP (Flag-GFP) or Flag-tagged GFP that was also tagged with the KPLQG peptide sequence at both the N- and C-termini (Flag-GFP-KPLQG) were generated by PCR amplification of GFP from the pEGFPC1 vector and insertion into the p3XFLAG-CMV-7.1 vector. The KPLQG peptide sequence 5′- GATAAACCTTTACAAGGAGAT-3′ was added by PCR amplification. The sgRNA constructs and the donor templates were generated following strategies similar to those described previously (Gao et al., 2019). The sgRNA sequences for mouse Ranbp2 were 5′- GCGGTACATCGCCTCGGTGC-3′ (Ranbp2-N1), 5′- GTAGCAAGGCCGACGTGGAG-3′ (Ranbp2-N3), 5′- GGAGACTCAAAACGCAAGGG-3′ (Ranbp2-I1), 5′- GAAGCGTGAGCTACAAGTGG-3′ (Ranbp2-I2), 5′- GTGAGAAAGATTGAATCATT-3′ (Ranbp2-C1) and 5′- TTACTATCCATGGCCAATTA-3′ (Ranbp2-C2). All DNA constructs were verified by sequencing (Genewiz).
Tissue and cell lysate preparation and immunoblotting
For tissue lysate preparation, whole mouse brains were sonicated three times for 5 s in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS and 1% NP-40 with protease inhibitor) on ice. Homogenates were then centrifuged at 21,130 g for 10 min at 4°C. The supernatants were collected, and protein concentration was measured. For cell lysate preparation, cells were lysed in lysis buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% Triton X-100 and 1 mM EDTA with protease inhibitor], and the lysates were centrifuged at 14,000 rpm for 10 min at 4°C. The supernatants were collected, and protein concentration was measured. Those lysates were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (GE Healthcare), blocked with 5% skimmed milk and immunoblotted with each primary antibody, and in turn with peroxidase-conjugated secondary antibodies.
Immunofluorescence microscopy
Cultured rat or mouse primary neurons, HEK293T cells, HeLa cells or sciatic nerves were fixed in 4% paraformaldehyde (pH 7.2) for 30 min (nerves) or 10 min (cells). Cells or nerve sections were immunostained with the appropriate antibodies in phosphate buffer with 0.3% Triton X-100 and 10% goat serum. Images of immunofluorescence were captured using an Axio-imager Z1 fluorescence microscope (Zeiss), an Axio-observer Z1 fluorescence microscope (Zeiss) or an Eclipse Ni-E microscope (Nikon). Zeiss 20× (0.8 NA) Plan Apochromat, 40× (0.75 NA) Plan Neofluar, 63× (1.4 NA) Plan Apochromat objectives or Nikon 20× (0.75 NA), 40× (0.95 NA) and 60× (1.4 NA) Plan Apochromat objectives were used to acquire images. Images were collected at room temperature using Zen acquisition software (Zeiss) or Nikon NIS-Elements software. Fluorophores included Alexa Fluor 488, Alexa Fluor 594 and AMCA conjugated to secondary antibodies and Hoechst. Figures were made using Adobe Photoshop and Adobe Illustrator.
Plasmid transfection and drug treatment
HEK293T cells and HeLa cells were transfected with plasmid DNA using PEI Max (Polysciences, 24765) according to the manufacturer's instructions. The media were replaced after 16-20 h of transfection. Taxol (Paclitaxel; Cayman chemicals) was added with medium change at 20 nM to stabilize microtubules in HeLa cells. The cells were fixed or lysed at 40-48 h of transfection for immunostaining or immunoblotting, respectively. Primary cultured hippocampal neurons were transfected with Lipofectamine 2000 (Invitrogen). The medium was replaced after 5 h of transfection.
Adeno-associated virus vector production and infection for neurons
Small-scale adeno-associated virus (AAV) supernatants were produced as described previously (Gao et al., 2019). Briefly, HEK293T cells were transfected with AAV plasmid, helper plasmid and serotype PHP.S plasmid with PEI Max (Polysciences, 24765). Cells were then incubated for 3-5 days, and the AAV-containing supernatant medium was collected. The supernatants were centrifuged at 9391 g for 2 min to remove debris, and stored at 4°C until use. An aliquot of 500 µl of the combinations of small-scale AAV supernatants (100 µl of Cas9, 200 µl of gRNA and 200 µl of donor in a well of a 12-well plate) were added to cultures at 0-1 DIV. The medium was replaced after 2 days of infection.
shRNA adenovirus
The neurofascin shRNA adenoviral construct was made as described previously (Hedstrom et al., 2007).
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Y.O., M.N.R.; Methodology: Y.O.; Investigation: Y.O.; Writing - original draft: Y.O., M.N.R.; Writing - review & editing: Y.O., M.N.R.; Visualization: Y.O.; Supervision: M.N.R.; Project administration: M.N.R.; Funding acquisition: M.N.R.
Funding
This work was supported by the National Institute of Neurological Disorders and Stroke (NIH NS044916), and by the Dr Miriam and Sheldon G. Adelson Medical Research Foundation. Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.256180.reviewer-comments.pdf
References
- Buffington, S. A., Sobotzik, J. M., Schultz, C. and Rasband, M. N. (2012). IκBα is not required for axon initial segment assembly. Mol. Cell. Neurosci. 50, 1-9. 10.1016/j.mcn.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, K.-J., Susuki, K., Dours-Zimmermann, M. T., Zimmermann, D. R. and Rasband, M. N. (2010). Oligodendrocyte myelin glycoprotein does not influence node of ranvier structure or assembly. J. Neurosci. 30, 14476-14481. 10.1523/JNEUROSCI.1698-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, K.-J., Zollinger, D. R., Susuki, K., Sherman, D. L., Makara, M. A., Brophy, P. J., Cooper, E. C., Bennett, V., Mohler, P. J. and Rasband, M. N. (2014). Glial ankyrins facilitate paranodal axoglial junction assembly. Nat. Neurosci. 17, 1673-1681. 10.1038/nn.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J. Q., Lambert, S. and Bennett, V. (1996). Molecular composition of the node of Ranvier: identification of ankyrin-binding cell adhesion molecules neurofascin (mucin+/third FNIII domain−) and NrCAM at nodal axon segments. J. Cell Biol. 135, 1355-1367. 10.1083/jcb.135.5.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., Hisey, E., Bradshaw, T. W. A., Erata, E., Brown, W. E., Courtland, J. L., Uezu, A., Xiang, Y., Diao, Y. and Soderling, S. H. (2019). Plug-and-play protein modification using homology-independent universal genome engineering. Neuron 103, 583-597.e8. 10.1016/j.neuron.2019.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, M. W. (2017). Nuclear pore complex tethers to the cytoskeleton. Semin. Cell Dev. Biol. 68, 52-58. 10.1016/j.semcdb.2017.06.017 [DOI] [PubMed] [Google Scholar]
- Hamdan, H., Lim, B. C., Torii, T., Joshi, A., Konning, M., Smith, C., Palmer, D. J., Ng, P., Leterrier, C., Oses-Prieto, J. A.et al. (2020). Mapping axon initial segment structure and function by multiplexed proximity biotinylation. Nat. Commun. 11, 100. 10.1038/s41467-019-13658-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom, K. L., Xu, X., Ogawa, Y., Frischknecht, R., Seidenbecher, C. I., Shrager, P. and Rasband, M. N. (2007). Neurofascin assembles a specialized extracellular matrix at the axon initial segment. J. Cell Biol. 178, 875-886. 10.1083/jcb.200705119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J. and Dasso, M. (2008). The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett. 582, 190-196. 10.1016/j.febslet.2007.11.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf, B., Roncador, A., Pischedda, F., Casini, A., Thomas, S., Piccoli, G., Kiebler, M. and Macchi, P. (2019). Ankyrin-G induces nucleoporin Nup358 to associate with the axon initial segment of neurons. J. Cell Sci. 132, jcs222802. 10.1242/jcs.222802 [DOI] [PubMed] [Google Scholar]
- Kuijpers, M., van de Willige, D., Freal, A., Chazeau, A., Franker, M. A., Hofenk, J., Rodrigues, R. J. C., Kapitein, L. C., Akhmanova, A., Jaarsma, D.et al. (2016). Dynein regulator NDEL1 controls polarized cargo transport at the axon initial segment. Neuron 89, 461-471. 10.1016/j.neuron.2016.01.022 [DOI] [PubMed] [Google Scholar]
- Leterrier, C. (2018). The axon initial segment: an updated viewpoint. J. Neurosci. 38, 2135-2145. 10.1523/JNEUROSCI.1922-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.-H., Seo, R., Ho, T. S.-Y. ., Stankewich, M., Mohler, P. J., Hund, T. J., Noebels, J. L. and Rasband, M. N. (2020). β spectrin-dependent and domain specific mechanisms for Na+ channel clustering. eLife 9, e56629. 10.7554/eLife.56629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay, S. L., Sotelo, C., Peters, A. and Orkand, P. M. (1968). The axon hillock and the initial segment. J. Cell Biol. 38, 193-201. 10.1083/jcb.38.1.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, K. J. and Trimmer, J. S. (2006). Antibodies as valuable neuroscience research tools versus reagents of mass distraction. J. Neurosci. 26, 8017-8020. 10.1523/JNEUROSCI.2728-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K. and Izpisua Belmonte, J. C. (2018). In vivo genome editing via the HITI method as a tool for gene therapy. J. Hum. Genet. 63, 157-164. 10.1038/s10038-017-0352-4 [DOI] [PubMed] [Google Scholar]
- Tait, S., Gunn-Moore, F., Collinson, J. M., Huang, J., Lubetzki, C., Pedraza, L., Sherman, D. L., Colman, D. R. and Brophy, P. J. (2000). An oligodendrocyte cell adhesion molecule at the site of assembly of the paranodal axo-glial junction. J. Cell Biol. 150, 657-666. 10.1083/jcb.150.3.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, T., Ogawa, Y., Liu, C.-H., Ho, T. S.-Y., Hamdan, H., Wang, C.-C., Oses-Prieto, J. A., Burlingame, A. L. and Rasband, M. N. (2020). NuMA1 promotes axon initial segment assembly through inhibition of endocytosis. J. Cell Biol. 219, e201907048. 10.1083/jcb.201907048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa, E., Adolfs, Y., Fukata, M., Pasterkamp, R. J., Kapitein, L. C. and Hoogenraad, C. C. (2017). Dynamic palmitoylation targets MAP6 to the axon to promote microtubule stabilization during neuronal polarization. Neuron 94, 809-825.e7. 10.1016/j.neuron.2017.04.042 [DOI] [PubMed] [Google Scholar]
- van Beuningen, S. F. B., Will, L., Harterink, M., Chazeau, A., van Battum, E. Y., Frias, C. P., Franker, M. A. M., Katrukha, E. A., Stucchi, R., Vocking, K.et al. (2015). TRIM46 controls neuronal polarity and axon specification by driving the formation of parallel microtubule arrays. Neuron 88, 1208-1226. 10.1016/j.neuron.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Willems, J., de Jong, A. P. H., Scheefhals, N., Mertens, E., Catsburg, L. A. E., Poorthuis, R. B., de Winter, F., Verhaagen, J., Meye, F. J. and MacGillavry, H. D. (2020). ORANGE: A CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLoS Biol. 18, e3000665. 10.1371/journal.pbio.3000665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, T., Stevens, S. R., Leterrier, C., Stankewich, M. C. and Rasband, M. N. (2017). Developmental changes in expression of βIV spectrin splice variants at axon initial segments and nodes of ranvier. Front. Cell Neurosci. 10, 304. 10.3389/fncel.2016.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A., Desmazieres, A., Zonta, B., Melrose, S., Campbell, G., Mahad, D., Li, Q., Sherman, D. L., Reynolds, R. and Brophy, P. J. (2015). Neurofascin 140 is an embryonic neuronal neurofascin isoform that promotes the assembly of the node of Ranvier. J. Neurosci. 35, 2246-2254. 10.1523/JNEUROSCI.3552-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D., Lambert, S., Malen, P. L., Carpenter, S., Boland, L. M. and Bennett, V. (1998). AnkyrinG is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J. Cell Biol. 143, 1295-1304. 10.1083/jcb.143.5.1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.