Abstract
Background
The research analyzed a group of patients to develop a statistical nomogram and a web-based survival rate predictor for the comprehensive estimate of the overall survival (OS) of children with acute myeloid leukemia.
Methods
Between 1999 to 2015, we used the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) database to evaluate and randomly divide 440 children diagnosed with AML into the population of training (n = 309) and validation (n = 131). The analysis of Lasso Cox was used to identify separate predictive variables. We have used essential forecasting considerations to construct a nomogram and a web-based calculator focused on Cox regression analysis. Nomogram validation was tested through discrimination and calibration.
Results
Compared to the multivariate training cohort models, a nomogram integrating gender, age of diagnose, WBC at diagnosis, bone marrow leukemic blast percentage, and chromosomal abnormalities [t(8; 21), inv(16)] were designed for the prediction of OS. We also developed a predictive survival nomogram and a web-based calculator. C-indexes validated internally and checked externally were 0.747 and 0.716. The calibration curves have shown that the nomogram might accurately forecast 3-year and 5-year OS.
Conclusions
A nomogram effectively predicts survival in children with AML. This prognostic model can be used in clinical practice.
1. Background
Acute myeloid leukemia (AML) is a disease of the hematopoietic stem cells (HSC), marked by irregular development and immature blast cell proliferation in the bone marrow [1, 2]. Childhood AMLs account for nearly 20% of childhood leukemia and >50% of fatalities in the aforementioned populations [3, 4]. Several contributing factors, including toxic exposures, chemotherapy or radiation treatment, myelodysplastic syndrome, and genetic factors, have led to AML pathogenesis [5–7]. Despite considerable progress in discovering pediatric AML pathophysiology, survival rates in patients have not significantly improved and nearly more than half of children diagnosed with AML suffering from recurrence.
In recent decades, AML has improved diagnosis and treatment but the overall survival rate (OS) is still low, less than 50% [8, 9]. The cytogenetic karyotype and molecular defects in the diagnosis are known to be the most important predictive factors for OS. Forecast probability stratification should be improved as it can establish successful diagnostic and therapeutic approaches.
Nomograms are accepted as a viable substitute method that can allow clinicians to accurately predict individuals [10, 11]. By adding clinically relevant variables, the survival rate can be measured accurately [12, 13]. Nevertheless, nomograms for the estimation of children's survival with AML have not yet been fully established.
In this research, we established a prognostic nomogram based on the TARGET population data to predict individualized survival in children with AML.
2. Methods
2.1. Patient Selection
We downloaded clinical information from the TARGET project database for AML patients (http://ocg.cancer.gov/). The criteria for AML were identified between 1999 and 2015. Criteria for exclusion were as follows: unclear gender, uncertain age of diagnose, unknown WBC at diagnosis, unknown bone marrow leukemic blast percentage (BM blast percentage), and unclear chromosomal results including t(8; 21), inv(16). As our mathematical research method, we used R software (3.5.2). The optimum diagnostic age cutoff value was 3489 days. 70% of all patients have been randomly chosen to form the Nomogram Construction Training Cohort, and the remaining 30% have been validated.
2.2. Ethical Approval
Because the identified patient information is not included in the TARGET database, no ethical consent is needed.
2.3. Training and Validation Cohort
The whole population was divided by the random sample process into the training or validation population (ratio, ~ 3 : 1). The training population was used to assess the predictive model and the statistical probability stratification. The validation population was used to verify the model prediction.
2.4. Statistical Analysis
Categorical measurements were represented as counts and percentages. Continuous measurements were represented as mean and range. The t-test was used to continuous measurements, while the Chi-square was used to compare ones. Statistically significant was P < 0.05. The primary endpoint was overall survival (OS). OS was described as an interval from diagnostic to death or last follow-up, regardless of the cause of death. The optimum age cutoff value was calculated by the R program package “survminer”.
We used the Lasso Cox regression model [14], defining individual operating system risk factors, as well as the “glmnet” package. Using the “rms package” program, nomograms and calibration plots were developed. We used the “shiny” and “DynNom” packages to create a web-based survival rate calculator that estimated overall survival rates (http://www.shinyapps.io/) dynamically. The nomogram measurement was carried out using the concordance index (C-index) and calibration curves. The C-index represents the nomogram's capacity to discriminate. The larger the C-index, the more accurate the model. For the analysis of the observed and predicted nomogram probabilities, calibration plots were used. The precision of the 3-year and 5-year nomogram survival was assessed by the ROC (receiver operating characteristic) curve.
3. Results
3.1. Patient Characteristics
In the TARGET database, we found 440 eligible patients (1999-2015). The median OS was 1547 days (range 1-3113 days). The OS rates for 3 years and 5 years were 70.9% and 44.3%, respectively. In Table 1, the demographic and clinical features of the population in training (n = 309), the validation population (n = 131), and all patients (n = 440) are illustrated.
Table 1.
Clinical characteristics of patients with AML.
| Variables | All patients | Training cohort | Validation cohort |
|---|---|---|---|
| (N = 440) | (n = 309) | (n = 131) | |
| Age | |||
| <14 | 312 (70.9%) | 224 (72.5%) | 88 (67.2%) |
| ≥14 | 128 (29.1%) | 85 (27.5%) | 43 (32.8%) |
| Gender | |||
| Male | 231 (52.5%) | 161 (52.1%) | 70 (53.4%) |
| Female | 209 (47.5%) | 148 (47.9%) | 61 (46.6%) |
| White blood cell | |||
| <100 | 363 (82.5%) | 254 (82.2%) | 109 (83.2%) |
| ≥100 | 77 (17.5%) | 55 (17.8%) | 22 (16.8%) |
| BM leukemic blast percentage | |||
| <90% | 333 (75.7%) | 255 (82.5%) | 78 (59.5%) |
| ≥90% | 107 (24.3%) | 54 (17.5%) | 53 (40.5%) |
| Chromosomal abnormalities | |||
| t(8; 21) | 79 (17.9%) | 58 (18.8%) | 21 (16.1%) |
| Inv(16) | 57 (12.9%) | 39 (12.6%) | 18 (13.7%) |
3.2. Identification of Independent Risk Factors
Lasso Cox also used the training population to evaluate regression and classify individual risk factors impacting the OS (Figure 1). With increases in λ, the coefficient of variables decreased. The excluded variable parameters were compressed to 0 when λ was optimum. Variables < 0 have therefore been chosen. As a result, a total of 6 predictive factors (gender, age of diagnose, WBC at diagnosis, BM blast percentage, chromosomal abnormalities [t(8; 21), inv(16)]) were included in the predictive model. These factors were applied to the nomogram.
Figure 1.
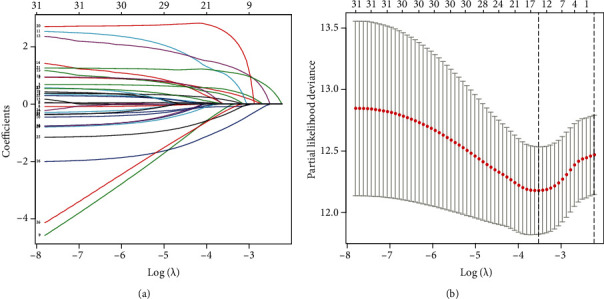
Identification of predictive factors using the Lasso Cox regression. (a) The vertical line was plotted at the given λ, selected by cross-validation. For the optimal λ, 6 features of them with a nonzero coefficient were selected. (b) The penalization coefficient λ in the Lasso model was adjusted using cross-validation and the minimum criterion. The vertical black lines define the optimal λ.
3.3. Nomogram
A nomogram that incorporates all the relevant independent factors was established for estimation of 3 years and 5 years OS, based on the reduced multivariate models of the training population (Figure 2). This model revealed the inv(16) translocation mainly contributed to the prognosis, followed by t(8; 21) translocation, WBC at diagnosis, age of diagnose, etc. Each factor received a score on the scale of points. By applying the scores to the overall scale, we might estimate the probability of 3-year and 5-year survival.
Figure 2.
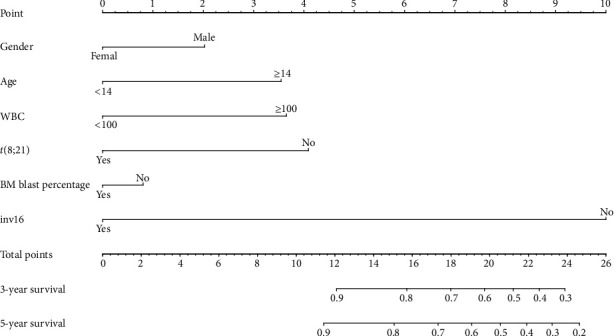
Predictive nomogram for OS estimation.
The calculator predicted patients' survival based on their clinical characteristics and based on these findings, we developed a dynamic web-based calculator (https://dxyjiang.shinyapps.io/AMLpredict/), to predict OS in AML patients by nomogram (Figure 3). For e.g., the OS rate for 5 years was approximately 96% for one patient aged 14, diagnosed with WBC of approximately 100 ≥ 109/L, BM blast percentage ≥ 90%, and t(8; 21) and inv(16) positive.
Figure 3.
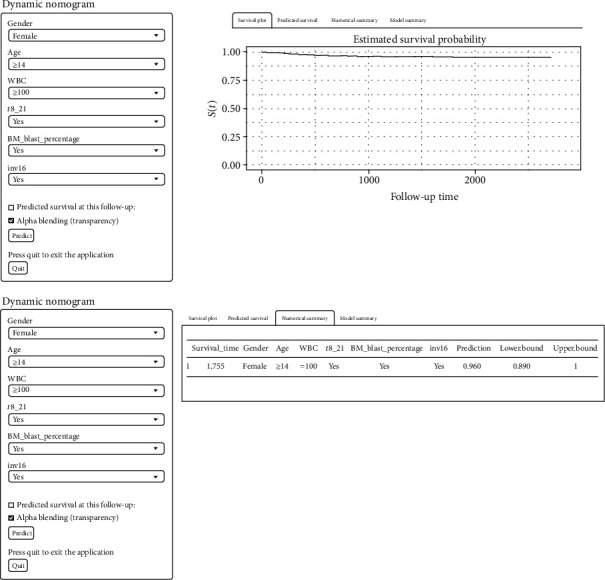
A dynamic web-based calculator b 95%confidence interval according to the web survival rate calculator.
3.4. Nomogram Validation
Internal analysis shows that the nomogram can estimate the OS correctly with a 0.747 C-index. Similarly, the external validation of the C-index was 0.716. The calibration statistics showed an outstanding correlation between the values for the 3-year and 5-year OS predicted and observed in both the population of training and the validation cohort (Figure 4).
Figure 4.
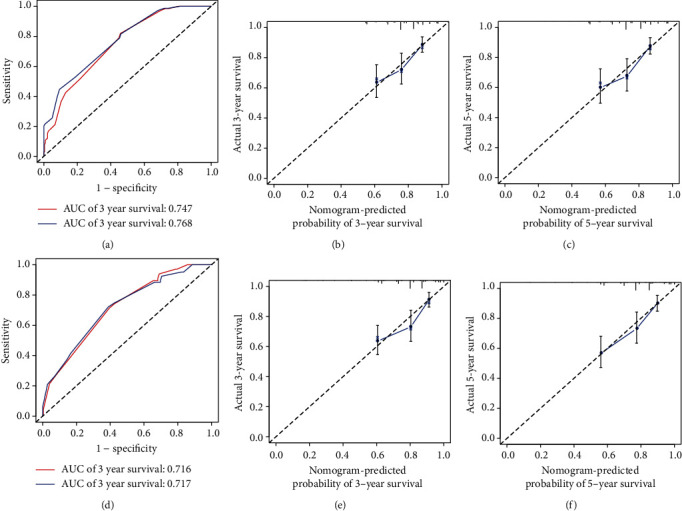
ROC curves and calibration plots of the nomogram in training and validation cohorts. (a) ROC curves for discrimination in the training set. (b) Calibration plot of observed and predicted probabilities for the nomogram in the 3-year training set. (c) Calibration plot of observed and predicted probabilities for the nomogram in the 5-year training set. (d) ROC curves for discrimination in the validation set. (e) Calibration plot of observed and predicted probabilities for the nomogram in the 3-year validation set. (f) Calibration plot of observed and predicted probabilities for the nomogram in the 5-year validation set.
3.5. Survival Curves for Prognostic Factors
Finally, we examined and developed curves of survival between the prognostic variables in the nomogram and the OS (Figure 5). We observed that age of diagnose, WBC at diagnosis, inv(16), and calculated risk scores wascorrelated with overall survival.
Figure 5.
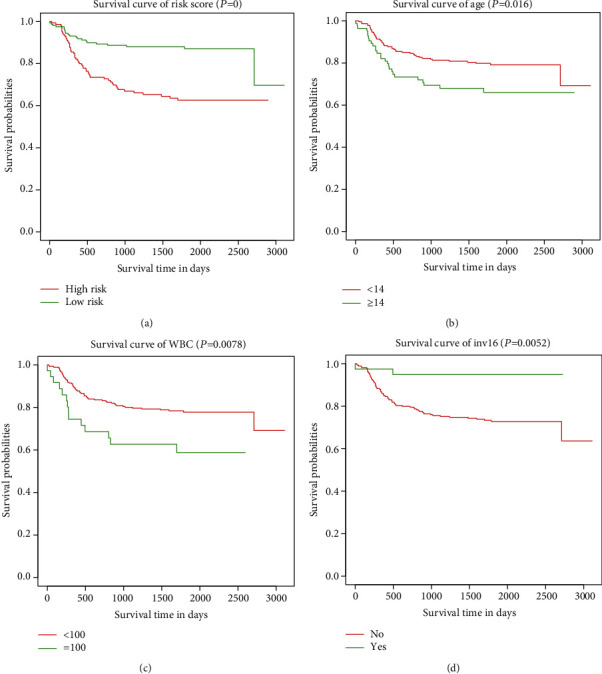
(a) Kaplan-Meier survival curves risk score. (b) Kaplan-Meier survival curves of age. (c) Kaplan-Meier survival curves of WBC. (d) Kaplan-Meier survival curves of inv16.
4. Discussion
As precision medicine develops quickly, physicians may create personalized diagnosis and follow-up plans for patients who need more accurate and easy models of survival [15, 16]. As a predictive tool, the nomogram can offer the most precise forecasts by means of a simple, easy to understand and easy to use in clinical procedure [17, 18]. The long-term survival of patients with various malignancies has been regularly estimated by demographic and clinical characteristics in a simple nomogram [19, 20].
AML is one of the most severe malignancies in childhood with different kinds of molecular and cellular heterogeneity [21, 22]. The standard cure of AML is the hematopoietic stem cell transplantation and chemotherapy, but the prognosis of childhood AML is suboptimal because of its elevated recurrence and mortality [23, 24]. Nomograms in recent research are typically more precise and convenient compared with conventional staging systems. In addition, web-based survival rate calculators were used to improve predictive model approachability. Recently, several reports have demonstrated nomograms for estimating AML adult patients' long-term survival outcomes [25, 26]. To our knowledge, few nomograms have been recorded to estimate the OS for children with AML. Hence, 440 patients were studied for the detection of OS-impact variables and a nomogram dependent calculator to effectively forecast prognostics for children with AML. Successful statistic evaluation was used to provide data for clinical consultation, pretreatment decision-making, and follow-up approaches.
The association between variables named multicollinearity has become a major issue in multivariate regression analysis [27]. We used the Lasso Cox regression approach rather than the conventional step-by-step process to tackle possible collinearity. The regression of Lasso Cox minimizes and reduces correlations thus offering a conclusive final model [28, 29].
The statistical model, which contained the aforementioned 13 individual risk factors, was extremely reliable in its survival prediction. C-index and validation plots have been used to test the predictive preciseness of the model and ensure the predictive precision of the nomogram. Both C-indices were >0.7 and demonstrated outstanding accuracy between predicted and actual survival. Nonetheless, given its high precision, the inconvenience of this model can limit its clinical use. We therefore have built a web-based survival risk calculator based on children AML's prediction nomograms. This tool accomplished a successful visualization, and the OS of children with AML was statistically predicted. Finally, an addition to the nomogram was created to differentiate patients at various mortality rates through prognostic risk stratification.
We identified 13 clinicopathological characteristics capable of predicting OS for AML children including gender, age of diagnose, WBC at diagnosis, BM blast percentage, and chromosomal abnormalities [t(8; 21), inv(16)]. Several studies have shown that age of diagnose and WBC at diagnosis were important predictive factors [30, 31], while the mechanism remains clear. Further studies are required to identify the mechanisms.
The TARGET database required detailed clinical information such as chemotherapy, which restricted our study of therapeutic modalities' impacts and prognostic complications. Furthermore, due to the retrospective aspect of the analysis, there was a data selection element. The online calculator set up in this thesis can be updated and serves as a foundational resource for further analysis.
5. Conclusion
Large data analysis is an important source of clinical prognostic indicators. Some of the latest analyzes of clinical data were focused on the SEER database. The TARGET database is part of the National Cancer Institute project and is barely published on its clinical data. The prognostic nomogram of childhood AML was developed based on the clinical evidence of the patients. Precise assessments of childhood AML will help physicians determine the current state of the individual, choose effective care choices, and establish better follow-up plans.
Contributor Information
Guoping Zhou, Email: gpzhou20@163.com.
Jimei Wang, Email: wjm821920@163.com.
Data Availability
The data analyzed were acquired from the Therapeutically Applicable Research To Generate Effective Treatments (TARGET) database (https://ocg.cancer.gov/programs/target).
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Authors' Contributions
Feng Jiang and Xiang Yu contributed equally to this work.
References
- 1.Lim S. H., Dubielecka P. M., Raghunathan V. M. Molecular targeting in acute myeloid leukemia. Journal of Translational Medicine. 2017;15(1):p. 183. doi: 10.1186/s12967-017-1281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pombo-de-Oliveira M. S., Andrade F. G., Brisson G. D., Dos S. B. F., Cezar I. S., Noronha E. P. Acute myeloid leukaemia at an early age: reviewing the interaction between pesticide exposure and KMT2A-rearrangement. Ecancermedicalscience. 2017;11:p. 782. doi: 10.3332/ecancer.2017.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prada-Arismendy J., Arroyave J. C., Rothlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Reviews. 2017;31(1):63–76. doi: 10.1016/j.blre.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Kogan A. A., Lapidus R. G., Baer M. R., Rassool F. V. Exploiting epigenetically mediated changes: acute myeloid leukemia, leukemia stem cells and the bone marrow microenvironment. Advances in Cancer Research. 2019;141:p. 213. doi: 10.1016/bs.acr.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Miller K. D., Nogueira L., Mariotto A. B., et al. Cancer treatment and survivorship statistics, 2019. CA: A Cancer Journal for Clinicians. 2019;69(5):363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 6.Jacobsohn D. A., Loken M. R., Fei M., et al. Outcomes of measurable residual disease in pediatric acute myeloid leukemia before and after hematopoietic stem cell transplant: validation of difference from normal flow cytometry with chimerism studies and Wilms tumor 1 gene expression. Biology of Blood and Marrow Transplantation. 2018;24(10):2040–2046. doi: 10.1016/j.bbmt.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu R., Lin W., Zhao W., Fan F., Tang L., Hu Y. A 4-micro RNA signature for survival prognosis in pediatric and adolescent acute myeloid leukemia. Journal of Cellular Biochemistry. 2019;120, article 3958 doi: 10.1002/jcb.27679. [DOI] [PubMed] [Google Scholar]
- 8.van der Linden M. H., Creemers S., Pieters R. Diagnosis and management of neonatal leukaemia. Seminars in Fetal & Neonatal Medicine. 2012;17(4):192–195. doi: 10.1016/j.siny.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Eppert K., Takenaka K., Lechman E. R., et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nature Medicine. 2011;17(9):1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 10.Cao J., Yuan P., Wang L., et al. Clinical nomogram for predicting survival of esophageal cancer patients after esophagectomy. Scientific Reports. 2016;6(1, article 26684) doi: 10.1038/srep26684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu J., Wu L., Jiang M., et al. Clinical nomogram for predicting survival outcomes in early mucinous breast cancer. PLoS One. 2016;11, article e164921 doi: 10.1371/journal.pone.0164921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Ju J., Liu X., et al. Nomograms for predicting long-term overall survival and cancer-specific survival in patients with major salivary gland cancer: a population-based study. Oncotarget. 2017;8(15):24469–24482. doi: 10.18632/oncotarget.14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinbach D., Bader P., Willasch A., et al. Prospective validation of a new method of monitoring minimal residual disease in childhood acute myelogenous leukemia. Clinical Cancer Research. 2015;21(6):1353–1359. doi: 10.1158/1078-0432.CCR-14-1999. [DOI] [PubMed] [Google Scholar]
- 14.Tibshirani R. The lasso method for variable selection in the cox model. Statistics in Medicine. 1997;16(4):385–395. doi: 10.1002/(SICI)1097-0258(19970228)16:4<385::AID-SIM380>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Kattan M. W., Leung D. H., Brennan M. F. Postoperative nomogram for 12-year sarcoma-specific death. Journal of Clinical Oncology. 2002;20(3):791–796. doi: 10.1200/JCO.2002.20.3.791. [DOI] [PubMed] [Google Scholar]
- 16.International Bladder Cancer Nomogram Consortium, Bochner B. H., Kattan M. W., Vora K. C. Postoperative nomogram predicting risk of recurrence after radical cystectomy for bladder cancer. Journal of Clinical Oncology. 2006;24, article 3967 doi: 10.1200/JCO.2005.05.3884. [DOI] [PubMed] [Google Scholar]
- 17.White R. R., Kattan M. W., Haney J. C., et al. Evaluation of preoperative therapy for pancreatic cancer using a prognostic nomogram. Annals of Surgical Oncology. 2006;13(11):1485–1492. doi: 10.1245/s10434-006-9104-y. [DOI] [PubMed] [Google Scholar]
- 18.Vernerey D., Huguet F., Vienot A., et al. Prognostic nomogram and score to predict overall survival in locally advanced untreated pancreatic cancer (PROLAP) British Journal of Cancer. 2016;115(3):281–289. doi: 10.1038/bjc.2016.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamada T., Nakai Y., Yasunaga H., et al. Prognostic nomogram for nonresectable pancreatic cancer treated with gemcitabine-based chemotherapy. British Journal of Cancer. 2014;110(8):1943–1949. doi: 10.1038/bjc.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abelson S., Collord G., Ng S., et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400–404. doi: 10.1038/s41586-018-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deschler B., de Witte T., Mertelsmann R., Lubbert M. Treatment decision-making for older patients with high-risk myelodysplastic syndrome or acute myeloid leukemia: problems and approaches. Haematologica. 2006;91, article 1513 [PubMed] [Google Scholar]
- 22.Leong S. R., Sukumaran S., Hristopoulos M., et al. An anti-CD3/anti-CLL-1 bispecific antibody for the treatment of acute myeloid leukemia. Blood. 2017;129(5):609–618. doi: 10.1182/blood-2016-08-735365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochtler T., Granzow M., Stolzel F., et al. Marker chromosomes can arise from chromothripsis and predict adverse prognosis in acute myeloid leukemia. Blood. 2017;129(10):1333–1342. doi: 10.1182/blood-2016-09-738161. [DOI] [PubMed] [Google Scholar]
- 24.Bhatt V. R., Shostrom V., Giri S., et al. Early mortality and overall survival of acute myeloid leukemia based on facility type. American Journal of Hematology. 2017;92(8):764–771. doi: 10.1002/ajh.24767. [DOI] [PubMed] [Google Scholar]
- 25.Dohner H., Estey E., Grimwade D., et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng S. W., Mitchell A., Kennedy J. A., et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 27.Rose P. G., Java J., Whitney C. W., et al. Nomograms predicting progression-free survival, overall survival, and pelvic recurrence in locally advanced cervical Cancer developed from an analysis of identifiable prognostic factors in patients from NRG oncology/gynecologic oncology group randomized trials of chemoradiotherapy. Journal of Clinical Oncology. 2015;33(19):2136–2142. doi: 10.1200/JCO.2014.57.7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon N., Friedman J., Hastie T., Tibshirani R. Regularization paths for Cox's proportional hazards model via coordinate descent. Journal of Statistical Software. 2011;39:p. 1. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alizadeh A. A., Gentles A. J., Alencar A. J., et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood. 2011;118(5):1350–1358. doi: 10.1182/blood-2011-03-345272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi J., Zhou L. Y., Yi Y. Y., et al. Low expression of pseudogene POU5F1B affects diagnosis and prognosis in acute myeloid Leukemia (AML) Medical Science Monitor. 2019;25:4952–4959. doi: 10.12659/MSM.914352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Lima M. C., Da S. D., Freund A. P., et al. Leucemia Mieloide Aguda: analise do perfil epidemiologico e taxa de sobrevida. Jornal de Pediatria. 2016;92(3):283–289. doi: 10.1016/j.jped.2015.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data analyzed were acquired from the Therapeutically Applicable Research To Generate Effective Treatments (TARGET) database (https://ocg.cancer.gov/programs/target).


