Abstract
CD46 is well known to be involved in diverse biological processes. Although several splice variants of CD46 have been identified, little is known about the contribution of alternative splicing to its tumorigenic functions. In this study, we found that exclusion of CD46 exon 13 is significantly increased in bladder cancer (BCa) samples. In BCa cell lines, enforced expression of CD46-CYT2 (exon 13-skipping isoform) promoted, and CD46-CYT1 (exon 13-containing isoform) attenuated, cell growth, migration, and tumorigenicity in a xenograft model. We also applied interaction proteomics to identify exhaustively the complexes containing the CYT1 or CYT2 domain in EJ-1 cells. 320 proteins were identified that interact with the CYT1 and/or CYT2 domain, and most of them are new interactors. Using an internal ribosome entry site (IRES)-dependent reporter system, we established that CD46 could regulate mRNA translation through an interaction with the translation machinery. We also identified heterogeneous nuclear ribonucleoprotein (hnRNP)A1 as a novel CYT2 binding partner, and this interaction facilitates the interaction of hnRNPA1 with IRES RNA to promote IRES-dependent translation of HIF1a and c-Myc. Strikingly, the splicing factor SRSF1 is highly correlated with CD46 exon 13 exclusion in clinical BCa samples. Taken together, our findings contribute to understanding the role of CD46 in BCa development.
Keywords: bladder cancer, CD46, hnRNPA1, translation, tumorigenesis
Graphical Abstract
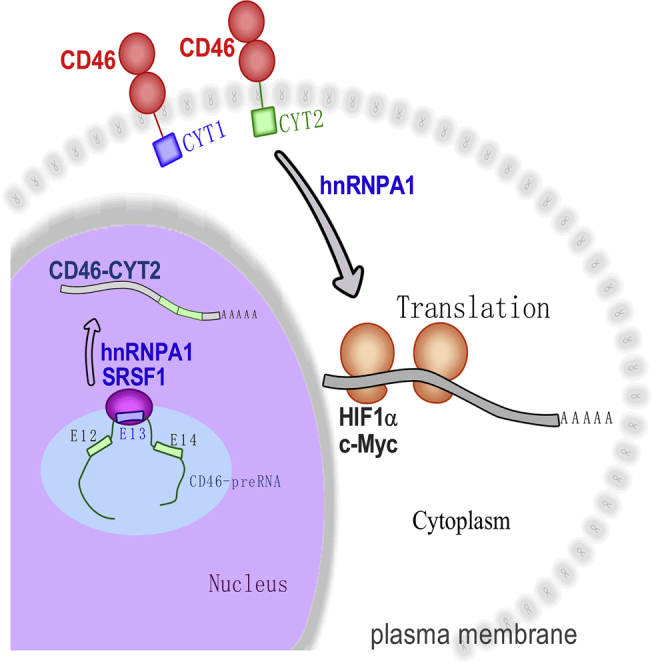
Aberrant alternative splicing is one of the molecular hallmarks of cancer. In this study, we investigated the contribution of CD46 alternative splicing to bladder cancer development and defined a new role for CD46 in regulation of translation. Next, we showed that SRSF1, by regulating the splicing of CD46, reinforces an aggressive phenotype.
Introduction
Alternative splicing (AS) is a pivotal mechanism for generating diversity at the RNA and protein levels in eukaryotes, and aberrant splicing events are frequently observed in many types of cancers.1 Many genes that regulate critical biological processes, such as cell division, apoptosis, and differentiation, are subjected to AS, and changes in the isoform ratios of these genes provide an important contribution to tumor development and progression.2 In addition, AS changes also play a key role in resistance to cancer therapy (reviewed in Siegfried and Karni3). Since the high-throughput technique came into being, its application in the identification of aberrant AS events of cancers has made great advancements.1,4 However, the function of most AS events is unknown.
CD46, also named membrane cofactor protein (MCP), has been reported to be involved in diverse biological processes. The best documented function of CD46 is to inhibit complement activation.5 Increasing evidence indicates that CD46 has many other functions. For example, CD46 plays a central role in cellular entry by human pathogens, fertilization processes, innate and acquired immunity, and T helper (Th)1 biology.5, 6, 7 This ubiquitously expressed protein is a human type I transmembrane glycoprotein, composed of four N-terminal conserved complement control protein (CCP) domains and a heavily O-glycosylated STP domain followed by a transmembrane hydrophobic anchor and a short cytoplasmic tail.5 CD46 has 14 exons, and multiple exons (exons 7, 8, 9, and 13) are subject to AS regulation, which generates different isoforms dependent on inclusion/exclusion of these regulated exons.8,9 AS at exons 7, 8, and 9 gives rise to the generation of at least four STP isoforms.8 Exon 13 inclusion or exclusion contributes to the generation of two distinct intracellular domains of the CD46 cytoplasmic tail, CYT1 and CYT2, which mediate differentially important signaling events.7,10, 11, 12, 13, 14 Marie et al.7 found that CD46-CYT1 and CD46-CYT2 have opposite effects on T cell immune responses. Similarly, CD46-CYT1 is required for interferon (IFN)-γ production and Th1 cell polarization in CD4+ T cells.6,15 In addition, Hirano et al.13 found that CD46-CYT1 and CD46-CYT2 have the opposite effect on nitric oxide (NO) production. Furthermore, Kolev et al.12 demonstrated that CD46-CYT1, but not CYT2, is crucial for amino acid and hexose uptake, as well as LAMTOR5 expression, to potentiate mTORC1 signaling. Taken together, these data suggest that the two CD46 cytoplasmic domains contain signaling motifs and have distinct cellular functions.
The dysregulation of CD46 pathways has been described in many human diseases, including asthma,16 rheumatoid arthritis,17 multiple sclerosis (MS),18 lupus,19 and atypical hemolytic uremic syndrome (aHUS).20,21 The molecular basis of CD46-mediated activities can at least partially be explained by its interaction with various proteins. For example, some pathogens that bind to the CD46 ectodomain can induce autophagy dependent on CD46-CYT1/GOPC interaction.22 In addition, CD46 associates with other molecules, including lipid rafts,23 DLG4,24 Lck,25 SHP-1,26 SPAK,17 α-E-catenin, and E-cadherin,27 which regulate T cell receptor (TCR) stimulation, cell polarity, morphological changes, NO production, T cell differentiation, and epithelial cell barrier integrity and repair, respectively. Notably, CD46 is subjected to further enzymatic processing by matrix metallopeptidase (MMP) and γ-secretase,15,28 which generates cytoplasmic tails with signaling abilities and is pivotal to ensure T cell function.29 Additionally, the cleaved CYT1 and CYT2 can translocate to the nucleus and regulate downstream gene expression.12 Recently, CD46 glycosylation was shown to be required for the enzymatic processing of CD46, and its defect may contribute to MS.30 Although great progress has been made, the functions of CD46 and relevant molecular mechanisms remain largely unknown.
CD46 also plays a key role in multiple cancers. Numerous studies have demonstrated that CD46 increases expression in tumors relative to the corresponding normal tissue, which may serve as a mechanism for cancer cells to block killing by complement.31, 32, 33, 34, 35, 36 Notably, the CD46 locus is frequently co-amplified with 1q21 in most relapsed myeloma patients, and it is highly expressed in myeloma cell lines.37 Sherbenou et al.37 have shown that antibody-drug conjugate targeting CD46 (CD46-ADC) inhibited proliferation in myeloma cell lines with little effect on normal cells, suggesting that CD46 is a promising target for the treatment of multiple myeloma. However, the molecular mechanisms responsible for the pro-tumorigenesis functions of the CD46 pathway remain ill-defined.
In this study, we observed that the exclusion of CD46 exon 13 is significantly increased in bladder cancer (BCa) samples. CD46-CYT1 inhibits, and CD46-CYT2 promotes, bladder tumor tumorigenesis. Of interest, we demonstrated that the splicing factor SRSF1 promotes the exclusion of exon 13 of the CD46 gene, and we assessed the role of SRSF1 in BCa progression. Importantly, SRSF1 is upregulated in BCa and highly correlated with CD46 exon 13 exclusion (p = 0.0276, r = 0.4238). We also systematically screened CYT1 and CYT2 domain protein interactors and identified heterogeneous nuclear ribonucleoprotein (hnRNP)A1 as a novel CYT2 binding partner, and this interaction facilitates the interaction of hnRNPA1 with internal ribosome entry site (IRES) RNA to promote IRES-dependent translation. These findings help understanding the roles and molecular mechanisms of CD46 in tumor development and progression.
Results
Dysregulated CD46 exon 13 splicing in clinical BCa samples
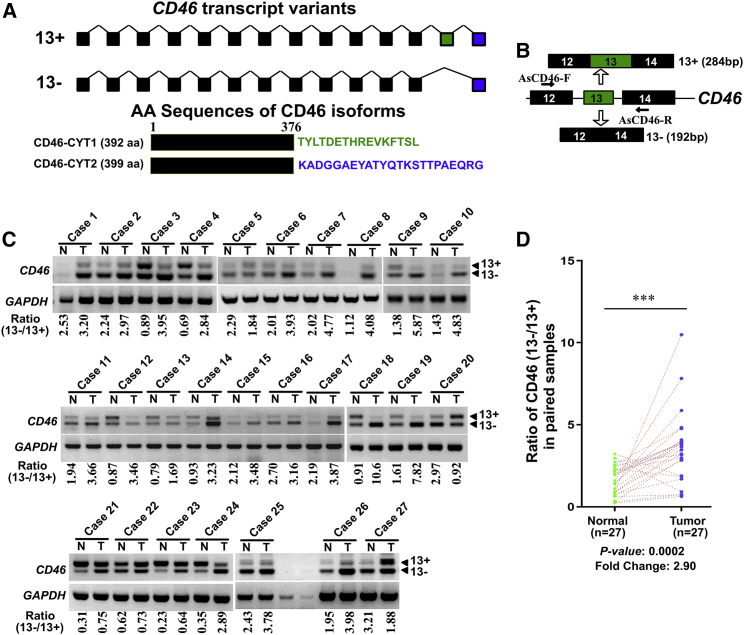
To identify aberrant AS events in BCa, we performed a systematic analysis of RNA sequencing (RNA-seq) profiles from a human BCa gene expression dataset (GEO: GSE31617). SpliceSeq software analysis38 and then manual proofreading identified 33 AS events that were deregulated in BCa as compared with normal bladder tissues (Table S1). We focused our attention on the differential splicing of CD46 because the function of CD46 in cancer remains largely unknown, and there was a huge difference (a fold change >5) in the CD46 exon 13 inclusion between human bladder tumors and matched normal tissues. Translation of the exon 13-included mRNA generates a CYT1 intracellular domain-containing protein (CD46-CYT1), while the exon 13-skipping transcript generates a CYT2 intracellular domain-containing protein (CD46-CYT2) (Figure 1A). To confirm that CD46 exon 13 splicing is dysregulated in BCa, we designed a primer pair that could differentiate alternative exon 13 isoforms (Figure 1B). RT-PCR analysis revealed that the CYT2-to-CYT1 ratio of CD46 was significantly higher in BCa tissue as compared to adjacent normal tissue (fold change = 2.9, p = 0.0002; Figures 1C and 1D). These data suggested that the dysregulation of CD46 exon 13 AS may play an important role in BCa development.
Figure 1.
CD46 exon 13 skipping is upregulated in bladder cancers
(A) Schematic representation of the CD46 exons structure to highlight the alternative splicing between exons 12 and 14 generating CD46-CYT1 and CD46-CYT2 variants. (B) Diagrams for detection of CD46-CYT1 and CD46-CYT2 mRNA. Primer pairs and product sizes for the two variants are shown. (C) Expression of CD46-CYT1 and CD46-CYT2 mRNA in 27 paired bladder cancer tissues (T) and adjacent nontumor tissues (N) by RT-PCR. GAPDH transcript level was used as the loading control. The ratio for 13−/13+ is listed below the panel. (D) Quantification of data from (C) for the exon 13 exclusion inclusion ratio. The data were analyzed by a paired Student’s t test. ∗∗∗p < 0.001.
CD46-CYT2 promotes the tumorigenic features of BCa cells
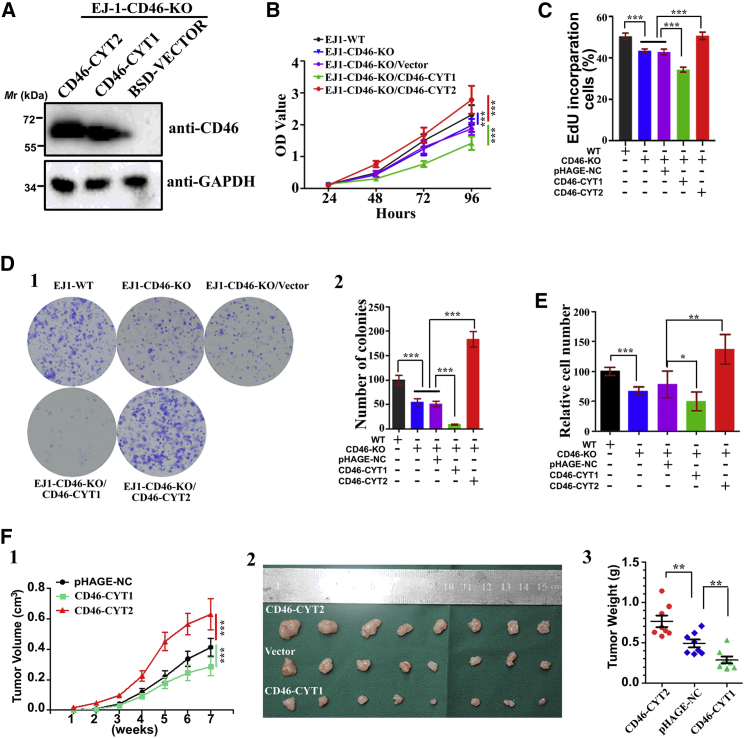
We next assessed the roles of these two isoforms in bladder tumorigenesis. To this end, we applied the efficient CRISPR-Cas genome-editing system targeting exon 1 and intron 1 boundaries of CD46 to knock out this gene in EJ-1 cells (Figures S1A–S1C). We found that CRISPR-Cas9-mediated knockout (KO) of CD46 attenuated cell proliferation, DNA synthesis, and colony formation of EJ-1 cells. In EJ-1 cells with CD46 KO, re-expression of CD46-CYT2 without exon 13 remarkably accelerated cancer cell proliferation, DNA synthesis, colony formation, and cell migration, while re-expression of CD46-CYT1 with exon 13 had the opposite effect of decreasing all of these activities (Figures 2A–2E; Figures S2A and S2B). A similar phenomenon was found when CD46-CYT1 or CD46-CYT2 was stably overexpressed in 5637 BCa cells (Figures S3A–S3D). Furthermore, in vivo tumorigenic analyses revealed that the restoration of CD46-CYT2 in EJ-1-CD46-KO cells resulted in an accelerated tumor growth and a more serious tumor burden, but CD46-CYT1 restoration suppressed tumor growth rate and tumor size in a subcutaneous xenograft model (Figure 2F). Taken together, these results indicate that AS variants of CD46 exon 13 can play distinct roles in the regulation of BCa cell growth and migration, with CD46-CYT2 being a tumorigenic factor in vitro and in vivo.
Figure 2.
CD46-CYT1 and CD46-CYT2 have opposite roles in bladder cancer development
(A) Generation of CD46-knockout (CD46-KO) cells that were engineered to re-express CD46-CYT1 or CD46-CYT2. CD46-KO cells were infected with lentiviruses expressing vector control, CD46-CYT1, or CD46-CYT2. Immunoblotting was performed to evaluate the expression of CD46. GAPDH is an internal control. (B) A CCK-8 kit was utilized to quantify cell viability at each time point. The data represent mean ± SD and were analyzed by a two-way ANOVA (n = 3). ∗∗∗p < 0.001. (C) Quantification of 5-ethynyl-2′-deoxyuridine (EdU)-incorporated cells in indicated engineered cell lines. The data represent mean ± SD and were analyzed by an unpaired two-tailed Student’s t test (n = 4). ∗∗∗p < 0.001. (D) A colony formation assay and quantification were performed with EJ-1 cells and CD46-KO cells expressing CD46-CYT1 or CD46-CYT2 as described in (B). The data represent mean ± SD and were analyzed by an unpaired two-tailed Student’s t test (n = 3). ∗∗∗p < 0.001. (E) Transwell cell migration assay for EJ-1 cells. Numbers of migrated cells were quantified in four random images from each treatment group. The data represent mean ± SD and were analyzed by an unpaired two-tailed Student’s t test (n = 4). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. (F) (1) Time course of xenograft growth. Mean tumor volume was measured by caliper on the indicated weeks. The data represent mean ± SD and were analyzed by a two-way ANOVA (n = 8). ∗∗∗p < 0.001. (2) Photographs of tumors excised 7 weeks after the inoculation of stably transfected EJ-1 cells into nude mice. (3) The tumor weight of CD46-CYT1- or CD46-CYT2-overexpressed EJ-1 cells in nude mice at the end of 7 weeks after transplantation. The data represent mean ± SD and were analyzed by an unpaired two-tailed Student’s t test (n = 8). ∗∗p < 0.01.
Identification of CD46 cytoplasmic tail interaction proteins by tandem affinity purification (TAP) and mass spectrometry (MS) analysis
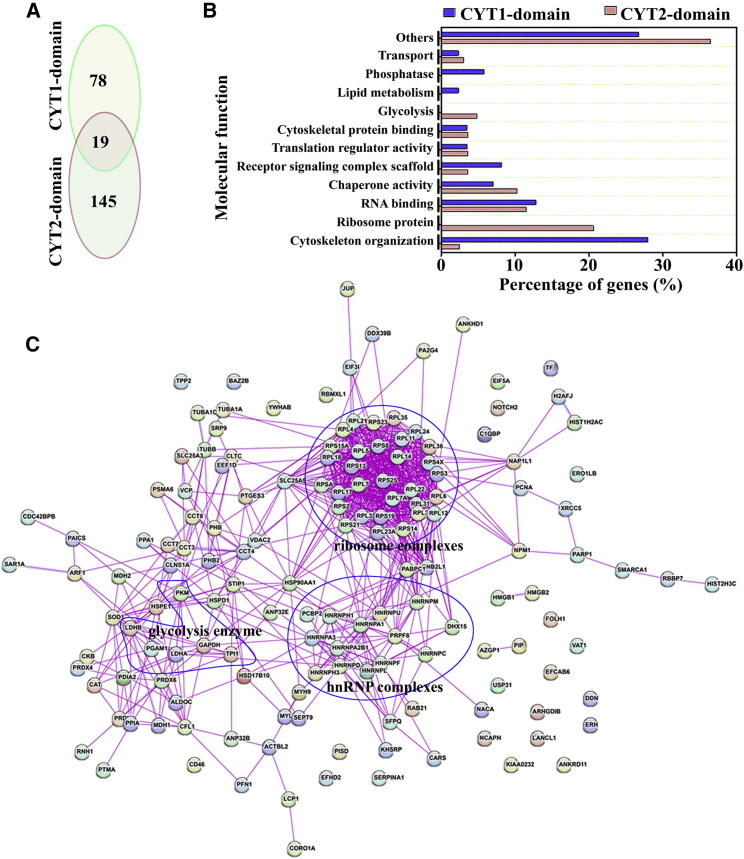
Because CD46-CYT1 and CD46-CYT2 isoforms differ only in their cytoplasmic tails, we reasoned that different tails must have different binding partners, which may contribute to their opposite roles in bladder tumor development. We next sought to screen for proteins interacting with the C-terminal cytosolic tail of CD46. To facilitate the purification of CYT1 or CYT2 interaction proteins, we established a stable cell line expressing Twin-Strep and the glutathione S-transferase (GST)-tagged CYT1 and CYT2 domains of CD46 (Figure S4A). Interaction proteins were recovered using a two-step purification that involved sequential binding to Strep-Tactin and glutathione agarose (Figure S4B). After elution from beads, all proteins identified by MS analysis corresponded to three different type interaction proteins, that is, CYT1-specific binding (n = 78), CYT2-specific binding (n = 145), and both CYT1/CYT2 binding proteins (n = 19) (Figure 3A). A compilation of the CYT1 domain- and CYT2 domain-associated proteomes is given in Table S2. We next proceeded to validate a subset of these potential CYT1/2 interaction partners by co-immunoprecipitation (coIP) experiments. To this end, five previously unknown interacting proteins were selected. CoIP assays of SNX27, PTPN3, HMGB1, EIF5A, and RPL17 with CD46-CYT1 and CD46-CYT2 in CD46-KO EJ-1 cells showed that CD46-CYT1 coprecipitated efficiently with SNX27 and PTPN3, while CD46-CYT2 coprecipitated efficiently with HMGB1 and RPL17, which is to a large extent consistent with the TAP-MS identification (Figures S5A and S5B). The interaction of these proteins with the CYT1 or CYT2 domain of CD46 was determined by reciprocal coprecipitation experiments (Figures S5C–S5G). Interestingly, consistent with previous reports that the PDZ domain can specifically bind to the CYT1 domain,22,24 we also identified four PDZ domain-containing proteins (SNX27, PTPN3, GOPC, and SLC9A3R2) as potential CYT1 domain-interaction partners. Among the four proteins, SNX27 and PTPN3 are unknown binding proteins (which is verified in Figure S5), while GOPC is a known CYT1 interaction partner.22 Taken together, these results confirm that our approach provided credible global partners of the CYT1 and CYT2 domains.
Figure 3.
Classification of identified CYT1 and CYT2 domain-interacting proteins
(A) The number of CYT1- and CYT2-binding partners in the EJ-1 cells. (B) Function classification of the 320 proteins identified as CYT1 and/or CYT2 domain partners in the EJ-1 cell line. (C) Network analysis of associated proteins identified through liquid chromatography-tandem mass spectrometry (LC-MS/MS) of CYT2 domain-interacting proteins. Three main complexes (ribosome complexes, the glycolysis enzyme complex, and hnRNP complexes) are boxed.
The identified CYT1 and CYT2 domain partners were grouped according to their molecular function based on literature investigations and functional enrichment analysis using FunRich software.39 Many of them belonged to distinct protein families and were known to participate in many cellular processes, including glycolysis, translation regulation, cytoskeleton organization, and nucleic acid metabolism (Figure 3B; Tables S3 and S4). Interestingly, we found that many proteins involved in glycolysis (n = 8) and ribosome (n = 34) complexes are specifically coprecipitated with the CYT2 domain, and proteins involved in lipid metabolism (n = 2) and protein phosphatase activity (n = 5) are specifically coprecipitated with the CYT1 domain (Figure 5B; Tables S3 and S4). A functional network analysis was also performed to test for possible complex formation between several CYT1 or CYT2 domain partners. Among these interactions, some well-known complexes such as ribosome complexes, the glycolysis enzyme complex, and hnRNP complexes were detected in CYT2 domain partners, but not in CYT1 domain partners (Figure 3C).
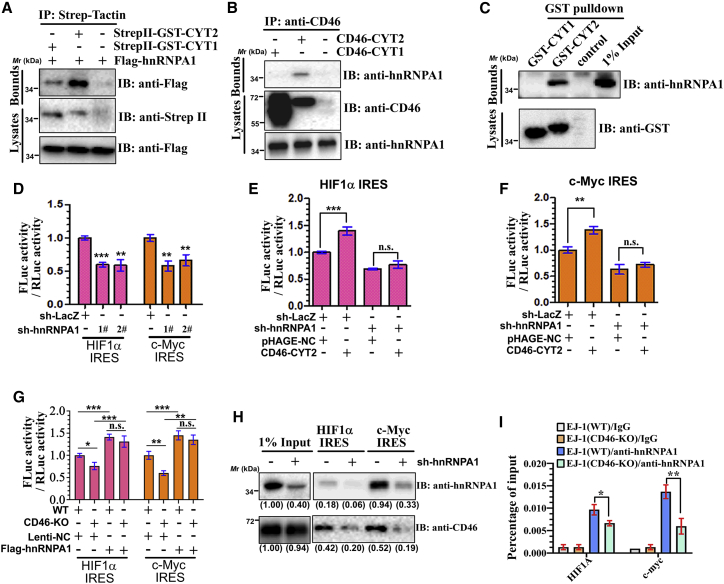
Figure 5.
CD46-CYT2 regulates IRES-dependent translation via hnRNPA1
(A) EJ-1 cells were transiently transfected with StrepII-GST-CYT1/2 and/or FLAG-hnRNPA1. The cell lysates were precipitated with StrepII-Tactin and immunoblotted with an anti-FLAG antibody. (B) Co-immunoprecipitation experiments were conducted with an CD46 antibody in CD46-KO cells stably expressing CD46-CYT1 or CD46-CYT2, respectively. (C) Lysates from 293T cells transfected with FLAG-hnRNPA1 were incubated with either purified protein GST-CYT1 or GST-CYT2. Bound FLAG-hnRNPA1 proteins were immunoblotted by anti-FLAG. (D) EJ-1 cells expressing sh-LacZ or sh-hnRNPA1 were transfected with indicated IRES-dependent reporters, respectively. The firefly and Renilla luciferase activities were measured. (E and F) EJ-1 cells expressing sh-LacZ or sh-hnRNPA1 were transfected with CD46-CYT2 or pHAGE-negative control (NC) (control/empty vector) and the indicated IRES-dependent reporter plasmids. The firefly and Renilla luciferase activities were measured. (G) Relative luciferase activity of wild-type (WT) or CD46-KO EJ-1 cells transfected with Lenti-NC (control) or FLAG-hnRNPA1 with the indicated tethering reporter. (H) CD46-KO cells stably expressing CD46-CYT2, MS2-GST together with sh-LacZ, or sh-hnRNPA1 were transfected with the 3′ MS2 stem-loop-tagged IRES sequence of HIF1a or c-Myc. After 48 h of culture, cells were lysed and incubated overnight with glutathione Sepharose beads. Precipitates were subjected to western blotting with anti-hnRNPA1 or anti-CD46 antibodies. Levels of hnRNPA1 and CD46 are normalized against input and expressed as fold change relative to base expression determined using control sh-LacZ. (I) Cell lysates from WT or CD46-KO cells were incubated with IgG or anti-hnRNPA1 antibody and immunoprecipitated with protein A/G-conjugated beads. Bound RNAs were then eluted, purified, and subjected to qPCR for CCND1 and c-Myc mRNAs.
CD46 cytoplasmic tails regulate translation
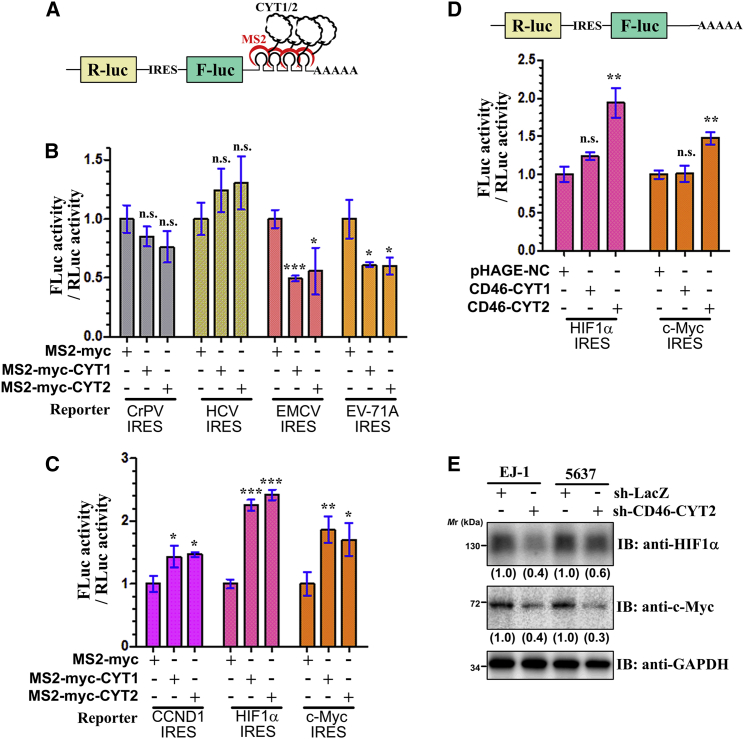
As described above, many CYT1/2 domain partners were components of ribosome complexes, the translation initiation complex, and hnRNP complexes, such as RPL17, eEF1D, EIF3I, EIF3S9, EIF4B, EIF5A, PABPC1, PCBP2, PDCD4, and hnRNPA1 (Table S2), which play essential roles in regulation of protein synthesis and translational initiation. Considering that translation initiation is typically the rate-limiting step of protein synthesis,40 we tested whether CD46 might play a role in translational initiation. To this end, we performed tethering experiments using IRES-dependent bicistronic reporter plasmids (Figure 4A). As shown in Figure 4A, the bicistronic reporter plasmid containing an internal IRES between Renilla luciferase (Rluc) and firefly luciferase (Fluc) and four MS2-binding sites (MS2bs) is located just downstream of the stop codon of the Fluc gene. We then tested this possibility using four virus IRESs from encephalomyocarditis virus (EMCV), enterovirus A71 (EV-A71), cricket paralysis virus (CrPV), and hepatitis C virus (HCV). We found that direct tethering of both CYT1 and CYT2 to the 3′ UTR of the above reporters specifically decreased the translation of EMCV and EV-A71 IRES-dependent translation of Fluc compared to Rluc translation. In contrast, neither CYT1 nor CYT2 tethering showed a statistically significant effect on CrPV and HCV IRES-dependent Fluc translation (Figure 4B; Figure S6). EMCV IRES and EV-A71 IRES require all translation initiation factors except for eIF4E, whereas CrPV IRES- and HCR IRES-dependent translation does not need any translation initiation factors, which indicated that CD46’s cytosolic tails might play an important role in regulating translation initiation.
Figure 4.
The CYT1 and CYT2 domain of CD46 regulates protein translation
(A) Schematics of the tethering reporter assay. (B) The tethering of CYT1 or CYT2 to the EMCV and EV-71A IRES reporters led to a decrease in translation in comparison with the control. The relative luciferase activity is shown of EJ-1 cells transfected with MS2-myc (control), MS2-myc-CYT1, or MS2-myc-CYT2 with the indicated tethering reporter plasmids. (C) The tethering of CYT1 or CYT2 to CCND1, HIF1a, or c-Myc IRES led to an increase in translation. The relative luciferase activity is shown of EJ-1 cells transfected with MS2-myc (control), MS2-myc-CYT1, or MS2-myc-CYT2 with the indicated tethering reporter. (D) CD46-CYT2 promotes the IRES-dependent translation of HIF1a and c-Myc. Top: schematic representation of bicistronic reporter constructs with different IRESs. Bottom: EJ-1 cells were transfected with the indicated plasmids. The firefly and Renilla luciferase activities were measured, and the ratios of firefly luciferase activity over Renilla luciferase activity were calculated. (E) EJ-1 and 5637 cells transfected with sh-CD46-CYT2 or sh-LacZ were cultured in Met-free DMEM for 1 h in the presence of AHA to capture newly synthesized proteins and immunoblotted using the indicated antibodies. Levels of HIF1a and c-Myc are normalized against GAPDH and expressed as fold change relative to base expression determined using control shRNA. All data represent mean ± SD and were analyzed by an unpaired two-tailed Student’s t test (n = 3). ∗p < 0.05 versus control; ∗∗p < 0.01, ∗∗∗p < 0.001. n.s., not significant.
We next investigated whether endogenous human IRES can be regulated by tethering of CD46’s cytosolic tails to downstream of the stop codon of the Fluc gene. CCND1, HIF1a, and c-Myc are known to be oncogenic drivers for many human cancers, and all of them harbor an IRES structure in the 5′ UTR of their mRNA. We thus investigated the effects of CYT1/CYT2 tethering on CCND1, HIF1a, and c-Myc IRES-dependent translation. Unexpectedly, both CYT1 and CYT2 tethering enhanced these three IRES-dependent Fluc translations (Figure 4C). Taken together, the results indicated that tethering the CD46’s cytosolic tails to the 3′ UTR have both translational repression and enhancement functions depending on the specific IRES.
The data described above led us to further explore the impact of CD46-CYT1/CYT2 overexpression on the IRES-dependent translation of HIF1a and c-Myc. Overexpression of CD46-CYT2 significantly increased the translation activity of HIF1a and c-Myc IRESs, while CD46-CYT1 overexpression had little effect on these activities (Figure 4D), suggesting that CD46-CYT2 might be a translational regulator of HIF1a and c-Myc. To further confirm these results, the levels of newly synthesized HIF1a and c-Myc protein in CD46-CYT2 knockdown and control cells were monitored by an l-azidohomoalanine (AHA) incorporation assay. As expected, AHA-labeled newly synthesized HIF1a and c-Myc proteins were markedly reduced in CD46-CYT2 knockdown cells compared with control cells (Figure 4E). We next wanted to determine whether CD46 was regulating the global translation. Using a surface sensing of translation (SUnSET) method to monitor protein synthesis, we demonstrated that neither CD46-CYT1 nor CD46-CYT2 has any effect on the translation in 293T and EJ-1 cells (Figures S7A and S7B). Taken together, our findings are highly supportive that CD46-CYT2 has an important role in regulating the translation of a subset of oncogenes, such as HIF1a and c-Myc.
CD46-CYT2 regulates IRES-dependent translation via hnRNPA1
Many IRES trans-acting factors (ITAFs) such as hnRNPA1, PABPC1, and PCBP2 were specifically immunoprecipitated by CYT2, but not by CYT1 (Table S2). Moreover, the hnRNPA1 protein is involved in promoting the IRES-dependent translation of a number of genes, including HIF1a and MYC.41 We thus decided to investigate the relationship between CD46-CYT2, hnRNPA1, and IRES-dependent translation in more detail. Reciprocal coIP assays confirmed the interaction of CD46-CYT2 with hnRNPA1 in 293T and EJ-1 cells (Figures 5A and 5B). Additionally, the bacterially expressed GST-CYT2 domain was associated with FLAG-hnRNPA1 (Figure 5C), indicating that CD46-CYT2 directly interacts with hnRNPA1.
To determine whether hnRNPA1 is required for CD46-CYT2’s translational regulatory function, we first confirmed the role of hnRNPA1 in the IRES-dependent translation. Control (short hairpin RNA [shRNA] LacZ [sh-LacZ]) or hnRNPA1 knockdown EJ-1 cells were transfected with HIF1a and c-Myc IRES reporters, and the IRES-dependent translation level of each gene was tested. Consistent with previously reported findings, knockdown of hnRNPA1 significantly suppressed the activities of HIF1a and c-Myc IRESs (Figure 5D; Figure S8).41 We next investigated the effects of CD46-CYT2 on IRES-dependent translation in hnRNPA1 depleted cells. The effect of CD46-CYT2 overexpression on IRES-dependent translation was partially reversed by concomitant hnRNPA1 depletion (Figures 5E and 5F). Furthermore, CD46-CYT2 knockdown decreased the activities of HIF1a and c-Myc IRESs, and the effect of CD46-CYT2 silencing on these activities was reversed by concomitant hnRNAPA1 overexpression (Figure 5G). In addition, the RNA pull-down assay was performed in hnRNPA1 depleted cells. As shown in Figure 5H, both the c-Myc IRES and HIF1a IRES precipitated considerably less CD46-CYT2 in hnRNPA1-knockdown EJ-1 cells compared with the sh-LacZ cells, indicating that the IRES-CYT2 interaction was hnRNPA1-dependent. Taken together, the results suggest that the effect of CD46-CYT2 on IRES-dependent translation is mediated, at least in part, through hnRNPA1.
Based on the above findings, our next aim was to clarify the mechanism of CYT2 binding on the regulation of hnRNPA1 activity. The binding affinity of hnRNPA1-IRES is thought to play a critical role in hnRNPA1’s regulation of IRES-dependent translation.41 Thus, we next performed RNA immunoprecipitation (RIP) analysis in CD46 wild-type or KO EJ-1 cells, using hnRNPA1 antibodies. Figure 5I demonstrates that hnRNPA1 precipitated considerably less HIF1a and c-Myc mRNAs in CD46 knocked out cells, indicating that CD46 enhances the IRES-hnRNPA1 interaction.
CD46-CYT2 promotes tumorigenesis via hnRNPA1
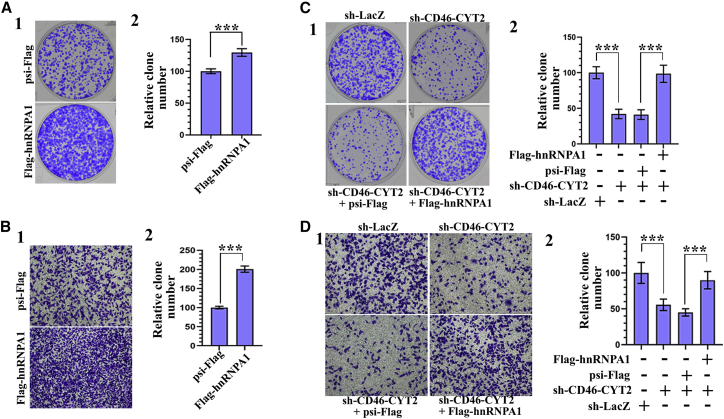
Given that hnRNPA1 has been proposed to function as an oncogene in various cancers,42 we wanted to determine the effect of hnRNPA1 on the tumorigenicity of BCa cells. In support of a pro-tumorigenic role, overexpression of hnRNPA1 increased colony formation and cell migration capability (Figures 6A and 6B; Figure S9A). Next, we sought to determine whether hnRNPA1 is required for CD46-CYT2 to regulate the proliferation, migration, and invasion abilities of BCa cells. To this end, we tested whether exogenous hnRNPA1 expression could compensate for CD46-CYT2 knockdown. We found that the effect of CD46-CYT2 silencing on colony formation, migration, and invasion was reversed by concomitant hnRNPA1 overexpression (Figures 6C and 6D; Figure S9B). Overall, these findings show that hnRNPA1 function is regulated by CYT2 binding, and they provide the rationale to explore the role of hnRNPA1 in mediating the tumorigenicity of CD46-CYT2.
Figure 6.
CD46-CYT2 regulates human bladder cancer cell migration via hnRNPA1
(A) (1) Representative images of cell culture plates following staining for colony formation of EJ-1 cells expressing FLAG-hnRNPA1 or control plasmid psi-FLAG. (2) Number of colonies was quantified. (B) Migration assay for bladder cancer cells. The number of migrated cells was quantified in five random images from each treatment group. The data represent mean ± SD and were analyzed by an unpaired two-tailed Student’s t test (n = 5). ∗∗∗p < 0.001 versus control. (C and D) EJ-1 cells were infected with different combinations of lentivirus encoding sh-LacZ, sh-CD46-CYT2, psi-FLAG, and/or FLAG-hnRNPA1 as indicated. After 72 h of culture, a colony formation assay and transwell migration assay were performed. ∗∗∗p < 0.001 versus control.
SRSF1 promotes BCa tumorigenesis in part via regulating CD46-CYT2 levels
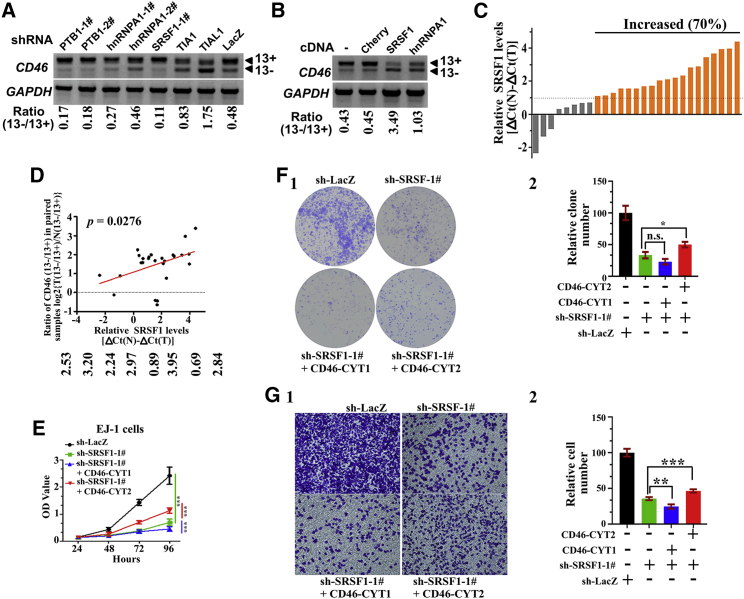
Previous studies have indicated that CD46 exon 13 splicing is regulated by multiple trans-acting splicing factors such as hnRNPA1, SRSF1, PTBP1, TIA1, and TIAL1 in HEK293T cells.8 We thus examined whether CD46 exon 13 inclusion is regulated by these splicing factors in BCa cells. To this end, we established knockdown EJ-1 cell lines stably expressing shRNA constructs against these genes. The result showed that knockdown of TIA1 or TIAL1 induced exon 13 skipping while PTBP1 and SRSF1 knockdown had the opposite effect (Figure 7A; Figures S10A–S10C). As a previous study showed that hnRNPA1 interacts with the exonic splicing enhancer (ESE) of CD46 exon 13,8 we speculated that other splicing factors (such as hnRNPA2B1) could compensate for the hnRNPA1 depletion. We thus investigated a role of hnRNPA1 overexpression in exon 13 splicing. As expected, both SRSF1 and hnRNPA1 overexpression promoted exon 13 skipping (Figure 7B). Taken together, these results suggested that CD46 exon 13 splicing is also regulated by hnRNPA1, SRSF1, PTBP1, TIA1, and TIAL1 in EJ-1 cells.
Figure 7.
SRSF1 promotes bladder cancer tumorigenesis in part via regulating CD46-CYT2 levels
(A) EJ-1 cells were infected with lentivirus expressing several indicated shRNAs to establish stably expressing cell lines. Semiquantitative RT-PCR analysis was performed to detect the CD46 exon 13 alternative splicing. The ratio for 13−/13+ is listed below the panel. (B) CD46 exon 13 splicing was measured by RT-PCR in EJ-1 cells stably expressing FLAG-Cherry, FLAG-SRSF1, or FLAG-hnRNPA1. (C) Relative expression of SRSF1 mRNA expression levels were evaluated by real-time PCR in 27 paired case specimens. Expression levels of SRSF1 were normalized to that of GAPDH. A bar value <1 indicates that SRSF1 is decreased in tumors. A bar value >1 indicates that SRSF10 is increased in tumors. (D) The positive correlation between the CD46 13−/13+ ratio and expression levels of SRSF1 was observed in bladder cancer samples. Relationships between these two variables were determined by Pearson’s correlation coefficients. The correlation was analyzed using GraphPad Prism 5 software. (E) A CCK-8 assay was utilized to quantify cell viability at each time point. Data are plotted as the mean ± SD of three independent experiments and were analyzed by two-way ANOVA. ∗∗∗p < 0.001. (F) (1) Representative images of cell culture plates following staining for colony formation of the indicated cell lines. (2) The number of colonies was quantified. The data represent mean ± SD and were analyzed by an unpaired two-tailed Student’s t test (n = 3). ∗p < 0.05. n.s., not significant. (G) (1) Migration assay for the indicated cell lines. (2) Number of migrated cells was quantified in five random images from each treatment group. The data represent mean ± SD and were analyzed by an unpaired two-tailed Student’s t test (n = 5). ∗∗p < 0.01, ∗∗∗p < 0.001.
As described in Figure 1, the CYT2-to-CYT1 ratio of CD46 was significantly higher in BCa tissue as compared to adjacent normal tissue, and thus we investigated whether a correlation between the expression of splicing factors and the 13−/13+ ratio of CD46 exists in clinical BCa samples. We found that 19 of 27 primary BCa specimens had upregulated (>2-fold) SRSF1 mRNA expression (Figure 7C). Furthermore, SRSF1 expression levels positively correlated with the ratio of CD46 splice variants (13−/13+) in these clinical samples (p = 0.0276, r = 0.4238, Spearman correlation) (Figure 7D). We thus decided to investigate whether SRSF1 regulates BCa cell proliferation and migration by promoting CD46 exon 13 skipping. SRSF1 functions as a key oncodriver in numerous solid tumors by regulating AS of many cancer-related genes.43, 44, 45 However, the role of SRSF1 in BCa is not well established. As expected, knockdown of SRSF1 significantly decreased cancer cell proliferation, colony formation ability, and migration in vitro (Figures S11A–S11E). Furthermore, in vivo tumorigenic analyses showed that depleted expression of SRSF1 in EJ-1 cells resulted in a decrease in tumor growth rate and tumor size (Figures S11A–S11E). We conclude that SRSF1 promotes tumorigenicity of BCa cells in both in vitro and in vivo assays. We next sought to determine whether the exogenous CD46 expression can compensate for SRSF1 knockdown effects. We found that forced expression of the CD46-CYT2 isoform in SRSF1-knockdown cells increased cancer cell proliferation, colony formation ability, and migration, although the growth impairment and migration defects were not fully restored compared with the control (Figures 7E–7G; Figure S12). However, cells expressing SRSF1 shRNA/CD46-CYT1 showed an opposite effect of decreasing these three activities compared with cells expressing SRSF1 shRNA alone (Figures 7E–7G), indicating that such phenotypical rescue is specific for CD46-CYT2. These results demonstrate that CD46 is a critical target of the SRSF1-mediated splicing program in BCa.
Discussion
Aberrant splicing of many genes plays a critical role in tumor development and progression. In this study, we demonstrated that CD46 exon 13 exclusion is a frequent event in BCa. We performed a full screening of the C-terminal cytosolic tail of CD46 interactome in BCa cells and found that many ribosome proteins and eukaryotic translation factors are CYT2, but not CYT1, binding partners. Importantly, we defined a critical role for CD46-CYT2 in regulating IRES-mediated translation; this regulation is mediated partially through interaction with hnRNPA1.
CD46 was originally reported to function as an inhibitor of complement activation, which may contribute to its pro-tumor activities in several tumors.31, 32, 33, 34, 35, 36 Increasing evidence indicates that CD46 is also involved in signal transduction pathways, and both of CD46’s cytoplasmic domains, CYT1 and CYT2, can transmit distinct intracellular signals, which may relate to their different function in tumors. Indeed, we also provide functional evidence that CD46-CYT1 inhibits, and CD46-CYT2 promotes, cancer cell growth, migration, and colony formation in vitro and tumorigenesis in vivo. A number of studies demonstrated that various CD46-related signaling pathways (e.g., the Notch signaling pathway6 and SPAK-mitogen-activated protein kinase [MAPK] pathway5,11,17,27) also play a prominent role in the regulation of tumor development/progression. However, the molecular basis for their opposite functions in BCa remains to be defined.
Notably, in the present study we indicate that CD46 is a translational regulator. The mechanism by which CD46 transmits intracellular signals remains largely unknown. Protein function is frequently regulated by its interaction with other proteins. This aspect is particularly important for transmembrane receptors, which transmit extracellular signals into the cells to influence cellular functions. Indeed, many studies have shown that the cytoplasmic tail of CD46 plays a critical role in multiple signaling pathways mediated by interacting with various intracellular proteins. Although several partners of CYT1 or CYT2 have already been described,5,6,11,17,22, 23, 24, 25, 26, 27,29 no global interactome analysis of the cytoplasmic tail of CD46 proteins is currently available. This study presents the first high-throughput analysis of the two intracellular tails of CD46 interactome by TAP-MS in a human cell line. Most CYT1 domain partners identified are involved in lipid metabolism, protein phosphatase activity, and cytoskeleton organization. As mentioned earlier, only the CD46-CYT1 isoform affects cell morphology and polarity by interacting with DLG4.23,24 It seems that CD46-CYT1 can regulate cell morphology through multiple other cytoskeleton-associated proteins. The C-terminal tetrapeptide FTSL of CD46-CYT1 has been shown to bind to the PDZ domain of DLG4 and GOPC.22,24 Of interest, our TAP-MS study revealed three other PDZ domain-containing proteins, that is, SNX27, PTPN3 and SLC9A3R2, as binding partners for the CYT1 domain. Furthermore, both SNX27 and SLC9A3R2 mediate many cellular processes by binding to and regulating endosome-to-membrane trafficking and endosomal recycling of plasma membrane receptors,46, 47, 48 suggesting that SNX27 and SLC9A3R2 may act as adaptors for CD46-CYT1 during its transport between endosomes and the cell surface. Importantly, we also found that ribosome complexes, the glycolysis enzyme complex, and hnRNP complexes were associated with the CYT2 domain, but not with the CYT1 domain. Significantly, we found that hnRNPA1 is a critical CYT2-interaction protein mediating the translational regulatory function of CD46-CYT2. It is well known that protein synthesis occurs primarily at both the cytosol and the cytoplasmic surface of the endoplasmic reticulum (ER). However, there is evidence at present in support of the idea that protein synthesis occurs at the plasma membrane.49 For example, RACK1 is a plasma membrane-associated protein that is stably associated with both membrane-bound receptors and the ribosome, which leads to the stimulation of local translation of specific mRNAs at the plasma membrane.50 In addition, the ER forms physical contacts with the plasma membrane, and they have many properties in common.51 Considering that CD46 is also a plasma membrane protein, it is highly likely that CD46 regulates the translation that occurs at the cytoplasmic surface of the plasma membrane. It was of significant interest that CD46 is subjected to further enzymatic processing and is cleaved on the cell surface, which releases CYT1 and CYT2 into the cytoplasm;15,28 therefore, another possibility is that released CYT1/CYT2 peptides in cytoplasm can interact with the translational machine and regulate the translation of a subset of oncogenes. The specific intracellular compartments where CD46 is involved in the regulation of translation are currently unknown.
Previous evidence revealed the importance of hnRNPA1 for translation control of many oncogenes, including HIF1a, CCND1, and c-Myc.41,52 Our discovery that CD46-CYT2 promotes hnRNPA1 binding to mRNA may, at least in part, provide an explanation for its translational regulatory function. Currently, there are >100 human genes predicted to contain IRES elements. Thus, our results suggest that dysregulation of the CD46-CYT2/hnRNPA1 translational network may fine-tune a large portion of them post-transcriptionally and drive tumorigenesis of BCa. Apart from hnRNPA1, several proteins that play key roles in translational control have been identified as interactors of the CD46-CYT2 complex. For example, PDCD4 has been demonstrated to be a translation inhibitor, which can inhibit the translation of p53 and Sin1.53, 54, 55, 56 These results led us to propose a model for regulation of translation by CD46-CYT2; that is, hnRNP proteins might function as adaptors of their target mRNA and CD46-CYT2, which regulate ribosome complexes binding for enhanced or decreased protein translation. Dysregulation of mRNA translation is a hallmark of many cancers. It will be interesting to explore the global effects of CD46-CYT2 on translation.
CD46 undergoing AS, which affects the cytoplasmic tail, has been known for a long time, and Tang et al.8 demonstrated that exon 13 inclusion and exclusion are subject to splicing regulation by multiple trans-acting splicing factors, including hnRNPA1, PTBP1, TIA1/TIAL1, and SRSF1.57 Consistent with previous reports, we showed that both SRSF1 and PTBP1 act as repressors of exon 13 inclusion, while TIA1 and TIAL1 promote exon 13 inclusion in BCa cells. Tang et al.8 also showed that hnRNPA1 interacts with the ESE of CD46 exon 13. We found that hnRNPA1 knockdown has little effect on exon 13 inclusion, while hnRNPA1 overexpression promotes the exon 13 exclusion, and we thus speculated that other splicing factors (such as hnRNPA2B1) could compensate for the hnRNPA1 depletion. Thus, we found that hnRNPA1 also promotes a CYT1-to-CYT2 splice switch, and that CD46-CYT2 also promotes hnRNPA1 activity, revealing a cross-regulation between CD46 and hnRNPA1 activities. Importantly, we found that SRSF1 is significantly upregulated in BCa tissues in comparison with matching adjacent normal tissues. Importantly, the level of SRSF1 mRNA is positively correlated with exon 13-lacking CD46 transcripts in clinical BCa samples. We thus propose a model for the crosstalk between AS regulation of CD46 exon 13 and translational regulation (Figure S13).
SRSF1 is the first splicing factor shown to be directly involved in cancer, and its expression was increased in many types of cancer.43,44 In addition, our results demonstrated that the exon 13-lacking CD46 (CD46-CYT2) isoform is upregulated, and the exon 13-containing CD46 (CD46-CYT1) isoform is downregulated, in BCa tissues, respectively, and CD46-CYT2 and CD46-CYT1 isoforms have opposite roles in the tumorigenesis of human BCa. Thus, it is reasonable to propose that SRSF1 regulates tumorigenesis of BCa through AS regulation of CD46 precursor (pre-)mRNA. Because we found that CD46-CYT2 was not sufficient to rescue the growth defects following SRSF1 depletion, the effects of SRSF1 knockdown on cell growth and migration likely involves a number of other genes. Furthermore, SRSF1 also has other biological functions; for example, SRSF1 has been shown to play critical roles in the regulation of mRNA stability and translation, miRNA processing, protein sumoylation, and stress response (reviewed in Chen et al.58).59 Further elucidation of the role of CD46 isoforms and other candidate SRSF1-regulated oncogenes will likely provide valuable insights into their mechanistic pathway in cancer cell growth.
Materials and methods
Human samples
Twenty-eight pairs of BCa and their corresponding adjacent normal tissues were obtained from BCa patients treated at the Department of Urology at Tongji Hospital (Wuhan, P.R. China) after their written informed consent was obtained. All samples were kept in liquid nitrogen before RNA extraction.
Antibodies
The following antibodies were used in the experiments: anti-FLAG (F3165) from Sigma-Aldrich; anti-Myc (562), anti-Strep-tag II (M211-3), and anti-hemagglutinin (HA) antibody (M180-3) from Medical & Biological Laboratories (MBL); anti-CD46 (#13241), anti-hnRNPA1 (#8443), anti-c-Myc (#9402), and anti-SRSF1 (#14902) from Cell Signaling Technology; anti-puromycin (MABE343, clone 12D10) purchased from Millipore; anti-HIF1α (NB100-105) purchased from Novus Biologicals; anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (sc-32233) purchased from Santa Cruz Biotechnology; goat anti-mouse immunoglobulin G (IgG) horseradish peroxidase (HRP)-linked whole antibody (31430) and goat anti-rabbit IgG HRP-linked whole antibody (31460) purchased from Thermo Scientific.
Western blot
Western blot analysis was performed to detect the expression levels of various proteins in cells. Cells were harvested, washed, with cold 1× PBS, and lysed with NETN buffer consisting of 50 mM Tris-HCl at pH 8.0, 0.15 M NaCl, 1 mM EDTA, 0.5% Nonidet P-40 (NP-40), and a 1× protease inhibitor cocktail (Roche) for 30 min on ice, then centrifuged at 12,000 × g for 15 min at 4°C. The total protein concentration was determined by a bicinchoninic acid (BCA) protein assay kit (Beyotime). Equal amounts (20 μg per load) of protein samples were subjected to SDS-PAGE electrophoresis and transferred onto polyvinylidene fluoride (PVDF) membranes (Roche). The blots were blocked in 5% nonfat milk (Becton Dickinson [BD]) and incubated with primary antibodies, followed by incubation with secondary antibodies conjugated with HRP. The protein bands were developed with the chemiluminescent reagents.
Plasmids and molecular cloning
The shRNA plasmids of hnRNPA1, PTBP1, and SRSF1 have been described previously.58 Human SRSF1, hnRNPA1, HMGB1, EIF5A, PTPN3 (500–901 aa), SNX27, and RPL17 cDNA were generated by PCR and cloned into BamHI and XhoI sites of psi-FLAG expression plasmid. Mammalian expression plasmids for human CD46-CYT1 and CD46-CYT2 were generated by PCR and cloned into NheI and XhoI sites of pHAGE-cytomegalovirus (CMV) (a gift from Prof. Xiaodong Zhang, Wuhan University, P.R. China) plasmids. PCDH-strepII-GST-C1, which contains fusion tags including the Strep-tag-II and GST-tag, was constructed by subcloning the MSCV promoter, Strep-tag-II, and GST-tag into the XbaI and NotI sites of PCDH-CD513B-1 (System Biosciences, USA). Human CYT1 and CYT2 domains of CD46 were PCR amplified, digested by BamHI and XhoI, and then ligated into PCDH-strepII-GST-C1 to create PCDH-strepII-GST-CYT1 and PCDH-strepII-GST-CYT2, respectively. psi-MCS-EGFP plasmid and psi-Rluc-MCS-Fluc plasmid were previously described.58,60 PCDH-Rluc-MCS-Fluc was constructed by subcloning the encoding region of Rluc-MCS-Fluc of psi-Rluc-MCS-Fluc into the BamHI and NheI sites of PCDH-MSCV. For tethering assays, a luciferase reporter containing four copies of the MS2 binding site was generated by cloning the MS2 stem loop region of single guide RNA (sgRNA) (MS2) plasmids (Addgene, #61424) into the NheI and EcoRI cut PCDH-Rluc-MCS-Fluc. CrPV IRES, HCV IRES, EMCV IRES, and EV-71A IRES were synthesized and cloned into BamHI and XhoI sites of PCDH-Rluc-MCS-Fluc-4xMS2 plasmid. CCND1 IRES, HIF1A IRES, and c-Myc IRES were generated by PCR and cloned into BamHI and XhoI sites of PCDH-Rluc-MCS-Fluc and PCDH-Rluc-MCS-Fluc-4xMS2 plasmids, respectively. EF1a promoter and the MS2-N55K open reading frame (ORF) was PCR amplified from MS2-P65-HSF1_GFP (Addgene, #61423) and cloned into the XbaI and BamHI sites of psi-MCS-T2A-puro to produce psi-EF1a-MS2-N55K-myc. Human CYT1 and CYT2 domains of CD46, as well as GST, were PCR amplified and ligated into psi-EF1a-MS2-N55K-myc to create MS2-N55K-myc-CYT1, MS2-N55K-myc-CYT2, and MS2-N55K-myc-GST, respectively. The PCDH-H1 shRNA cloning vector was constructed by digesting out the CMV promoter of the PCDH-CD513B-1 plasmid, and then inserting the H1 promoter of pSilencer5.1-H1 Retro plasmid (Thermo Fisher Scientific, USA). All of the splicing factors of RNAi plasmids used in the present study were previously described.58 Gene-specific shRNA target sequences were synthesized, annealed, and cloned into the BamH and NotI sites of the PCDH-H1 plasmid. For application of Cas9 for site-specific genome editing in human cells, CD46-specific sgRNA was synthesized and cloned into lentiCRISPR v2 containing two expression cassettes, hSpCas9 and the chimeric guide RNA (Addgene, #52961). The primers for making these constructs are shown in Table S5. All plasmids were verified by sequencing.
TAP and MS
EJ-1 cells (approximately 200 million) stably expressing StrepII-GST, StrepII-GST-CYT1, or StrepII-GST-CYT2 were lysed in NETN buffer consisting of 50 mM Tris-HCl at pH 8.0, 0.15 M NaCl, 1 mM EDTA, 0.5% NP-40, and a 1× protease inhibitor cocktail (Roche). Prior to complex purification, avidin (A9275, Sigma, 20 μg/1 mL extracts) was added to the extracts to remove biotinylated molecules that may bind unspecifically to the Strep-Tactin XT superflow resin (2-4010, IBA). Then, the Strep-Tactin XT superflow resin (80 μL) was added and incubated for 4 h at 4°C. Beads were washed three times with 6 mL of washing buffer 1 (50 mM Tris-HCl at pH 8.0, 0.3 M NaCl, 1 mM EDTA, 0.5% NP-40) and then two times with 6 mL of washing buffer 2 (50 mM Tris-HCl at pH 8.0, 0.5 M NaCl, 1 mM EDTA, 0.5% NP-40). Beads were equilibrated in NETN buffer and complexes were eluted with biotin (50 mM in elution buffer) in two steps of 10–15 min at 4°C. Eluates were pooled, cleared by centrifugation, and used for the second purification step. Glutathione Sepharose beads (GE Healthcare/Dharmacon) were incubated with the eluates for 4 h at 4°C. The beads were washed four times with washing buffer 2. The retained proteins were eluted by NuPAGE lithium dodecyl sulfate (LDS) sample buffer (NP0007, Thermo Fisher Scientific) and then separated by SDS-PAGE (NP0322BOX, Thermo Fisher Scientific).
The separated proteins were cut out from Coomassie blue-stained gels into three portions according to their molecular weights (10–35, 35–70, and 70–180 kDa). Then, the in-gel proteins were reduced, alkylated, and trypsin digested as previously described.61 Protein mixtures were then analyzed by a high-performance liquid chromatography (HPLC)-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). The resulting RAW files were searched against the NCBI nr using the Mascot search algorithm to identify the corresponding proteins using the Proteome Discoverer 2.1 software package (Thermo Scientific). The detailed results of the experiments are shown in Table S2. Functional enrichment analysis was performed using FunRich software (http://www.funrich.org).39
RT-PCR and real-time PCR analysis
Total RNAs were extracted from 1–5 × 106 cells by TRIzol (Invitrogen), and cDNAs were synthesized using ReverTra Ace qPCR RT Kit (Toyobo). Real-time PCR was performed using SYBR Green real-time PCR master mix (Roche) and the ABI ViiA7 qPCR system (Applied Biosystems). GAPDH was the housekeeping gene used as a control. The primers and the PCR conditions used for RT-PCR and real-time PCR are provided in Table S5. For semiquantitative RT-PCR, the PCR products were run on a 1.5% agarose gel and visualized under UV light, after which densitometric analysis was performed using ImageJ software for the quantification analysis.
CoIP
To analyze protein interactions, coIP experiments were performed using HEK293T or EJ-1 cells after 48-h transfection according to previously published protocols.35 To analyze protein interactions, HEK293T or EJ-1 cells (about 5 × 106 cells) were transfected with the indicated plasmids. After a 48-h transfection, the cells were lysed in NETN buffer consisting of 50 mM Tris-HCl at pH 8.0, 0.15 M NaCl, 1 mM EDTA, 0.5% NP-40, and a 1× protease inhibitor cocktail (Roche). Specified antibody and protein G agarose (Roche) were incubated with the cell lysates overnight at 4°C. The resins were washed four times with wash buffer (50 mM Tris-HCl at pH 8.0, 0.5 M NaCl, 1 mM EDTA, 0.5% NP-40). After elution by loading buffer, the bound proteins were separated by SDS-PAGE.
RIP assays
To analyze RNA molecules associated with CD46 and hnRNPA1, RIP experiments were performed as described.58 The cells (about 1 × 107 cells) were scraped and lysed in 0.5 mL of cold RIP lysis buffer supplemented with RNase inhibitor. After centrifugation, the specified antibody and magnetic beads were incubated with the cell lysates for 4 h at 4°C. The beads were washed five times with RIP wash buffer. The RIP RNA fraction was extracted using TRIzol reagent (Invitrogen). The subsequent steps were the same as in RT-PCR and real-time PCR analysis described in this section.
Cell culture
HEK293T, EJ-1, and 5637 cell lines were purchased from the National Infrastructure of Cell Line Resource (Beijing, P.R. China). These cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) with 10% fetal bovine serum (HyClone). All cells were maintained in the presence of 5% CO2 at 37°C.
AHA incorporation assay
Newly synthesized proteins were analyzed by AHA incorporation. Cells were cultured in methionine (Met)-free DMEM (Sigma) supplemented with AHA (Click-iT AHA, Life Technologies). Approximately 1 h after AHA incubation, cells were lysed and subjected to click reaction using biotin-alkyne according to the manufacturer’s instructions (Invitrogen, catalog #B10184). Labeled proteins were then desalted with Zeba spin desalting columns (7 kDA molecular weight cutoff [MWCO], 2 mL). Then, the biotin-labeled de novo synthesized proteins were affinity purified using streptavidin resin (Sigma). The newly synthesized proteins were subjected to immunoblot analysis.
MS2 pull-down assay
The cells were transfected with the plasmid encoding MS2-GST and the indicated 3′ MS2 stem-loop-tagged IRES sequence of HIF1a or c-Myc and harvested 48 h post-transfection. Cytoplasmic proteins were extracted as described61 and then incubated with fusion MS2-GST bound glutathione Sepharose 4B beads for 3 h at 4°C. The beads were subsequently washed three times with wash buffer (20 mM HEPES at pH 7.9, 200 mM KCl, and 1 mM EDTA) and once with ice-cold PBS. Bound proteins were dissociated by boiling with 1× SDS-PAGE sample buffer and resolved on 10% SDS-PAGE.
AS analysis
SRA datasets were downloaded from https://www.ncbi.nlm.nih.gov/sra/. Datasets were divided into two groups as normal (SRX093212, SRX093210, SRX093208) and tumor (SRX093211, SRX093209, SRX093207) and analyzed by SpliceSeq.38 Genes with differentially splicing patterns between two groups were confirmed by manual proofreading.
Colony formation, cell proliferation, cell migration, and invasion assays
Colony formation was measured 2 weeks after seeding 1,000 cells per well in six-well plates. Cell proliferation was estimated using the Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories) according to the manufacturer’s instructions. Migration and invasion assays were performed using uncoated and Matrigel-coated transwell inserts according to the manufacturer’s instructions. All experiments were performed in triplicate.
Animal experiments
Tumorigenesis in nude mice was determined as described previously. Five mice each were injected subcutaneously with prepared cells at a single site. Tumor onset was measured with calipers at the site of injection weekly at different times on the same day. All experiments were approved by the Animal Care and Use Committee of Tongji Medical College of Huazhong University of Science and Technology.
Statistical analysis
The data are presented as the mean ± SD. Differences among groups were determined by a two-way ANOVA followed by a Tukey post hoc test. Comparisons between two groups were performed using an unpaired Student’s t test. A p value of <0.05 was considered significant.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant nos. 81772721, 81470935, 81670645, 81602236, and 81402105); the Natural Science Foundation of Jiangxi Province of China (20202BABL216060); the Chenguang Program of Wuhan Science and Technology Bureau (2015070404010199 and 2015071704021644); and by The National High Technology Research and Development Program 863 (2014AA020607). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
H.X., K.C., and J.Z. designed the study. K.C., J.Z., and H.X. wrote the manuscript. J.H. and L.W. revised the manuscript for important intellectual content. J.Z., K.C., Y.S., and H.Z. performed experiments. C.H., Y.Z., and Z.C. performed the statistical analysis. K.C., C.H., J.X., T.W., and Z.Y. performed the data analysis. J.Z., H.X., G.Y., W.Y., and W.X. prepared the clinical samples. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.02.019.
Supplemental information
References
- 1.Sveen A., Kilpinen S., Ruusulehto A., Lothe R.A., Skotheim R.I. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016;35:2413–2427. doi: 10.1038/onc.2015.318. [DOI] [PubMed] [Google Scholar]
- 2.Climente-González H., Porta-Pardo E., Godzik A., Eyras E. The functional impact of alternative splicing in cancer. Cell Rep. 2017;20:2215–2226. doi: 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Siegfried Z., Karni R. The role of alternative splicing in cancer drug resistance. Curr. Opin. Genet. Dev. 2018;48:16–21. doi: 10.1016/j.gde.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Jayasinghe R.G., Cao S., Gao Q., Wendl M.C., Vo N.S., Reynolds S.M., Zhao Y., Climente-González H., Chai S., Wang F., Cancer Genome Atlas Research Network Systematic analysis of splice-site-creating mutations in cancer. Cell Rep. 2018;23:270–281.e3. doi: 10.1016/j.celrep.2018.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto H., Fara A.F., Dasgupta P., Kemper C. CD46: the “multitasker” of complement proteins. Int. J. Biochem. Cell Biol. 2013;45:2808–2820. doi: 10.1016/j.biocel.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Le Friec G., Sheppard D., Whiteman P., Karsten C.M., Shamoun S.A., Laing A., Bugeon L., Dallman M.J., Melchionna T., Chillakuri C. The CD46-Jagged1 interaction is critical for human TH1 immunity. Nat. Immunol. 2012;13:1213–1221. doi: 10.1038/ni.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marie J.C., Astier A.L., Rivailler P., Rabourdin-Combe C., Wild T.F., Horvat B. Linking innate and acquired immunity: divergent role of CD46 cytoplasmic domains in T cell induced inflammation. Nat. Immunol. 2002;3:659–666. doi: 10.1038/ni810. [DOI] [PubMed] [Google Scholar]
- 8.Tang S.J., Luo S., Ho J.X., Ly P.T., Goh E., Roca X. Characterization of the Regulation of CD46 RNA alternative splicing. J. Biol. Chem. 2016;291:14311–14323. doi: 10.1074/jbc.M115.710350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen A.S., Bundgaard B.B., Møller B.K., Höllsberg P. Non-random pairing of CD46 isoforms with skewing towards BC2 and C2 in activated and memory/effector T cells. Sci. Rep. 2016;6:35406. doi: 10.1038/srep35406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell S.M., Loveland B.E., Johnstone R.W., Thorley B.R., McKenzie I.F. Functional characterisation of alternatively spliced CD46 cytoplasmic tails. Transplant. Proc. 1992;24:2329–2330. [PubMed] [Google Scholar]
- 11.Cope A., Le Friec G., Cardone J., Kemper C. The Th1 life cycle: molecular control of IFN-γ to IL-10 switching. Trends Immunol. 2011;32:278–286. doi: 10.1016/j.it.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Kolev M., Dimeloe S., Le Friec G., Navarini A., Arbore G., Povoleri G.A., Fischer M., Belle R., Loeliger J., Develioglu L. Complement regulates nutrient influx and metabolic reprogramming during Th1 cell responses. Immunity. 2015;42:1033–1047. doi: 10.1016/j.immuni.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano A., Yang Z., Katayama Y., Korte-Sarfaty J., Wong T.C. Human CD46 enhances nitric oxide production in mouse macrophages in response to measles virus infection in the presence of gamma interferon: dependence on the CD46 cytoplasmic domains. J. Virol. 1999;73:4776–4785. doi: 10.1128/jvi.73.6.4776-4785.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kemper C., Atkinson J.P. T-cell regulation: with complements from innate immunity. Nat. Rev. Immunol. 2007;7:9–18. doi: 10.1038/nri1994. [DOI] [PubMed] [Google Scholar]
- 15.Ni Choileain S., Weyand N.J., Neumann C., Thomas J., So M., Astier A.L. The dynamic processing of CD46 intracellular domains provides a molecular rheostat for T cell activation. PLoS ONE. 2011;6:e16287. doi: 10.1371/journal.pone.0016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y.Q., Gao Y.D., Yang J., Guo W. A defect of CD4+CD25+ regulatory T cells in inducing interleukin-10 production from CD4+ T cells under CD46 costimulation in asthma patients. J. Asthma. 2010;47:367–373. doi: 10.3109/02770903.2010.481340. [DOI] [PubMed] [Google Scholar]
- 17.Cardone J., Le Friec G., Vantourout P., Roberts A., Fuchs A., Jackson I., Suddason T., Lord G., Atkinson J.P., Cope A. Complement regulator CD46 temporally regulates cytokine production by conventional and unconventional T cells. Nat. Immunol. 2010;11:862–871. doi: 10.1038/ni.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Astier A.L., Meiffren G., Freeman S., Hafler D.A. Alterations in CD46-mediated Tr1 regulatory T cells in patients with multiple sclerosis. J. Clin. Invest. 2006;116:3252–3257. doi: 10.1172/JCI29251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Buanec H., Gougeon M.L., Mathian A., Lebon P., Dupont J.M., Peltre G., Hemon P., Schmid M., Bizzini B., Künding T. IFN-α and CD46 stimulation are associated with active lupus and skew natural T regulatory cell differentiation to type 1 regulatory T (Tr1) cells. Proc. Natl. Acad. Sci. USA. 2011;108:18995–19000. doi: 10.1073/pnas.1113301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khandelwal P., Birla S., Bhatia D., Puraswani M., Saini H., Sinha A., Hari P., Sharma A., Bagga A. Mutations in membrane cofactor protein (CD46) gene in Indian children with hemolytic uremic syndrome. Clin. Kidney J. 2018;11:198–203. doi: 10.1093/ckj/sfx078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardone J., Le Friec G., Kemper C. CD46 in innate and adaptive immunity: an update. Clin. Exp. Immunol. 2011;164:301–311. doi: 10.1111/j.1365-2249.2011.04400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joubert P.E., Meiffren G., Grégoire I.P., Pontini G., Richetta C., Flacher M., Azocar O., Vidalain P.O., Vidal M., Lotteau V. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Ludford-Menting M.J., Crimeen-Irwin B., Oliaro J., Pasam A., Williamson D., Pedersen N., Guillaumot P., Christansen D., Manie S., Gaus K., Russell S.M. The reorientation of T-cell polarity and inhibition of immunological synapse formation by CD46 involves its recruitment to lipid rafts. J. Lipids. 2011;2011:521863. doi: 10.1155/2011/521863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludford-Menting M.J., Thomas S.J., Crimeen B., Harris L.J., Loveland B.E., Bills M., Ellis S., Russell S.M. A functional interaction between CD46 and DLG4: a role for DLG4 in epithelial polarization. J. Biol. Chem. 2002;277:4477–4484. doi: 10.1074/jbc.M108479200. [DOI] [PubMed] [Google Scholar]
- 25.Wang G., Liszewski M.K., Chan A.C., Atkinson J.P. Membrane cofactor protein (MCP; CD46): isoform-specific tyrosine phosphorylation. J. Immunol. 2000;164:1839–1846. doi: 10.4049/jimmunol.164.4.1839. [DOI] [PubMed] [Google Scholar]
- 26.Kurita-Taniguchi M., Fukui A., Hazeki K., Hirano A., Tsuji S., Matsumoto M., Watanabe M., Ueda S., Seya T. Functional modulation of human macrophages through CD46 (measles virus receptor): production of IL-12 p40 and nitric oxide in association with recruitment of protein-tyrosine phosphatase SHP-1 to CD46. J. Immunol. 2000;165:5143–5152. doi: 10.4049/jimmunol.165.9.5143. [DOI] [PubMed] [Google Scholar]
- 27.Cardone J., Al-Shouli S., Kemper C. A novel role for CD46 in wound repair. Front. Immunol. 2011;2:28. doi: 10.3389/fimmu.2011.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weyand N.J., Calton C.M., Higashi D.L., Kanack K.J., So M. Presenilin/γ-secretase cleaves CD46 in response to Neisseria infection. J. Immunol. 2010;184:694–701. doi: 10.4049/jimmunol.0900522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni Choileain S., Astier A.L. CD46 processing: a means of expression. Immunobiology. 2012;217:169–175. doi: 10.1016/j.imbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni Choileain S., Hay J., Thomas J., Williams A., Vermeren M.M., Benezech C., Gomez-Salazar M., Hugues O.R., Vermeren S., Howie S.E.M. TCR-stimulated changes in cell surface CD46 expression generate type 1 regulatory T cells. Sci. Signal. 2017;10:eaah6163. doi: 10.1126/scisignal.aah6163. [DOI] [PubMed] [Google Scholar]
- 31.Fishelson Z., Donin N., Zell S., Schultz S., Kirschfink M. Obstacles to cancer immunotherapy: expression of membrane complement regulatory proteins (mCRPs) in tumors. Mol. Immunol. 2003;40:109–123. doi: 10.1016/s0161-5890(03)00112-3. [DOI] [PubMed] [Google Scholar]
- 32.Hakulinen J., Junnikkala S., Sorsa T., Meri S. Complement inhibitor membrane cofactor protein (MCP; CD46) is constitutively shed from cancer cell membranes in vesicles and converted by a metalloproteinase to a functionally active soluble form. Eur. J. Immunol. 2004;34:2620–2629. doi: 10.1002/eji.200424969. [DOI] [PubMed] [Google Scholar]
- 33.Lu Z., Zhang C., Cui J., Song Q., Wang L., Kang J., Li P., Hu X., Song H., Yang J., Sun Y. Bioinformatic analysis of the membrane cofactor protein CD46 and microRNA expression in hepatocellular carcinoma. Oncol. Rep. 2014;31:557–564. doi: 10.3892/or.2013.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui W., Zhao Y., Shan C., Kong G., Hu N., Zhang Y., Zhang S., Zhang W., Zhang Y., Zhang X., Ye L. HBXIP upregulates CD46, CD55 and CD59 through ERK1/2/NF-κB signaling to protect breast cancer cells from complement attack. FEBS Lett. 2012;586:766–771. doi: 10.1016/j.febslet.2012.01.039. [DOI] [PubMed] [Google Scholar]
- 35.Geis N., Zell S., Rutz R., Li W., Giese T., Mamidi S., Schultz S., Kirschfink M. Inhibition of membrane complement inhibitor expression (CD46, CD55, CD59) by siRNA sensitizes tumor cells to complement attack in vitro. Curr. Cancer Drug Targets. 2010;10:922–931. doi: 10.2174/156800910793357952. [DOI] [PubMed] [Google Scholar]
- 36.Cui W., Zhang Y., Hu N., Shan C., Zhang S., Zhang W., Zhang X., Ye L. miRNA-520b and miR-520e sensitize breast cancer cells to complement attack via directly targeting 3′UTR of CD46. Cancer Biol. Ther. 2010;10:232–241. doi: 10.4161/cbt.10.3.12277. [DOI] [PubMed] [Google Scholar]
- 37.Sherbenou D.W., Aftab B.T., Su Y., Behrens C.R., Wiita A., Logan A.C., Acosta-Alvear D., Hann B.C., Walter P., Shuman M.A. Antibody-drug conjugate targeting CD46 eliminates multiple myeloma cells. J. Clin. Invest. 2016;126:4640–4653. doi: 10.1172/JCI85856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan M.C., Cleland J., Kim R., Wong W.C., Weinstein J.N. SpliceSeq: a resource for analysis and visualization of RNA-seq data on alternative splicing and its functional impacts. Bioinformatics. 2012;28:2385–2387. doi: 10.1093/bioinformatics/bts452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 40.Lin S., Choe J., Du P., Triboulet R., Gregory R.I. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao G., Dhar S., Bedford M.T. PRMT5 regulates IRES-dependent translation via methylation of hnRNP A1. Nucleic Acids Res. 2017;45:4359–4369. doi: 10.1093/nar/gkw1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy R., Huang Y., Seckl M.J., Pardo O.E. Emerging roles of hnRNPA1 in modulating malignant transformation. Wiley Interdiscip. Rev. RNA. 2017;8:e1431. doi: 10.1002/wrna.1431. [DOI] [PubMed] [Google Scholar]
- 43.Karni R., de Stanchina E., Lowe S.W., Sinha R., Mu D., Krainer A.R. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anczuków O., Rosenberg A.Z., Akerman M., Das S., Zhan L., Karni R., Muthuswamy S.K., Krainer A.R. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 2012;19:220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Miguel F.J., Sharma R.D., Pajares M.J., Montuenga L.M., Rubio A., Pio R. Identification of alternative splicing events regulated by the oncogenic factor SRSF1 in lung cancer. Cancer Res. 2014;74:1105–1115. doi: 10.1158/0008-5472.CAN-13-1481. [DOI] [PubMed] [Google Scholar]
- 46.Shinde S.R., Maddika S. PTEN regulates glucose transporter recycling by Impairing SNX27 retromer assembly. Cell Rep. 2017;21:1655–1666. doi: 10.1016/j.celrep.2017.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hussain N.K., Diering G.H., Sole J., Anggono V., Huganir R.L. Sorting Nexin 27 regulates basal and activity-dependent trafficking of AMPARs. Proc. Natl. Acad. Sci. USA. 2014;111:11840–11845. doi: 10.1073/pnas.1412415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritter-Makinson S.L., Paquet M., Bogenpohl J.W., Rodin R.E., Chris Yun C., Weinman E.J., Smith Y., Hall R.A. Group II metabotropic glutamate receptor interactions with NHERF scaffold proteins: implications for receptor localization in brain. Neuroscience. 2017;353:58–75. doi: 10.1016/j.neuroscience.2017.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Browning K.S., Bailey-Serres J. Mechanism of cytoplasmic mRNA translation. Arabidopsis Book. 2015;13:e0176. doi: 10.1199/tab.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nilsson J., Sengupta J., Frank J., Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saheki Y., De Camilli P. Endoplasmic reticulum-plasma membrane contact sites. Annu. Rev. Biochem. 2017;86:659–684. doi: 10.1146/annurev-biochem-061516-044932. [DOI] [PubMed] [Google Scholar]
- 52.Shi Y., Yang Y., Hoang B., Bardeleben C., Holmes B., Gera J., Lichtenstein A. Therapeutic potential of targeting IRES-dependent c-myc translation in multiple myeloma cells during ER stress. Oncogene. 2016;35:1015–1024. doi: 10.1038/onc.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Suzuki C., Garces R.G., Edmonds K.A., Hiller S., Hyberts S.G., Marintchev A., Wagner G. PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc. Natl. Acad. Sci. USA. 2008;105:3274–3279. doi: 10.1073/pnas.0712235105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q., Zhu J., Wang Y.W., Dai Y., Wang Y.L., Wang C., Liu J., Baker A., Colburn N.H., Yang H.S. Tumor suppressor Pdcd4 attenuates Sin1 translation to inhibit invasion in colon carcinoma. Oncogene. 2017;36:6225–6234. doi: 10.1038/onc.2017.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Q., Yang H.-S. The role of Pdcd4 in tumour suppression and protein translation. Biol. Cell. 2018;110:169–177. doi: 10.1111/boc.201800014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wedeken L., Singh P., Klempnauer K.H. Tumor suppressor protein Pdcd4 inhibits translation of p53 mRNA. J. Biol. Chem. 2011;286:42855–42862. doi: 10.1074/jbc.M111.269456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aznarez I., Barash Y., Shai O., He D., Zielenski J., Tsui L.-C., Parkinson J., Frey B.J., Rommens J.M., Blencowe B.J. A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008;18:1247–1258. doi: 10.1101/gr.073155.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen K., Xiao H., Zeng J., Yu G., Zhou H., Huang C., Yao W., Xiao W., Hu J., Guan W. Alternative splicing of EZH2 pre-mRNA by SF3B3 contributes to the tumorigenic potential of renal cancer. Clin. Cancer Res. 2017;23:3428–3441. doi: 10.1158/1078-0432.CCR-16-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Das S., Krainer A.R. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol. Cancer Res. 2014;12:1195–1204. doi: 10.1158/1541-7786.MCR-14-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen K., Zeng J., Xiao H., Huang C., Hu J., Yao W., Yu G., Xiao W., Xu H., Ye Z. Regulation of glucose metabolism by p62/SQSTM1 through HIF1α. J. Cell Sci. 2016;129:817–830. doi: 10.1242/jcs.178756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen K., Huang C., Yuan J., Cheng H., Zhou R. Long-term artificial selection reveals a role of TCTP in autophagy in mammalian cells. Mol. Biol. Evol. 2014;31:2194–2211. doi: 10.1093/molbev/msu181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.