Abstract
Lupus erythematosus (LE) is an autoimmune disease that can be divided into two types. The cutaneous lupus erythematosus (CLE), such as discoid LE (DLE), affects only the skin. While the systemic lupus erythematosus (SLE) affects the hematopoietic, renal, and other systems. We previously found that IFI44L methylation could be a biomarker for SLE. Here, we detect the IFI44L methylation by high-resolution melting-quantitative polymerase chain reaction (HRM-qPCR) assay. The positive percentages of SLE, DLE and healthy controls (HC) are 96.00%, 27.45%, 2.00%, if the curve of 25% methylation was used as the threshold of SLE. And we determined the serum IFN-a1 level by enzyme-linked immunosorbent assay (ELISA) in SLE, DLE and HC. The serum concentration of IFN-a1 in patients with SLE was significantly higher than in the DLE (12.63 ± 6.38 pg/mL vs 7.99 ± 2.28 pg/mL, P < 0.05) and HC (12.63 ± 6.38 pg/mL vs 7.17 ± 1.86 pg/mL, P < 0.05). But the expression level of IFN-a1 in serum was not significantly different between DLE and HC (7.99 ± 2.28 pg/mL vs 7.17 ± 1.86 pg/mL, P = 0.5365). This suggests that methylation of IFI44L and serum concentration of IFN-a1 may be used as biomarkers to distinguish DLE from SLE.
Keywords: Discoid lupus erythematosus, Systemic lupus erythematosus, IFI44L, DNA methylation, HRM-qPCR, IFN-a1
Highlights
-
•
DNA methylation of the IFI44L promoter region in SLE patients was significantly decreased compared with DLE patients and HC.
-
•
The serum concentration of IFN-a1 in SLE patients was significantly higher than in the DLE patients and HC.
-
•
DNA methylation of IFI44L and serum concentration of IFN-a1 may be used as biomarkers to distinguish DLE from SLE.
1. Introduction
The lupus erythematosus (LE) is a complex autoimmune disease with a variety of clinical manifestations, only skin damage or damage to multiple systems. Cutaneous lupus erythematosus (CLE) is a subtype of LE which only involving the skin, and one of the most common subtypes of CLE is discoid LE (DLE), which belongs to chronic CLE [1, 2]. The overlap in clinical features between CLE and systemic lupus erythematosus (SLE) presents a challenge to correct diagnosis, and early correct diagnosis and treatment are critical to the prognosis of LE [3]. SLE and CLE can exist independently or simultaneously, and a portion of the CLE can also develop into SLE. The probability of progressing to SLE varies with the CLE subtype from 5% to 23% [4]. However, the current diagnosis of CLE mainly relies on skin lesion characteristics and clinical symptoms, while the diagnosis of SLE relies on diagnostic criteria. There are no laboratory indicators with both high sensitivity and specificity to distinguish SLE from CLE, although the new EULAR/ACR 2019 criteria has higher specificity for diagnosing SLE from CLE, which needs biopsy-proven [4]. Disseminated LE lesions, nonspecific lesions, autoantibodies and other laboratory indicator like leukopenia can be used as markers indicating disease progression from CLE to SLE [5]. It is reported that there appear to be unique genetic factors specific for CLE which is not clear yet [6]. Pathological biopsies can also help identify the CLE subtype, but not all patients are willing to undergo biopsies [7]. Epigenetic studies of CLE have also been reported. For example, miR-12, miR-150, and miR-1264 levels were downregulated in DLE [8].
Studies showed that genetic factors, environmental factors, and immune disorders are involved in the development of CLE into SLE [5, 9, 10]. Among them, type I interferon (IFN–I) are crucial [11]. We previously identified that DNA methylation of the Interferon-induced protein 44-like (IFI44L) promoter region is markedly downregulated in SLE and could be used as a biomarker for SLE [12]. Then, we have reported a high-resolution melting-quantitative polymerase chain reaction (HRM-qPCR) to semi-quantitative analyze the methylation of IFI44L promoter, that can be used as a biomarker for the diagnosis of SLE with relative high sensitivity and specificity [13]. IFI44L gene is a stimulator of type I-IFN [14]. It remains unclear whether methylation level of IFI44L is different in DLE from SLE, and whether type I-IFN genes can be used as biomarkers to identifying SLE and DLE?
Here, we detected the methylation level of the IFI44L promoter region using HRM-qPCR method in SLE, DLE and HC. The serum levels of IFN-a1 in SLE, DLE and HC were also examined. The results indicated that methylation of IFI44L promoter and serum levels of IFN-a1 may be potential biomarkers for DLE distinguishing from SLE.
2. Materials and methods
2.1. Sample characteristics
All peripheral blood used in this study were obtained from 50 patients with SLE, 51 patients with DLE and 50 HC at the Department of Dermatology, Second Xiangya Hospital and peripheral blood samples were collected in tubes with EDTA. The serums used in this study were collected from 26 patients with SLE, 23 patients with DLE and 6 HC. All participants have signed the informed consent, and their characteristics such as gender and age were listed in Table 1.
Table 1.
The characteristics of samplesa.
| Disease | SLE | DLE | HC |
|---|---|---|---|
| Samples for HRM-qPCR | |||
| Sample size | 50 | 51 | 50 |
| Age(y) (mean ± SD) | 34.56 ± 12.77 | 42.35 ± 12.13 | 41.64 ± 12.83 |
| Sex (%) Female | 84.00 | 72.55 | 70.00 |
| Samples for ELISA | |||
| Sample size | 26 | 23 | 6 |
| Age(y) (mean ± SD) | 33.30 ± 10.98 | 36.33 ± 9.84 | 44.00 ± 10.26 |
| Sex (%) Female | 92.31 | 86.96 | 100 |
SLE, systemic lupus erythematosus; DLE, discoid lupus erythematosus; HC, healthy controls.
2.2. DNA extraction and bisulfite conversion
GeneJET Whole Blood Genomic DNA Purification Mini Kit (Thermo Fisher Scientific) was used for DNA extraction. Then DNA concentration and purity were determined with the NanoDrop. Bisulfite conversion of the genomic DNA was performed with an EZ DNA Methylation™ Kit following the instruction manual. Finally, 5μl of bisulfite-treated DNA per sample was used to detect methylation using HRM-qPCR.
2.3. Conduct of qPCR-HRM to determine methylation of the IFI44L promoter
The 0%,25%,50%,75%,100% methylation standards were prepared according to the sequence and preparation method in our previous study and the primers also used the same sequence from the previous study [13]. The HRM-qPCR experiment was performed on a LightCycler 96® real-time PCR system (Roche). The kit used in this experiment is LightCycler® 480 High-Resolution Melting Master (Roche). These standards were used for the evaluation of the IFI44l methylation of samples.
2.4. Detection of expression of the IFN-αwith ELISA
The IFN-a1 concentrations in the serum were quantified with an ELISA kit (Proteintech® Human IFNA1 ELISA; ptglab®).
2.5. Statistical analysis
The raw data of HRM-qPCR were analyzed in LightCycler 96® software. All statistical analyses were done with GraphPad Prism. Differences between LE subtypes and HC were analyzed using the two-tailed unpaired t-test and a P < 0.05 was considered statistically significant.
3. Results
3.1. Methylation of IFI44L is lower in SLE than in DLE
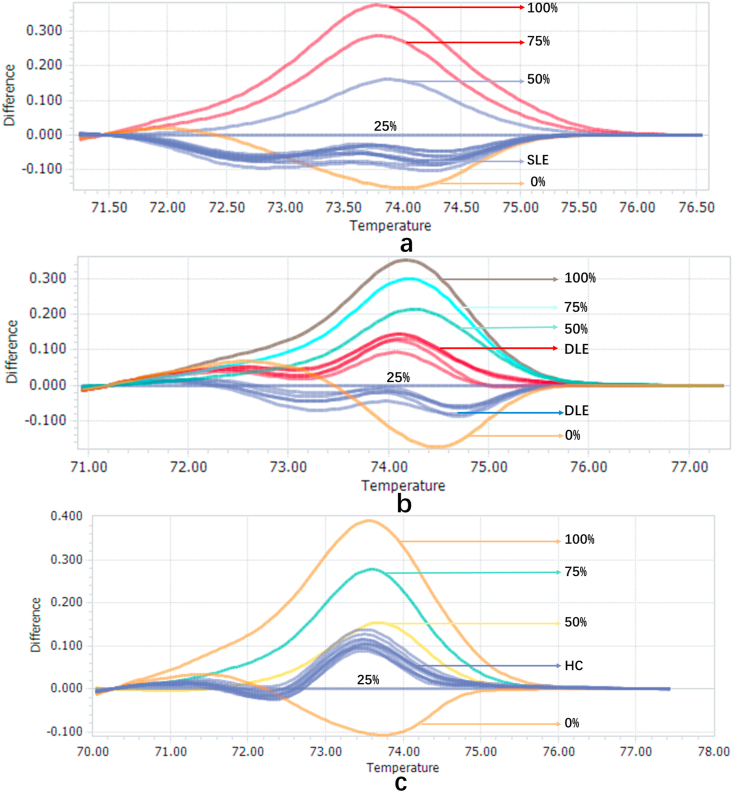
We previously found that IFI44L methylation was significantly down-regulated in SLE, compared with HC and disease controls including rheumatoid arthritis and other diseases. Using 25% methylation as the cutoff value, IFI44L is a biomarker for SLE with high sensitivity and specificity. To detect whether there was difference in methylation of IFI44L between SLE, DLE and HC, we conducted HRM-qPCR on 50 SLE, 51 DLE and 50 HC samples to determine the methylation level of the IFI44L promoter region (Fig. 1). We used 25% methylation as threshold of SLE, which has been shown to be optimal in our previous study(13). The sample with lower than 25% is positive and higher than 25% is negative. The positive rate of SLE, DLE and HC is 96.00%, 27.45% and 2.00%. The negative rate of SLE, DLE and HC is 4.00%, 72.55%, and 98.00%. We calculated the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) to evaluate the diagnostic value of the methylation of IFI44L for diagnosing SLE from DLE or HC. The sensitivity was 96.00%, specificity was 72.55%, PPV was 77.42% and NPV was 94.87% when 25% methylation as cutoff value for distinguishing SLE from DLE (Table 2).
Fig. 1.
a. Melting curves of 10 SLE samples when using the melting curve of 25% methylation standard as the cut-off value. b. Melting curves of 10 DLE samples when using the melting curve of 25% methylation standard as the cut-off value. c. Melting curves of 10 HC samples when using the melting curve of 25% methylation standard as the cut-off value.
Table 2.
The diagnostic value of the methylation of IFI44L for diagnosing SLE from DLE or HC.
| HRM-qPCR | ||||||
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| SLE | 48 | 2 | Sensitivity | Specificity | PPV | NPV |
| DLE | 14 | 37 | 96.00 | 72.55 | 77.42 | 94.87 |
| HC | 1 | 49 | 96.00 | 98.00 | 97.96 | 96.08 |
HRM-qPCR, High resolution melting quantitative Polymerase chain reaction; SLE, systemic lupus
Erythematosus; DLE, discoid lupus erythematosus; HC, healthy control; PPV, Positive predictive value; NPV, Negative predictive value.
3.2. Expression of IFN-a1 in SLE serum is higher than DLE and HC
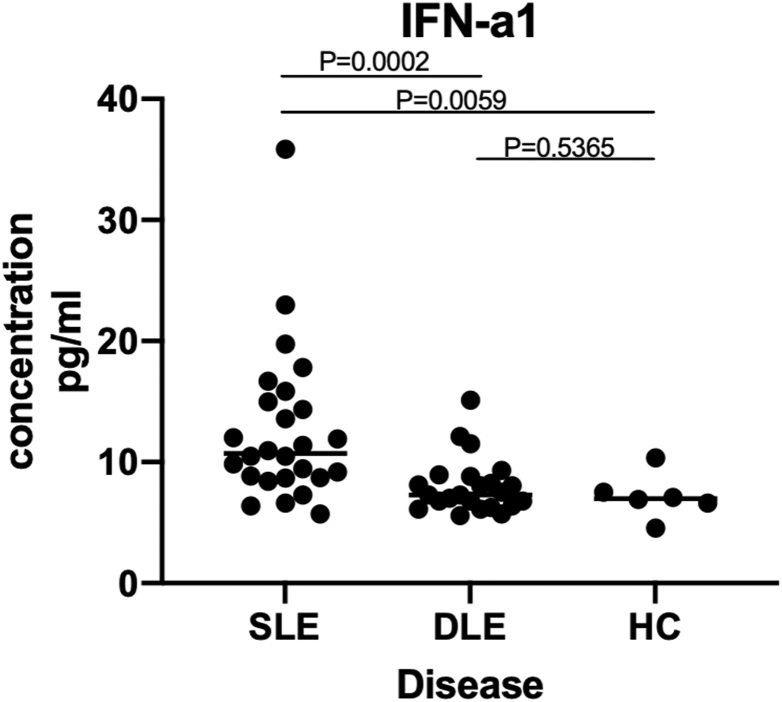
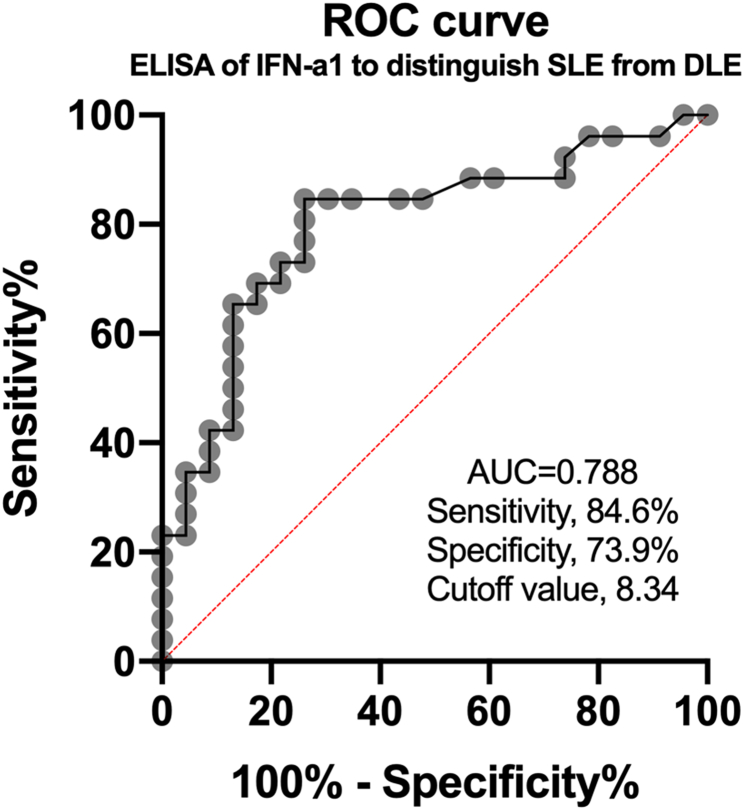
IFI44L is the regulator of IFN. Previous studies repeatedly confirmed that type-1 IFN level was upregulated in SLE [15]. To determine whether IFN expression is different in SLE and DLE, we detected the expression levels of IFN-a1 in serum of SLE, DLE and HC, all patients and HCs were matched for age and sex (Table 1). The serum concentration of IFN-a1 in patients with SLE was significantly higher than in the DLE (12.63 ± 6.38 pg/mL vs 7.99 ± 2.28 pg/mL, P < 0.05) and HC (12.63 ± 6.38 pg/mL vs 7.17 ± 1.86 pg/mL, P < 0.05). But the serum level of IFN-a1 was not significantly different between DLE and HC (7.99 ± 2.28 pg/mL vs 7.17 ± 1.86 pg/mL, P = 0.5365) (Table 3) (Fig. 2). The area under the receiver operating characteristic (ROC) curve (AUC) was calculated, which was 0.799(Fig. 3). And the maximum value of Youden’s index was used as cut-off point, taking 8.34 pg/mL as the cutoff value, if serum IFN-a1 is higher than 8.34 pg/mL, it is diagnosed as SLE, and if it is less than this value, it is diagnosed as DLE. Sensitivity and specificity are 84.6% and 73.9% respectively.
Table 3.
Serum concentration of IFN-γ compared between SLE, DLE and HCa.
| Disease | N | IFN-a1 (pg/ml) | SD | Minimum (pg/ml) | Maximum (pg/ml) |
|---|---|---|---|---|---|
| SLE | 26 | 12.63 | 6.38 | 6.37 | 35.85 |
| DLE | 23 | 7.99 | 2.28 | 5.72 | 15.12 |
| HC | 6 | 7.17 | 1.86 | 4.56 | 10.34 |
SLE, systemic lupus erythematosus; DLE, discoid lupus erythematosus; HC, healthy controls.
Fig. 2.
Scatter plots of IFN-a1 levels in serum of patients with SLE, DLE and HC. IFN-a1 levels in SLE patients were significantly higher than those in DLE and HC.
Fig. 3.
Receiver operating characteristic (ROC) curves for the serum IFN-a1 levels in patients with SLE compared with DLE.
4. Discussion
In this study, we examined IFI44L methylation level in SLE, DLE and HC by HRM-qPCR assay that we established previously. It was found that IFI44L methylation level was not significantly reduced in DLE, which is similar with HC, while significantly decreased in SLE. The expression levels of IFN-a1 in serum of patients with SLE, DLE and HC were then measured. The results showed that compared with DLE and HC, the IFN-a1 in serum of SLE patients was significantly increased, which may explain why SLE affects the multiple systems.
To investigate the effects of serological changes, disease activity and treatment on methylation and ELISA results, we collected clinical data on 40 SLE and 51 DLE samples for HRM-qPCR and 20 SLE and 20 DLE samples for ELISA. Clinical data included urine protein, Antinuclear antibody (ANA), anti-Smith antibody (Sm), anti-double-stranded DNA antibody (dsDNA), and use of glucocorticoids. Then we analyzed the effect of clinical data on the results of HRM-qPCR or ELISA. The results of showed that there was no significant influence of urine protein, Sm, dsDNA, glucocorticoids on the results of HRM-qPCR and ELISA. However, decreased methylation of IFI44L appeared to be parallel with ANA, possibly because SLE was more likely to present ANA positive than DLE patients (Table S1) [16].
LE is an autoimmune disease with strong heterogeneity. Different subtypes of LE have different prognoses, SLE can damage the nervous system, blood system and other systems and even cause death [17]. Early correct diagnosis and timely treatment are crucial for the prognosis of LE. There are several diagnostic criteria for SLE, while the diagnosis of CLE mainly relies on clinical skin lesion characteristics, history, and biopsy. Jin H etc. compared three sets of classification criteria for SLE, such as the 1997 American College of Rheumatology, in distinguishing SLE from CLE. They found none of them can accurately distinguish due to low specificity [2]. It is reported that fewer than 5% of DLE patients can progress into SLE [18]. A study showed the level of gene expression which is related to IFN correlates with cutaneous disease activity [19], and IFI44L is an IFN regulatory protein. Therefore, we supposed down-regulated methylation of IFI44L may work as an indicator of the progression of CLE into SLE, which requires more randomized controlled trial (RCT) and case follow-up. However, the accuracy of methods for detecting serum IFN-a1 levels may reduce the sensitivity and specificity of distinguishing between SLE and DLE, Mathian A etc. established an ultrasensitive single-molecule array digital immunoassay for detecting serum IFN- a1 level and found that abnormal serum IFN-a1 levels were associated with short-term recurrence [20]. Our previous work also demonstrated that reduced methylation levels of IFI44L seem to be associated with renal damage [12]. Both IFI44L methylation and IFN-a1 have the potential to work as prognostic indicators.
Although existing laboratory indicators, such as ANA and dsDNA, can also help in differentiating diagnoses CLE and SLE [21]. In clinical practice, some patients’ autoantibodies appear very late, or even do not appear in the course of disease. While changes in DNA methylation often occur early in the course of disease, prior to the general serological indicators [22]. IFI44L hypomethylation and serum levels of IFN-a1 may work as predictive biomarkers to distinguish SLE from DLE, which may guide follow-up and help improve the prognosis of LE patients.
5. Conclusions
In summary, we verified that the DNA methylation of IFI44L promoter region of SLE is lower than that of DLE and HC, in contrast, the serum level of IFN-a1 is higher than that of DLE and HC, which may be used as biomarkers for SLE and DLE diagnosis.
Author statements
Qianjin Lu and Ming Zhao gave the research idea and design the experiment. Bo Zhang and Tian Zhou conducted experiments. Haijing Wu conducted the statistical analyses. Bo Zhang an initial paper draft and Qianjin Lu and Ming Zhao revised it. Then all the authors contributed to preparing the final version and agreed to published it.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We acknowledge for the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2020-RC320-003), National Natural Science Foundation of China (No. 81874243, No. 81861138016, No. 81830097, No.82030097).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtauto.2021.100092https://doi.org/10.1016/j.jtauto.2021.100092.
Contributor Information
Ming Zhao, Email: zhaoming307@csu.edu.cn.
Qianjin Lu, Email: qianlu5860@pumcderm.cams.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Jin H., Zhang G., Zhou Y., Chang C., Lu Q. Old lines tell new tales: blaschko linear lupus erythematosis. Autoimmun. Rev. 2016;15:291–306. doi: 10.1016/j.autrev.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 2.Jin H., Huang T., Wu R., Zhao M., Wu H., Long H., Yin H., Liao J., Luo S., Liu Y., Zhang Q., Zhang P., Tan Y., Luo S., Huang X., Deng Y., Liao W., Duan L., Chen J., Zhou Y., Yin J., Qiu H., Yuan J., Wang Z., Li M., Wu X., Chen L., Cai L., Huang C., Li Q., Tang B., Yu B., Li X., Gao X., Hu Y., Ren X., Xue H., Wei Z., Chen J., Li F., Ling G., Luo H., Zhao H., Yang S., Cui Y., Lin Y., Yao X., Sun L., Guo Q., Fang H., Zeng K., Deng D., Zhang J., Li Y., Pu X., Liao X., Dang X., Huang D., Liang Y., Sun Q., Xie H., Zeng L., Huang C., Diao Q., Tao J., Yu J., Li Z., Xu H., Li H., Lai W., Liu X., Wu J., Li T., Lei T., Sun Q., Li Y., Zhang G., Huang X., Lu Q. A comparison and review of three sets of classification criteria for systemic lupus erythematosus for distinguishing systemic lupus erythematosus from pure mucocutaneous manifestations in the lupus disease spectrum. Lupus. 2020;29:1854–1865. doi: 10.1177/0961203320959716. [DOI] [PubMed] [Google Scholar]
- 3.Wieczorek I., Propert K., Okawa J., Werth V. Systemic symptoms in the progression of cutaneous to systemic lupus erythematosus. JAMA dermatology. 2014;150:291–296. doi: 10.1001/jamadermatol.2013.9026. [DOI] [PubMed] [Google Scholar]
- 4.Stec-Polak M., Matyja-Bednarczyk A., Wojas-Pelc A., Pastuszczak M. Higher specificity of the new EULAR/ACR 2019 criteria for diagnosing systemic lupus erythematosus in patients with biopsy-proven cutaneous lupus. Clin. Exp. Rheumatol. September 2019 doi: 10.55563/clinexprheumatol/syxbuz. [DOI] [PubMed] [Google Scholar]
- 5.Zhou W., Wu H., Zhao M., Lu Q. New insights into the progression from cutaneous lupus to systemic lupus erythematosus. Expet Rev. Clin. Immunol. 2020;16:829–837. doi: 10.1080/1744666X.2020.1805316. [DOI] [PubMed] [Google Scholar]
- 6.Hersh A., Arkin L., Prahalad S. Immunogenetics of cutaneous lupus erythematosus. Curr. Opin. Pediatr. 2016;28:470–475. doi: 10.1097/MOP.0000000000000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berthier C., Tsoi L., Reed T., Stannard J., Myers E., Namas R., Xing X., Lazar S., Lowe L., Kretzler M., Gudjonsson J., Kahlenberg J. Molecular profiling of cutaneous lupus lesions identifies subgroups distinct from clinical phenotypes. J. Clin. Med. 2019;8 doi: 10.3390/jcm8081244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Méndez-Flores S., Furuzawa-Carballeda J., Hernández-Molina G., Ramírez-Martinez G., Regino-Zamarripa N., Ortiz-Quintero B., Jiménez-Alvarez L., Cruz-Lagunas A., Zúñiga J. MicroRNA expression in cutaneous lupus: a new window to understand its pathogenesis. Mediat. Inflamm. 2019;2019:5049245. doi: 10.1155/2019/5049245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hile G., Kahlenberg J. Immunopathogenesis of skin injury in systemic lupus erythematosus. Curr. Opin. Rheumatol. 2020;332(2) doi: 10.1097/BOR.0000000000000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maz M., Michelle Kahlenberg J. Cutaneous and systemic connections in lupus. Curr. Opin. Rheumatol. 2020;32:583–589. doi: 10.1097/BOR.0000000000000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambers W., de Leeuw K., Doornbos-van der Meer B., Diercks G., Bootsma H., Westra J. Interferon score is increased in incomplete systemic lupus erythematosus and correlates with myxovirus-resistance protein A in blood and skin. Arthritis Res. Ther. 2019;21:260. doi: 10.1186/s13075-019-2034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao M., Zhou Y., Zhu B., Wan M., Jiang T., Tan Q., Liu Y., Jiang J., Luo S., Tan Y., Wu H., Renauer P., Del Mar Ayala Gutiérrez M., Castillo Palma M., Ortega Castro R., Fernández-Roldán C., Raya E., Faria R., Carvalho C., Alarcón-Riquelme M., Xiang Z., Chen J., Li F., Ling G., Zhao H., Liao X., Lin Y., Sawalha A., Lu Q. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann. Rheum. Dis. 2016;75:1998–2006. doi: 10.1136/annrheumdis-2015-208410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B., Liu L., Zhou T., Shi X., Wu H., Xiang Z., Zhao M., Lu Q. A simple and highly efficient method of IFI44L methylation detection for the diagnosis of systemic lupus erythematosus. Clinical immunology (Orlando, Fla. 2020;221:108612. doi: 10.1016/j.clim.2020.108612. [DOI] [PubMed] [Google Scholar]
- 14.Huang W., Tung S., Chen Y., Chen P., Chu P. IFI44L is a novel tumor suppressor in human hepatocellular carcinoma affecting cancer stemness, metastasis, and drug resistance via regulating met/Src signaling pathway. BMC Canc. 2018;18:609. doi: 10.1186/s12885-018-4529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niewold T. Interferon alpha as a primary pathogenic factor in human lupus. J. Interferon Cytokine Res. : the official journal of the International Society for Interferon and Cytokine Research. 2011;31:887–892. doi: 10.1089/jir.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao M., Zhou Y., Zhu B., Wan M., Jiang T., Tan Q., Liu Y., Jiang J., Luo S., Tan Y., Wu H., Renauer P., Del Mar Ayala Gutierrez M., Castillo Palma M.J., Ortega Castro R., Fernandez-Roldan C., Raya E., Faria R., Carvalho C., Alarcon-Riquelme M.E., Xiang Z., Chen J., Li F., Ling G., Zhao H., Liao X., Lin Y., Sawalha A.H., Lu Q. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann. Rheum. Dis. 2016;75:1998–2006. doi: 10.1136/annrheumdis-2015-208410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo S., Long H., Lu Q. Recent advances in understanding pathogenesis and therapeutic strategies of Systemic Lupus Erythematosus. Int. Immunopharm. 2020;89:107028. doi: 10.1016/j.intimp.2020.107028. [DOI] [PubMed] [Google Scholar]
- 18.Li Q., Wu H., Liao W., Zhao M., Chan V., Li L., Zheng M., Chen G., Zhang J., Lau C., Lu Q. A comprehensive review of immune-mediated dermatopathology in systemic lupus erythematosus. J. Autoimmun. 2018;93:1–15. doi: 10.1016/j.jaut.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Braunstein I., Klein R., Okawa J., Werth V. The interferon-regulated gene signature is elevated in subacute cutaneous lupus erythematosus and discoid lupus erythematosus and correlates with the cutaneous lupus area and severity index score. Br. J. Dermatol. 2012;166:971–975. doi: 10.1111/j.1365-2133.2012.10825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathian A., Mouries-Martin S., Dorgham K., Devilliers H., Yssel H., Garrido Castillo L., Cohen-Aubart F., Haroche J., Hié M., Pineton de Chambrun M., Miyara M., Pha M., Rozenberg F., Gorochov G., Amoura Z. Ultrasensitive serum interferon-α quantification during SLE remission identifies patients at risk for relapse. Ann. Rheum. Dis. 2019;78:1669–1676. doi: 10.1136/annrheumdis-2019-215571. [DOI] [PubMed] [Google Scholar]
- 21.Patsinakidis N., Gambichler T., Lahner N., Moellenhoff K., Kreuter A. Cutaneous characteristics and association with antinuclear antibodies in 402 patients with different subtypes of lupus erythematosus. J. Eur. Acad. Dermatol. Venereol. : JEADV. 2016;30:2097–2104. doi: 10.1111/jdv.13769. [DOI] [PubMed] [Google Scholar]
- 22.Vrba L., Futscher B. DNA methylation changes in biomarker loci occur early in cancer progression. F1000Research. 2019;8:2106. doi: 10.12688/f1000research.21584.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.