See “Intestinal host response to SARS-CoV-2 infection and COVID-19 outcomes in patients with gastrointestinal symptoms,” by Livanos AE, Jha D, Cossarini F, et al, on page 2435.
As we eclipse year 1 of the severe acute respiratory syndrome novel coronavirus-2 (SARS-CoV-2) pandemic, our understanding of this virus is only beginning to unfold despite an exponentially growing body of literature. Early descriptions of the novel respiratory illness coronavirus disease 2019 (COVID-19) were quickly followed by reports of a multisystem disease.1 Gastrointestinal (GI) symptoms are frequently reported in patients with COVID-19, which raised questions of gut infection and the possibility of fecal–oral transmission. Indeed, viral RNA was detected in stool of infected patients by quantitative reverse transcriptase polymerase chain reaction, which frequently remained positive beyond the duration of positivity of their nasopharyngeal samples.2, 3, 4 Multiple groups have reported GI epithelial coexpression of the host receptor ACE2 and TMPRSS2, a protease enabling cell entry through cleavage of the eponymous spike protein.5, 6, 7, 8 In line with the susceptibility of the GI epithelium to SARS-CoV-2 entry, viral RNA by in situ hybridization9 and viral nucleocapsid protein (NP) by immunofluorescence have been detected in infected patient intestinal biopsies,2 whereas viral particles have been demonstrated by transmission electron microscopy.9 Moreover, recent studies using ex vivo enteroid or organoid models convincingly demonstrated that GI epithelium is permissive to SARS-CoV-2 infection.6 , 9, 10, 11 Notably, human intestinal enteroids inoculated with the virus or nasopharyngeal aspirates from infected patients showed intracellular NP and increasing viral titers over time, indicating productive viral replication within intestinal epithelium.9, 10, 11 Enteroids also exhibited double-stranded RNA, a requisite intermediate produced during the replication of this positive-sense RNA virus.11 What happens in vivo remains unclear owing to the inherent logistical challenges of studying large cohorts of patients infected with this novel pathogen. Beyond the possibility of GI infection, viral interactions with GI tract tissues and the immune system, and how this influences clinical outcomes in COVID-19 remain largely unknown.
In this issue of Gastroenterology, Livanos et al12 attempt to correlate clinical observations in patients with COVID-19 with immunologic changes within the GI tract on histologic, transcriptomic, and proteomic levels. The authors analyzed endoscopic biopsy specimens from COVID-19–afflicted patients, finding no gross abnormalities when compared with matched controls.12 Although they found angiotensin-converting enzyme 2 (ACE2) diffusely expressed throughout the GI tract, viral NP was found more prominently in the ileum versus duodenum in the limited number of specimens studied.12 They attribute this finding to increased MUC2+ goblet cell prevalence in distal small intestine, which colocalized with NP immunostaining, and their transmission electron microscopy images showing viral particles present primarily in exit vesicles of goblet cells.12 This finding may corroborate findings from bronchial air–liquid interface cultures demonstrating tropism for goblet cells,13 but were nevertheless surprising given relatively low ACE2 expression in intestinal goblet cell clusters,5 evidence of goblet cell depletion in an infected patient’s colonic biopsies,9 and data from human intestinal organoids demonstrating enterocytes were susceptible to infection whereas goblet cells were not.6 , 11 Livanos et al12 also find evidence of NP staining in a nongoblet crypt base cell population but do not discern this population as Paneth, stem cells, or another population.12 This raises additional questions of cellular tropism and host entry determinants because enterocytes in the villus compartment express the highest levels of ACE2.
The authors also performed a mass cytometry immunophenotypic analysis of endoscopic biopsies and blood samples from these patients.12 They found that specific dendritic cell populations were diminished in the lamina propria of infected patients, possibly supporting some alteration in antigen presentation.12 Bulk RNA sequencing of these tissues found decreased expression of several inflammatory pathways in the lamina propria and a trend toward the up-regulation of antiviral pathway signatures within the epithelial compartment.12
Notably, Vero E6 cells inoculated with homogenized biopsy tissue obtained from patients did not show evidence of productive infection.12 This finding may simply result from sampling or technical error, but it may also reflect the biology of SARS-CoV-2 in the gut. There remains a striking lack of evidence of infectious virion isolation from the gut, especially given the many reports of viral RNA shed in patient stool, demonstrations of viral proteins in epithelial cells, and the permissiveness of gut epithelium organoids to SARS-CoV-2 infection ex vivo. Perhaps there are specific properties of GI epithelium or luminal contents impeding SARS-CoV-2 isolation in culture from presumably infectious tissue. A low multiplicity of infection in the biopsied samples, an explanation offered by the authors, seems plausible. The patients had positive nasopharyngeal swab tests, but upon review of the endoscopic biopsy timing relative to symptom onset and nasopharyngeal swab positivity, it is fair to wonder how infectious these patients were when biopsied. Only 3 of the patients had any GI symptoms at the time of the endoscopy, 37% had a negative nasopharyngeal swab test most recently before endoscopy, and the interval between symptom onset and biopsy extended beyond 100 days in 1 case.12 Data are limited, but these constraints are understandable given the prudent restrictions on nonurgent endoscopic procedures on patients with COVID-19 implemented by many hospitals during the pandemic.
Livanos et al12 also correlated patients having COVID-19 with GI symptoms of diarrhea, nausea, or vomiting at the time of hospital admission with a nearly 50% decreased odds of severe disease or mortality when compared to patients without GI symptoms. A similar analysis applied to an independent Italian cohort found patients with diarrhea had significantly lower rates of mortality or intensive care unit admission.12 Although the authors attribute this finding to immunomodulation, perhaps these patients differed in other ways as well (eg, presented earlier owing to more troubling or obvious GI symptoms). The authors partially dispel this explanation, reporting that rates of respiratory symptoms and oxygen requirements were similar regardless of GI symptoms and few patients overall had mild disease. They also found no significant difference in nasopharyngeal viral loads to explain these disparate outcomes.12 In support of an altered immune response in GI patients, they analyzed levels of circulating cytokines and found that those with GI symptoms had lower levels of IL-6 and IL-8, among other cytokines associated with poor outcomes in COVID-19.12 , 14 , 15
In its totality, this study identifies less severe clinical outcomes in patients with COVID-19 with GI symptoms across multiple cohorts, in agreement with prior studies.16 , 17 The authors attribute this observation to an attenuated immune response related to COVID-19 involvement of the GI tract (Figure 1 ). It is known that type III interferons (IFNs) are important for mucosal immunity and induce less systemic inflammation and tissue destruction compared with type I IFNs.18 The absence of IFN III in a genetically deficient colon epithelial line culture resulted in dramatically increased SARS-CoV-2 replication.19 A skew toward type III IFN was demonstrated in enteroids infected with SARS-CoV-2,11 and may explain the plausible link between mucosal immune regulation and systemic viral sequelae. In the lung, type III IFNs are more prevalent in lower airways, participate in mucosal injury along with type I IFNs and may impair epithelial regeneration.20 , 21 Further studies will need to investigate if similar effects occur in intestinal epithelium, because Livanos et al found increased type I and II IFNs in the epithelium despite no evidence of gross or histologic injury and downregulation of various cytokines in serum.12
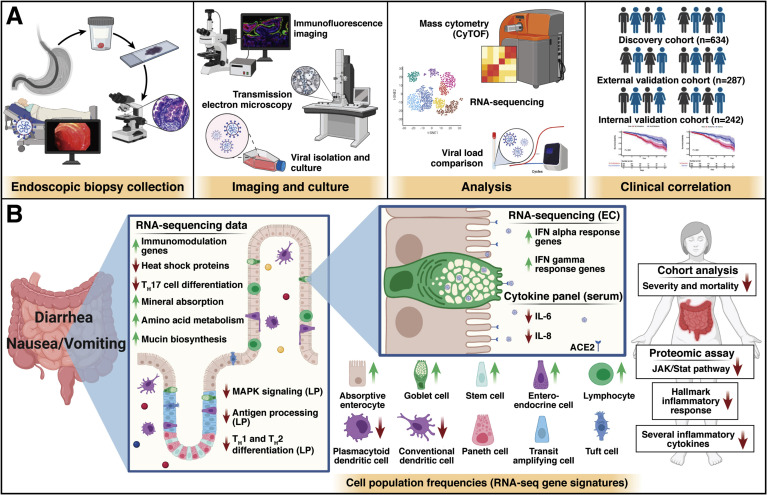
Figure 1.
Patients with COVID-19 and gastrointestinal (GI) symptoms exhibit an attenuated immune response. (A) Schematic of workflow adapted from Livanos et al. Endoscopic biopsies from infected and noninfected patients were analyzed histologically, homogenized to inoculate Vero E6 cells in culture, and were analyzed by mass cytometry. Large clinical cohorts demonstrated improved survival in patients with GI symptoms. (B) The cellular, transcriptomic, and proteomic changes observed in patients with COVID-19 with GI manifestations correlated with clinical observations of decreased severity and improved survival. EC, epithelial compartment; LP, lamina propria.
Major questions remain regarding how SARS-CoV-2 behaves in the GI tract, including the viability of the SARS-CoV-2 viral envelope in the harsh biochemical environment of the GI lumen.6 It remains unclear which cell types are actually infected in vivo. Other questions include the interaction of the virus with disease states that modify susceptibility, including potential infection via susceptible metaplastic cells in the esophagus, thus bypassing the gastric acidity or bile emulsification in those patients with Barrett’s esophagus.22 The rapid global spread of this virus has required equally rapid dissemination of biomedical information, but we are hopefully now witnessing the shift toward more rigorous study of SARS-CoV-2. Moving forward, as new variants arise and the population becomes increasingly vaccinated, interplay of the virus, GI epithelium, and the immune system will remain a topic of great interest to potentially inform novel anti-viral therapeutics or development of an oral vaccine strategy.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding K.S. Yan is supported by BWF Career Award for Medical Scientists, Irma T. Hirschl Trust, Louis V. Gerstner Foundation, Lisa Dean Moseley Foundation, NIH 1DP2DK128801 and 1R01AG067014. J.Miller is supported by NIH 5T32DK083256-12.
References
- 1.Gupta A., Madhavan M.V., Sehgal K., et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao F., Tang M., Zheng X., et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W., Xu Y., Gao R., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung K.S., Hung I.F.N., Chan P.P.Y., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J.J., Kopetz S., Vilar E., et al. Relative abundance of SARS-CoV-2 entry genes in the enterocytes of the lower gastrointestinal tract. Genes (Basel) 2020:11. doi: 10.3390/genes11060645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zang R., Gomez Castro M.F., McCune B.T., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang W., Feng Z., Rao S., et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 8.Zou X., Chen K., Zou J., et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han Y., Duan X., Yang L., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamers M.M., Beumer J., van der Vaart J., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J., Li C., Liu X., et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 12.Livanos A.E., Jha D., Cossarini F., et al. Intestinal host response to SARS-CoV-2 infection and COVID-19 outcomes in patients with gastrointestinal symptoms. Gastroenterology. 2021;160:2435–2450. doi: 10.1053/j.gastro.2021.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osan JK, Talukdar SN, Feldmann F, et al. Goblet cell hyperplasia increases SARS-CoV-2 infection in COPD. bioRxiv 2020 Nov 12 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 14.Zhou F., Yu T., Du R., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Valle D.M., Kim-Schulze S., Huang H.H., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nobel Y.R., Phipps M., Zucker J., et al. Gastrointestinal symptoms and coronavirus disease 2019: a case-control study from the United States. Gastroenterology. 2020;159:373–375.e2. doi: 10.1053/j.gastro.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aghemo A., Piovani D., Parigi T.L., et al. COVID-19 digestive system involvement and clinical outcomes in a large academic hospital in Milan, Italy. Clin Gastroenterol Hepatol. 2020;18:2366–2368.e3. doi: 10.1016/j.cgh.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreakos E., Zanoni I., Galani I.E. Lambda interferons come to light: dual function cytokines mediating antiviral immunity and damage control. Curr Opin Immunol. 2019;56:67–75. doi: 10.1016/j.coi.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanifer M.L., Kee C., Cortese M., et al. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32:107863. doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broggi A., Ghosh S., Sposito B., et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Major J., Crotta S., Llorian M., et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin R.U., Brown J.W., Li Q.K., et al. Tropism of SARS-CoV-2 for Barrett's esophagus may increase susceptibility to developing COVID-19. Gastroenterology. 2021;160:2165–2168. doi: 10.1053/j.gastro.2021.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]