Abstract
Circular RNAs (circRNAs) are a class of non-coding RNAs with covalently closed single-stranded structures lacking 5′–3′ polarity and a polyadenine tail. Over recent years, a growing body of studies have been conducted to explore the roles of circRNAs in human diseases. Systemic lupus erythematosus (SLE) is a severe autoimmune disorder characterized by the presence of autoantibodies and excessive inflammation, which impact multiple organs. Recent advances have begun to shed light on the roles of circRNAs in SLE, providing fresh insights into the pathogenesis of SLE and the latent capacity for translation into clinical applications. Here, we briefly introduce these “star molecules” and summarize their roles in SLE. In addition, we outline the limitations of the current studies and raise prospects for future research.
Keywords: circular RNA, systemic lupus erythematosus, expression, function, mechanism
Graphical abstract
This review provides an overview of the current knowledge of circRNA biology, with a particular emphasis on their roles and future perspectives in SLE. The research ideas about the expression, transmission, function, and mechanism of circRNAs are not only suitable for SLE but also generally suitable for other autoimmune diseases.
Introduction
Systemic lupus erythematosus (SLE) is a severe autoimmune disorder characterized by the presence of autoantibodies and excessive inflammation, which impacts multiple organs.1,2 This disease typically affects women of childbearing age, initiating from the interactions of genetic susceptibility, environmental stimuli, and hormone abnormalities.3 The pathogenesis of SLE is extraordinarily complex, involving the dysfunction of T and B lymphocytes and other immune cell subsets, which results in the production of large amounts of pathogenic autoantibodies and inflammatory cytokines, leading to the damage of organs accompanied by a variety of clinical manifestations.4,5
Circular RNAs (circRNAs) are a class of non-coding RNAs (ncRNAs) with covalently closed single-stranded structures lacking 5′–3′ polarity and a polyadenine tail.6,7 Although these molecules were discovered many years ago, their landscape was largely overlooked until the recent boom in research on ncRNA molecules.6 In fact, circRNAs were initially disregarded as artifacts of pre-mRNA splicing. However, advances in high-throughput RNA sequencing and bioinformatics have revealed large numbers of circRNAs in humans, thus re-igniting the interest of researchers in these molecules.7 Recently, an increasing number of studies have been conducted to clarify the role of RNA in human diseases, resulting in significant advances in this field.8, 9, 10
In this review, we provide an overview of the current knowledge of circRNA biology, with a particular emphasis on SLE. We also summarize the limitations of the previously reported studies and shed light on future research directions.
Characterization of circRNAs
Biogenesis and degradation of circRNAs
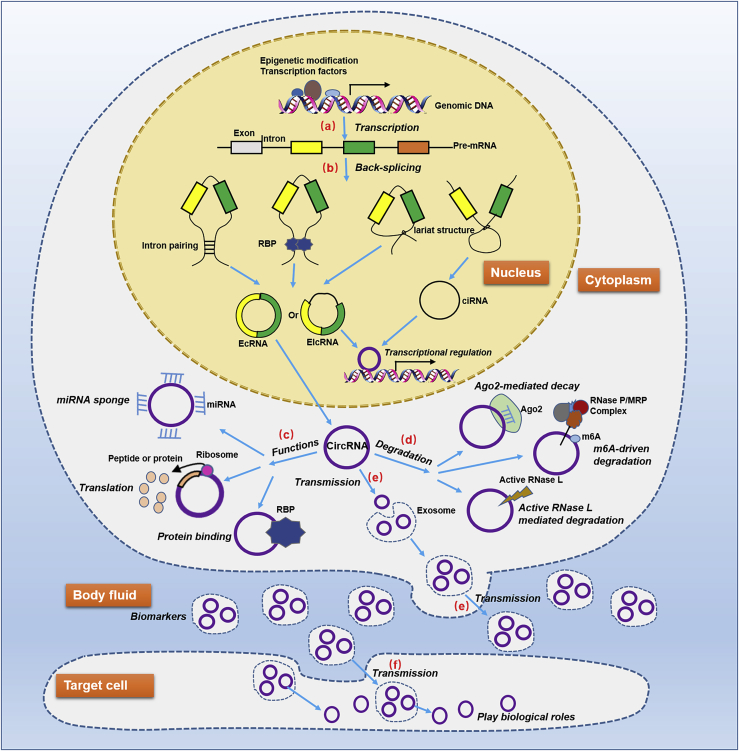
circRNAs are primarily generated by the back-splicing of pre-mRNA transcripts, a process in which a downstream 5′ splice site is connected to a 3′ splice site and which competes with canonical mRNA splicing.11 circRNAs, which are loop structures without free ends, are highly stable molecules that are resistant to exonucleases (RNase R).12 Based on biogenesis from different genomic regions, circRNAs can be classified into three main types: exonic circRNAs (EcRNAs), which contain one or more exons derived from alternative splicing and account for over 80% of all known circRNAs;13 intronic circRNAs (ciRNAs), which only contain introns and may rely on a consensus motif containing a specific base distribution;14 and exon-intron circRNAs (EIcRNAs), which are formed by both exon and intron elements (Figure 1).15
Figure 1.
Generation, function, transmission, and degradation of circRNAs
This process involves several important events. (A) The transcription of pre-mRNAs, which is modulated by epigenetic modifications and transcription factors. (B) The generation of circRNAs, which are a consequence of pre-mRNA back-splicing events such as lariat-driven circularization, intron pairing-driven circularization, and RBP-mediated circularization. (C) The biological functions of circRNAs, which are based on a variety of molecular regulatory mechanisms, including miRNA sponging, protein binding, transcription or splicing regulation, and peptide or protein coding. (D) The degradation of circRNAs, which is related to several molecular biological processes, such as Ago2-mediated RNA decay, m6A-driven circRNA degradation, and active RNase L triggered circRNA degradation. (E) The transmission of circRNAs from cells to body fluid, which endows them with the latent capacity to be biomarkers. (F) The transmission of circRNAs from body fluid to target cells, which is an important basis for mediating biological processes of target cells.
Since circRNAs are primarily generated by the back-splicing of pre-mRNAs, their expression is also determined by the transcription levels of pre-mRNAs, which depend on transcription factor activity and epigenetic modifications, such as methylation, acetylation, and ncRNAs.16, 17, 18, 19 circRNA expression is also modulated by the regulation of circularization, which involves three main hypothetical models: lariat-driven circularization, in which the formation of a lariat structure brings the upstream splice acceptor and downstream donor into close proximity before splicing; intron-pairing-driven circularization, in which the inverted complementary repeat sequences combine into RNA double strands and are subjected to alternative splicing; and RNA binding protein (RBP)-mediated circularization, in which RBPs join the upstream and downstream flanking introns to facilitate pre-mRNA circularization back-splicing (Figure 1).13,20, 21, 22 The type of circRNAs generated depends mainly on the elements involved in the three hypothetical models and the splicing pattern.
The current understanding of circRNA degradation is limited, and only several circRNA degradation modes have been reported (Figure 1). The first is Argonaute 2 (Ago2)-mediated RNA decay, in which small interfering RNAs (siRNAs) or microRNAs (miRNAs) are assembled into the RNA-mediated silencing complex RISC by Ago2 and guide RISC to cleave and degrade the circRNAs.10,23 Another mode is N6-methyladenosine (m6A)-driven circRNA degradation, in which mA-containing circRNA associates with YTHDF2 in an HRSP12-dependent manner and is selectively cleaved by RNase P/MRP complex.24 A third way is the active RNase L-triggered circRNA degradation, in which global circRNA degradation by RNase L is triggered by both poly(I:C) and viral double-stranded RNAs (dsRNAs).25
Distribution of circRNAs
circRNAs are generated in the nucleus and can appear in the nucleus and/or cytoplasm. In general, EcRNAs are located in the cytoplasm, while ciRNAs and EIcRNAs are more likely to exist in the nucleus.10,26,27 The mechanism underlying this phenomenon is not entirely clear. One possible mechanism is the length-dependent evolutionarily conserved pathway, in which the lengths of circRNAs are somehow involved in the mode of their export by association with DDX39B or DDX39A.28 In addition, RNA modification, such as m6A modification, widely occurs on circRNAs, and m6A may possibly affect RNA export by the m6A-binding protein YTHDC1, which may also be a possible way of regulating nuclear export of circRNAs; this remains to be determined.7
Recently, studies have demonstrated the enrichment and stability of circRNAs in exosomes; hence, these molecules are abundant in body fluids and able to be transmitted throughout the body.29, 30, 31 Therefore, the exosomes secreted by pathological cells can enter the body fluids and cause anomalous levels of specific circRNAs, thus providing valuable biomarkers. Similarly, exosomal circRNAs in body fluids can also integrate into target cells and exert corresponding biological roles, leading to pathological changes.29, 30, 31, 32
Biological functions of circRNAs
With advances in research, four main biological functions of circRNAs have been revealed: miRNA sponging, protein binding, transcription or splicing regulation, and peptide or protein coding (Figure 2).7, 8, 9, 10, 11 In general, the biological functions of circRNAs are closely related to their subcellular localization.
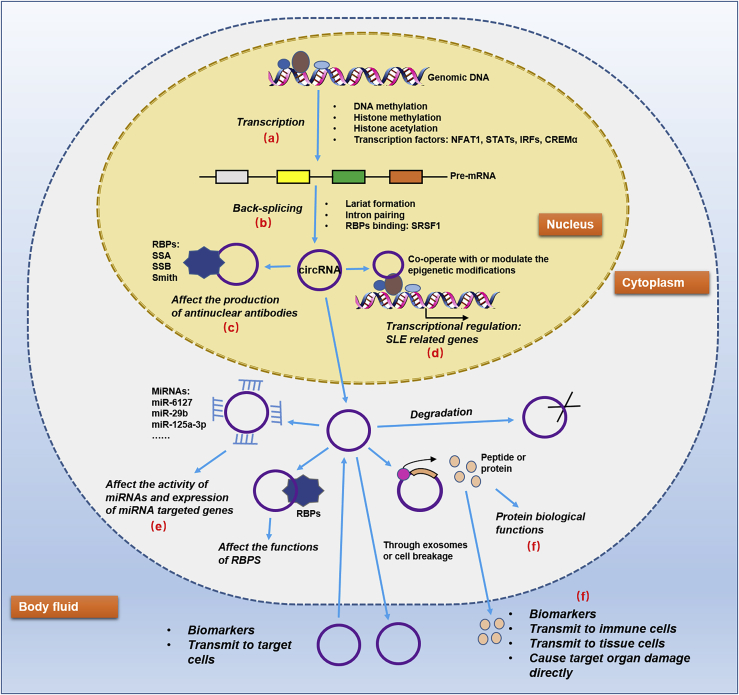
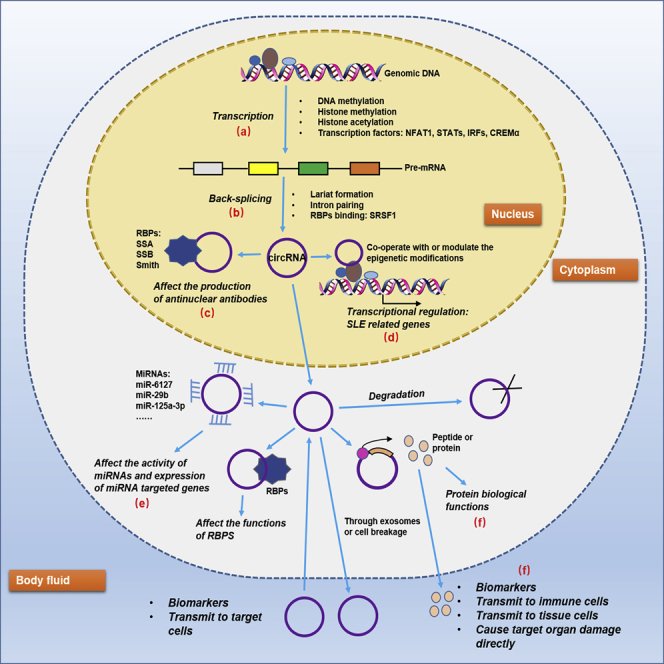
Figure 2.
Prospective regulatory network of circRNAs in SLE
There are several valuable and interesting points in the network. (A and B) The aberrant epigenetic modifications as well as anomalously expressed transcription factors and RBPs in SLE may be specific factors to explicate the dysregulation of circRNAs. (C) The binding between circRNAs and some nuclear RBPs may affect the production of antinuclear antibodies. (D) Co-operating with or modulating the epigenetic modifications of circRNAs in SLE may be responsible for the unusual level of some disease-related genes. (E) Acting as miRNA sponges is still the hottest topic on circRNAs currently. (F) Peptides or proteins encoded by circRNAs may either function in original cells or transmit to body fluid and target cells to exert roles.
The most frequently reported function of circRNAs is their capacity to act as miRNA sponges. This function is dependent on a circRNA-miRNA-mRNA regulation network, in which circRNAs may contain dozens of miRNA binding sites, thereby adsorbing target miRNAs in a manner that resembles a sponge and impairing miRNA-mediated gene suppression.33,34 For instance, the circRNA circITCH sponges miR-330-5p, and as a result upregulates the expression of miR-330-5p target genes, including SIRT6, Survivin, and SERCA2a, to ameliorate doxorubicin-induced cardiotoxicity.35 In another study, circRNA-ZNF532 was shown to regulate diabetes-induced retinal pericyte degeneration and vascular dysfunction by acting as a miR-29a-3p sponge and inducing increased expression of NG2, LOXL2, and CDK2.36 In general, the EcRNAs located in the cytoplasm exert their biological functions in this way.34,37,38
circRNAs are able to bind to specific RBPs and regulate the functions of RBPs. For example, the circRNA circFndc3b binds to the RBP Fused in the sarcoma to regulate VEGF expression and signaling, thereby regulating cardiac repair after myocardial infarction.39 The circRNA circZKSCAN1 binds competitively to FMRP to inhibit its interaction with the β-catenin-binding protein CCAR1, leading to inhibition of Wnt signaling.40
Unlike EcRNAs, ciRNAs and EIcRNAs are primarily enriched in the nucleus, increasing their probability of influencing gene expression by transcriptional regulation and splicing modification.10,13,33 For instance, circMRPS35 not only acts as a modular scaffold to recruit the histone acetyltransferase KAT7 to the promoters of FOXO1/3a genes but also directly binds to FOXO1/3a promoter regions, resulting in transcription trigger of FOXO1/3a genes.41 circRNAs such as circMbl and circSEP3 can also affect the content of their linear cognates by competing with linear splicing.42,43
Although most circRNAs lack protein-coding capacity and tend to be considered as ncRNAs, recent studies have demonstrated that several circRNAs carry open reading frames (ORFs) and may encode peptides or proteins in a cap-independent manner.12,13,44 Some peptides or proteins translated from circRNAs may modulate cellular biological processes. For example, a circRNA circβ-catenin facilitates liver cancer cell growth through activating the Wnt pathway by encoding a 370-amino acid β-catenin isoform.45 A 174 amino acid protein produced by circAKT3 competitively interacts with phosphorylated PDK1, thereby suppressing phosphorylation of AKT at the Thr308 site and acting as a negative regulator of the PI3K/AKT pathway.46 To the best of our knowledge, only circRNAs carrying exons and located in the cytoplasm have protein-encoding potential.44
circRNAs and diseases
An increasing number of studies have demonstrated that circRNAs are involved in various diseases, displaying a potentiality to serve as clinical biomarkers and therapeutic targets.47
circRNAs and cancers
The vast majority of studies on circRNAs in relation to human diseases have focused on cancers. Moreover, research in this area has covered almost all of the commonest malignant tumors. The abnormal expression of many circRNAs in the circulation or tumors of cancer patients is closely correlated with clinical factors such as tumor size, metastasis, differentiation, clinical stage, and pathological type, as well as the prognosis of patients.38,47,48 In addition, because of their stable presence in the blood, circRNAs are implicated as valuable diagnostic markers or clinical indicators of cancers.31,49,50
circRNAs also participate in various aspects of tumor biology, thus affecting malignant processes of cancers, such as metabolism, proliferation, differentiation, invasion, and metastasis, as well as angiogenesis, stemness, immune escape, and inflammation.10,33,51,52 On the basis of these features, circRNAs can function as tumor suppressors or tumor promoters in human cancers, indicating a clinical application potential of circRNA-based therapeutic strategies in cancer management.51,52
circRNAs and autoimmune diseases
Autoimmune diseases are a family of disorders originating from the pathological immune response to self-antigens, resulting in inflammation and damage to target organs. In addition to SLE, circRNAs have been reported to be aberrantly expressed and involved in the development of some other autoimmune diseases, including rheumatoid arthritis (RA), multiple sclerosis (MS), type 1 diabetes mellitus (T1DM), and Sjögren’s syndrome (SS) (Table 1). Most of these studies demonstrate that circRNAs represent potential biomarkers of autoimmune diseases. Moreover, several of these circRNAs are possible therapeutic targets due to their vital roles as disease process regulators. For example, hsa_circ_0088036 acts as a miR-140-3p sponge to upregulate SIRT1 expression and promotes the proliferation and migration of fibroblast-like synoviocytes in RA.53 In addition, ciRS-7 may reduce the inhibitory effect of miR-7 on mTOR by sponging miR-7 in RA.56
Table 1.
circRNAs in some autoimmune diseases
| circRNAs | Diseases | Samples | Expression | Roles | References |
|---|---|---|---|---|---|
| Hsa_circ_0088036 | RA | PBMCs | up | promotes the proliferation and migration of fibroblast-like synoviocytes | Zhong et al.53 |
| Hsa_circ_0002715 | RA | peripheral blood | up | correlated with SJC, TJC, disease duration, RF, ACPA, and hematologic disorder; diagnostic markers | Luo et al.54 |
| Hsa_circ_0035197 | RA | peripheral blood | up | correlated with hematologic disorder | Luo et al.54 |
| Hsa_circ_0000367 | RA | peripheral blood | up | diagnostic marker | Luo et al.54 |
| Hsa_circ_0001947 | RA | peripheral blood | up | – | Luo et al.54 |
| Hsa_circ_0000396, hsa_circ_0130438 | RA | PMBCs | down | diagnostic marker | Yang et al.55 |
| CiRS-7 | RA | PMBCs | up | diagnostic marker; regulates miR-7/mTOR axis | Tang et al.56 |
| Hsa_circ_0044235 | RA | peripheral blood | down | diagnostic marker | Luo et al.57 |
| Circ_0005402 and Circ_0035560 | MS | leukocytes | down | biomarker | Iparraguirre et al.58 |
| Hsa_circRNA_101062, hsa_circRNA_100332, hsa_circRNA_085129, hsa_circRNA_103845 | T1DM | plasma | up | – | Li et al.59 |
| Circ-IQGAP2, circ-ZC3H6 | SS | minor salivary gland; plasma exosomes | up | correlated with clinical features, serum IgG level, and MSG focus scores; diagnostic markers | Li et al.60 |
| Hsa_circRNA_001264, hsa_circRNA_104121, hsa_circRNA_045355 | SS | PBMCs | up | correlated with clinical, laboratory parameters, and disease activity index; diagnostic markers | Su et al.61 |
PBMCs, peripheral blood mononuclear cells; SJC, swollen joint count; TJC, tender joint count; RF, rheumatoid factor; ACPA, anticitrullinated protein antibodies; MS, multiple sclerosis; T1DM, type 1 diabetes mellitus; SS, Sjögren’s syndrome; IgG, immunoglobulin G; MSG, minor salivary gland.
Although studies on the biological functions and mechanisms underlying the effects of circRNAs in autoimmune diseases are finite, a considerable number of studies have revealed their effects on molecular biological processes and immune regulation of various immune cells.7,62, 63, 64 For instance, circ-ANRIL induces nucleolar stress and p53 activation, resulting in the apoptosis elevation and proliferation suppression in macrophages.65 circRNA-002178 can be delivered by exosomes into CD8 T cells to induce PD1 expression, leading to T cell exhaustion.66 circSnx5 may mediate dendritic cell (DC) activation and function via the miR-544/SOCS1 axis and regulation of PU.1 activity to modulate DC-driven immunity and tolerance.67 Circ_Malat-1 is involved in the increase in expression of immunosuppressive/inhibitory molecules in DCs, enhancement of T cell exhaustion induced by DCs, and regulatory T cell (Treg) generation promoted through indoleamine 2,3 dioxygenase signaling mediated by growth differentiation factor 15.68 These immunomodulatory effects of circRNAs underlie the ability of these molecules to participate in immune-related diseases.
circRNAs and SLE
To date, many aberrantly expressed circRNAs have been identified in SLE using microarrays and high-throughput sequencing, drawing circRNAs in SLE into the spotlight.69 Most of these circRNAs have not yet been verified, and only a few circRNAs have been studied in depth; details are presented in Table 2.
Table 2.
circRNAs in SLE
| circRNAs | Samples | Expression | Roles | References |
|---|---|---|---|---|
| Hsa_circ_0044235 | PBMCs | down | diagnostic marker; associated with anti-dsDNA and anti-ribosomal P antibodies | Luo et al.70 |
| Hsa_circ_0068367 | PBMCs | down | diagnostic marker | Luo et al.70 |
| CircPTPN22 | PBMCs | down | diagnostic marker and disease severity indicator | Miao et al.71 |
| Hsa_circ_0021372, hsa_circ_0075699 | peripheral blood | down | associated with C3 and C4 levels | Li et al.72 |
| Hsa_circ_0057762 | peripheral blood | up | positively associated with the SLEDAI-2K score; diagnostic marker | Li et al.72 |
| Hsa_circ_0003090 | peripheral blood | up | diagnostic marker | Li et al.72 |
| Hsa_circRNA_407176, hsa_circRNA_001308 | plasma, PBMCs | down | diagnostic marker | Zhang et al.73 |
| Hsa_circRNA_406567 | PBMCs | down | diagnostic marker | Zhang et al.73 |
| circRNA_002453 | Plasma | up | associated with the severity of renal involvement; diagnostic marker | Ouyang et al.74 |
| Hsa_circ_0049224, has_circ_0049220 | PBMCs | down | associated with DNMT1 expression, disease activity and clinical characteristics | Zhang et al.75 |
| Hsa_circ_0000479, | PBMCs | up | associated with clinical characteristics and therapeutic effect; diagnostic marker | Guo et al.,76 Luo et al.77 |
| Hsa_circ_0082689, hsa_circ_0082689 | PBMCs | up | associated with clinical characteristics and therapeutic effect | Luo et al.77 |
| Hsa_circ_0045272 | T cells | down | binding with hsa-miR-6127; negatively regulating apoptosis and interleukin-2 secretion of Jurkat cells | Li et al.78 |
| CircIBTK | PBMCs | down | disease marker; regulate DNA demethylation and AKT signaling pathway via binding to miR-29b | Wang et al.79 |
| Hsa_circ_0012919 | CD4+ T cells | up | associated with SLE characters; increase DNMT1 expression; regulate DNA hypomethylation and expression of CD70 and CD11a; regulate KLF13, RANTES and MDA5 by miR-125a-3p | Zhang et al.80,81 |
| dsRNA-containing circRNA | PBMCs | down | alleviate the aberrant PKR activation cascade | Kato et al.82 |
dsDNA, double-stranded DNA; SLEDAI, Lupus Erythematosus Disease Activity Index.
Expression of circRNAs in SLE
Analysis of circRNA expression in SLE may provide valuable diagnostic markers. By detecting the relative expression of circRNAs and consulting the receiver operating characteristic (ROC) curve, researchers endow circRNAs with the capacity of discriminating SLE patients from healthy individuals or patients with other diseases. For example, the area under curve (AUC) of the combination of hsa_circ_0042345 and hsa_circ_0068367 reached 0.876 (95% CI = 0.778–0.967), with a sensitivity of 70.00% and specificity of 100.00%.70 The combination of hsa_circRNA_407176, hsa_circRNA_406567, and hsa_circRNA_001308 may raise the AUC to 0.855 (95% CI = 0.748–0.962), with a sensitivity and specificity as high as 92.31% and 65.39%, respectively.73 circRNAs are also applied in combination with traditional SLE biomarkers in the diagnosis of this disease. For instance, the hsa_circ_0000479 + anti-dsDNA model can effectively distinguish SLE patients from other individuals with a sensitivity of 86.00%, a specificity of 100.00%, and an accuracy of 95.10%.77 These studies indicate that some circRNAs may be good markers for screening SLE, while some are specific markers for diagnosing SLE, similar to antinuclear antibody (ANA) and anti-double-stranded DNA (anti-dsDNA) antibody or anti-Sm antibody. Moreover, the combination of circRNA expression with traditional SLE biomarkers may improve the accuracy of diagnosis compared with either of these markers used individually.
circRNAs are also promising clinical disease indicators for SLE. Some circRNAs, such as circPTPN22, hsa_circ_0057762, hsa_circ_0049224, has_circ_0049220, circIBTK, and hsa_circ_0012919, have been found to be closely related to disease activity or SLEDAI score.72,75,79,80 Another important clinical significance of circRNAs is their relationship with different clinical manifestations, such as photosensitivity, Raynaud’s phenomenon arthritis, and lupus nephritis.75,80 Furthermore, circRNAs are more likely to display correlations with laboratory parameters, including anti-dsDNA antibody, anti-Sm antibody, anti-cardiolipin antibodies, leukocytes, platelets, hemoglobin, urinary protein, erythrocyte sedimentation rate, and complement level.75, 76, 77,79,80 Finally, efficacious clinical treatment may be associated with remarkable changes in the expression of circRNAs, indicating their potential value in the evaluation of therapeutic effect.76,79,80
Most importantly, these studies on the expression of circRNAs in SLE were performed using whole peripheral blood or partial blood components, including peripheral blood mononuclear cells (PBMCs), plasma, and T cells. The availability of the samples for testing and the stability of circRNAs in both cells and body fluids emphasize the practicability of the application of circRNAs as biomarkers of SLE.
Roles of circRNAs in SLE progression
Relatively few investigations on the biological functions of circRNAs in the onset and progression of SLE have been reported. Nevertheless, these studies also explored the roles of circRNAs in SLE from different perspectives. For example, the downregulated hsa_circ_0045272 in SLE T cells negatively regulates apoptosis and interleukin-2 secretion of T cells.78 The insufficient circIBTK level in PBMCs of SLE patients is responsible for DNA demethylation and activation of the AKT signaling pathway, in which a target miRNA of circIBTK, miR-29b, acts as an intermediate regulator.79 The increased expression of hsa_circ_0012919 in the CD4 T cells of SLE patients not only upregulates the expression of CD70 and CD11a by regulating DNMT1 and DNA methylation but also regulates KLF13, RANTES, and MDA5 by sponging miR-125a-3p in a competing endogenous RNA (ceRNA)-dependent manner.80,81 Thus, circRNAs may modulate SLE pathophysiological processes by inducing the activation and apoptosis of immune cells, cytokine secretion, epigenetic changes, and disease-related key genes and pathways, which have become a focus of research into the mechanism underlying SLE.4,5,82, 83, 84, 85 In addition, a recent high-quality study has expanded our understanding of the roles of circRNAs in SLE from a new perspective.25 In this study, it was shown that 16- to 26-bp imperfect RNA duplexes generated from endogenous circRNAs are present at reduced levels in PBMCs derived from SLE patients. The double-stranded RNA-containing circRNAs alleviate the aberrant dsRNA-activated protein kinase (PKR) activation cascade to suppress excessive innate immune responses in PBMCs or T cells in SLE patients. Overall, these existing studies have expanded our understanding of the pathogenesis of SLE to some extent and provided important foundations and directions for future research on SLE.
Incidentally, we found an interesting phenomenon that upregulated circRNAs with tumor-promoting roles were identified in most cancer studies, indicating that the pathophysiology of cancers is initiated by the abnormal activation of oncogenes. In contrast, in SLE, there seem to be more downregulated circRNAs than upregulated circRNAs validated (13 versus 7). Hence, the disease suppressors or regulators in SLE might be promising research targets.
Limitations and future perspectives
Since the research in this field is still in its infancy, there are many deficiencies. In view of these shortcomings, we present some suggestions for future research perspectives.
Limitations and future perspectives on circRNA expression in SLE
First, most of the anomalously expressed circRNAs identified by microarray or RNA sequencing analyses have not been verified in human samples. Although some circRNAs have already been preliminarily validated, the sample size used in the analysis was small, leading to a lack of confidence in the results due to type I and type II errors. This issue is not easy to resolve, because it is difficult to obtain relatively ideal blood samples, as SLE is often accompanied by hemolytic anemia, leukopenia, and thrombocytopenia, thus limiting the amount of blood components that can be collected.2,22 In addition, before diagnosis, SLE patients may receive drugs such as glucocorticoids and non-steroidal anti-inflammatory drugs to treat the prodromal symptoms and disease. To a greater or lesser extent, these drugs will impact the gene expression profile of the blood components.86,87 Therefore, it would be useful to establish a SLE or autoimmune disease database of gene expression profiles that is constantly updated, similar to the cancer-related databases that have emerged in recent years. This type of cooperation will enable us to better understand the gene expression profiles of the disease.
Second, at present, the main research objects are blood specimens, while other body fluids and lesions are ignored. Furthermore, the studies are conducted mainly on PBMCs. In consideration of the whole blood composition changes in SLE, other blood components such as plasma (or exosomes), erythrocytes, granulocytes, and platelets are also optional research subjects. Previous studies have focused on T cell or CD4+ T cell populations in PBMCs, while other cells such as B cells, natural killer cells, and monocytes are also good research subjects. Of course, it will be better if we can study subdivided cell subsets, such as helper T (Th) cells (Th1, Th2, and Th17), suppressor T (Ts) cells, cytotoxic T lymphocytes (CTLs), and Tregs. Detection of circRNAs in different blood constituents can not only contribute to a better understanding of the tissue or cell specificity of their expression but also clarify their potential transfer processes. The tissue or cell specificity and transfer processes are important foundations for studying their biological functions.
Third, the mechanism of circRNA deregulation in SLE represents a knowledge gap, which, as described above, could be interpreted by their formation and degradation. In terms of biogenesis, several important regulatory factors should be considered. First, it is an indisputable fact that SLE is associated with extensive dysregulated epigenetic modifications, such as aberrant DNA methylation, abnormal histone methylation, and acetylation.88, 89, 90, 91, 92 Second, many transcription factors have been shown to display anomalous transcriptional activity in SLE, including nuclear factor of activated T cells 1 (NFAT1), signal transducer and activator of transcription (STAT), interferon regulatory factors (IRFs), and cAMP response element modulator α (CREMα).93, 94, 95, 96 These two aspects can explain the discrepancies in the levels of circRNAs in SLE compared with healthy individuals from the transcriptional perspective. A third factor affecting the generation of circRNAs in SLE is the abnormal expression of RBPs, such as serine arginine-rich splicing factor 1 (SRSF1), which may mediate the back-splicing of pre-mRNAs.97,98 Lariat-driven circularization, intron-pairing-driven circularization, and degradation, which may also influence circRNA expression, have not been investigated in depth; therefore, we have not proposed any assumptions related to SLE here (Figure 2).
Importance of circRNA transmission in SLE
As mentioned previously, circRNAs are abundant in body fluids and transported throughout the body via exosomes and body fluids.29, 30, 31, 32 However, the transmission of circRNAs in SLE is a vital deficit in current research. Since SLE is a disease involving the blood and multiple organs, it is highly significant to study the transmission of circRNAs in SLE. Internal and/or external factors may cause aberrant expression of numerous circRNAs in blood or tissue cells, with some secreted into body fluids by exosomes. It is also possible that circRNAs enter body fluids directly from abnormal apoptotic or necrotic cells, because SLE is always accompanied by blood cell destruction and tissue damage. These circRNAs may then remain in the body fluids or integrate into blood and tissue cells, resulting in local or systemic enrichment. The divergent accumulation of circRNAs in SLE endows them with latent potential as biomarkers and is also a prerequisite for their participation in the pathophysiological process of SLE (Figure 3).
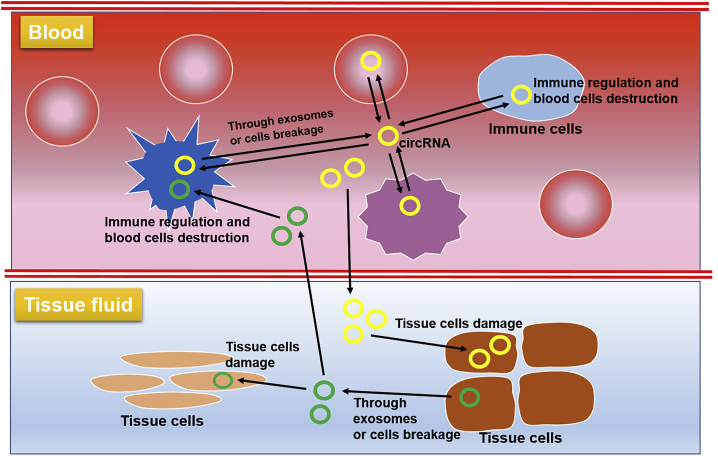
Figure 3.
Prospective transmission pathway of circRNAs in SLE
circRNAs generated in blood cells may enter serum via exosomes or cell breakage. These circRNAs then enter other blood cells or transmit to tissue fluid and enter tissue cells, causing immune response, inflammation, and cell damage. Accordingly, circRNAs generated in tissue cells may also influence other tissue cells and blood cells.
Limitations and future perspectives on circRNA roles in SLE
Some studies have revealed “associations” between circRNA expression and clinical factors, such as disease activity, or SLE-related genes or signaling pathways, drawing the hypothesis that these circRNAs regulate SLE progression. However, this theory is far from convincing. As described previously, the specific inflammatory, immune, and epigenetic environment of SLE can also affect the expression of circRNAs. Therefore, it is not clear that SLE alters the expression of these circRNAs or that these circRNAs regulate the pathophysiological processes of SLE. Indeed, not all the dysregulated circRNAs modulate the disease progression, with some merely exhibiting varied expression. To draw the conclusion that circRNAs impact the pathogenesis of SLE, rigorous functional and mechanistic investigations should be conducted.
Accordingly, some circRNAs may act as important regulators of the process of SLE. The current studies have only explored the roles of circRNAs in PBMCs or T cells; however, these investigations are insufficient to clarify the complexity of the functions of circRNAs in SLE. For example, the aberrant expression of circRNAs in a specific immune cell type can not only mediate the dysfunction or damage of these cells but also transfer to other immune cells and even tissue cells, thus mediating immune responses and causing tissue damage. Correspondingly, circRNAs produced by tissue injury can also lead to damage of other tissues and dysregulation of immune cells (Figure 3).
Research on the mechanisms of circRNAs in SLE is still very limited. In previous studies, mechanistic investigations of circRNAs have been focused mainly on the induction of immune cell abnormalities by regulating miRNAs. However, other mechanisms underlying the functions of circRNAs may also be of importance in SLE research. For instance, since circRNAs located in the nucleus may act as transcription manipulators by cooperating with or modulating the epigenetic modifications associated with SLE, their impact on gene expression will be more prominent. circRNAs can interact with RBPs to regulate the functions of RBPs, and many anti-RBP autoantibodies are produced in SLE.99, 100, 101 Therefore, it is possible that the binding of circRNAs to RBPs affects the production of anti-RBP antibodies by changing the characteristics of RBPs, such as configuration, function, stability, or subcellular localization. Furthermore, some circRNAs encode polypeptides or proteins, which may perform various functions. First, as proteins, they may possess and exert their functions in the original cells. Second, some enter the body fluids via exosomes (especially micromolecular ones) or after cell disruption, thus implicating them as biomarkers. Third, peptides or proteins released into body fluids may be transported to tissues or blood cells, inducing immune responses, inflammation, and cell damage. Fourth, the possibility that these peptides or proteins are deposited in target organs and cause direct damage should not be ignored. We think these presumptions are very intriguing and promising topics for future research (Figure 2).
Conclusions
Emerging evidence has demonstrated that circRNAs are important regulators in various diseases. In this work, we briefly introduce these “star molecules” and summarize their roles in SLE. Although a great deal of research has been conducted in this field, the limitations we have described remain to be addressed. More extensive and intensive studies are required to provide novel and valuable ideas in this area of interest. In summary, integrating a comprehensive understanding of functional circRNA biology into the area of SLE may clarify the pathogenesis and development of this disease. More importantly, such fundamental studies will provide a sound theoretical foundation for the application of circRNAs in the clinical diagnosis and treatment of SLE.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (grant nos. 81673050, 81773310, 81872522, and 82073429); the Innovation Program of Shanghai Municipal Education Commission (no. 2019-01-07-00-07-E00046); the Program of Science and Technology Commission of Shanghai Municipality (no. 18140901800); the Excellent Subject Leader Program of Shanghai Municipal Commission of Health and Family Planning (no. 2018BR30); the Clinical Research Program of Shanghai Hospital Development Center (nos. SHDC2020CR1014B and SHDC12018X06); and the Program of Shanghai Academic Research Leader (no. 20XD1403300).
Author Contributions
X.W. and R.M. wrote the manuscript, Z.W. and W.S. retrieved literature, and Y.S. critically revised the manuscript. All authors have read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Xin Wang, Email: wx739976260@foxmail.com.
Yuling Shi, Email: shiyuling1973@tongji.edu.cn.
References
- 1.Dörner T., Furie R. Novel paradigms in systemic lupus erythematosus. Lancet. 2019;393:2344–2358. doi: 10.1016/S0140-6736(19)30546-X. [DOI] [PubMed] [Google Scholar]
- 2.Durcan L., O’Dwyer T., Petri M. Management strategies and future directions for systemic lupus erythematosus in adults. Lancet. 2019;393:2332–2343. doi: 10.1016/S0140-6736(19)30237-5. [DOI] [PubMed] [Google Scholar]
- 3.Tsokos G.C. Autoimmunity and organ damage in systemic lupus erythematosus. Nat. Immunol. 2020;21:605–614. doi: 10.1038/s41590-020-0677-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsokos G.C., Lo M.S., Costa Reis P., Sullivan K.E. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2016;12:716–730. doi: 10.1038/nrrheum.2016.186. [DOI] [PubMed] [Google Scholar]
- 5.Mohan C., Putterman C. Genetics and pathogenesis of systemic lupus erythematosus and lupus nephritis. Nat. Rev. Nephrol. 2015;11:329–341. doi: 10.1038/nrneph.2015.33. [DOI] [PubMed] [Google Scholar]
- 6.Xiao M.S., Ai Y., Wilusz J.E. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol. 2020;30:226–240. doi: 10.1016/j.tcb.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 8.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 9.Aufiero S., Reckman Y.J., Pinto Y.M., Creemers E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019;16:503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 10.Li J., Sun D., Pu W., Wang J., Peng Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer. 2020;6:319–336. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Soghli N., Qujeq D., Yousefi T., Soghli N. The regulatory functions of circular RNAs in osteosarcoma. Genomics. 2020;112:2845–2856. doi: 10.1016/j.ygeno.2020.03.024. [DOI] [PubMed] [Google Scholar]
- 12.Wang X., Fang L. Advances in circular RNAs and their roles in breast Cancer. J. Exp. Clin. Cancer Res. 2018;37:206. doi: 10.1186/s13046-018-0870-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong P., Xu D., Xiong Y., Yue J., Ihira K., Konno Y., Watari H. The Expression, Functions and Mechanisms of Circular RNAs in Gynecological Cancers. Cancers (Basel) 2020;12:1472. doi: 10.3390/cancers12061472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Mehta S.L., Dempsey R.J., Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol. 2020;186:101746. doi: 10.1016/j.pneurobio.2020.101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Y., Ci C., Zhang X., Su M., Lv W., Chen C., Liu H., Zhang D., Zhang S., Zhang Y. Prediction of circRNAs Based on the DNA Methylation-Mediated Feature Sponge Function in Breast Cancer. Front. Bioeng. Biotechnol. 2019;7:365. doi: 10.3389/fbioe.2019.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davie J.R., Xu W., Delcuve G.P. Histone H3K4 trimethylation: dynamic interplay with pre-mRNA splicing. Biochem. Cell Biol. 2016;94:1–11. doi: 10.1139/bcb-2015-0065. [DOI] [PubMed] [Google Scholar]
- 18.Cheng N., Xiao J., Ge S., Li J., Huang J., Wu X., Zhang S., Xiang T. High-Throughput Sequencing Strategy for miR-146b-regulated circRNA Expression in Hepatic Stellate Cells. Med. Sci. Monit. 2018;24:8699–8706. doi: 10.12659/MSM.910807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welden J.R., Stamm S. Pre-mRNA structures forming circular RNAs. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:194410. doi: 10.1016/j.bbagrm.2019.194410. [DOI] [PubMed] [Google Scholar]
- 20.Zaiou M. The Emerging Role and Promise of Circular RNAs in Obesity and Related Metabolic Disorders. Cells. 2020;9:1473. doi: 10.3390/cells9061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beltrán-García J., Osca-Verdegal R., Nacher-Sendra E., Pallardó F.V., García-Giménez J.L. Circular RNAs in Sepsis: Biogenesis, Function, and Clinical Significance. Cells. 2020;9:1544. doi: 10.3390/cells9061544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu C., He J., Qi L., Ren X., Zhang C., Duan Z., Yang K., Wang W., Lu Q., Li Z. Emerging landscape of circular RNAs as biomarkers and pivotal regulators in osteosarcoma. J. Cell. Physiol. 2020;235:9037–9058. doi: 10.1002/jcp.29754. [DOI] [PubMed] [Google Scholar]
- 23.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park O.H., Ha H., Lee Y., Boo S.H., Kwon D.H., Song H.K., Kim Y.K. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell. 2019;74:494–507.e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Liu C.X., Li X., Nan F., Jiang S., Gao X., Guo S.K., Xue W., Cui Y., Dong K., Ding H. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell. 2019;177:865–880.e21. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 26.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang Q., Yang Z., Jia R., Ge S. The novel roles of circRNAs in human cancer. Mol. Cancer. 2019;18:6. doi: 10.1186/s12943-018-0934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C., Liang D., Tatomer D.C., Wilusz J.E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M., Yu F., Li P., Wang K. Emerging Function and Clinical Significance of Exosomal circRNAs in Cancer. Mol. Ther. Nucleic Acids. 2020;21:367–383. doi: 10.1016/j.omtn.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng J., Meng J., Zhu L., Peng Y. Exosomal noncoding RNAs in Glioma: biological functions and potential clinical applications. Mol. Cancer. 2020;19:66. doi: 10.1186/s12943-020-01189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi X., Wang B., Feng X., Xu Y., Lu K., Sun M. circRNAs and Exosomes: A Mysterious Frontier for Human Cancer. Mol. Ther. Nucleic Acids. 2020;19:384–392. doi: 10.1016/j.omtn.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai J., Su Y., Zhong S., Cong L., Liu B., Yang J., Tao Y., He Z., Chen C., Jiang Y. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020;5:145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong Y., He D., Peng Z., Peng W., Shi W., Wang J., Li B., Zhang C., Duan C. Circular RNAs in cancer: an emerging key player. J. Hematol. Oncol. 2017;10:2. doi: 10.1186/s13045-016-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 35.Han D., Wang Y., Wang Y., Dai X., Zhou T., Chen J., Tao B., Zhang J., Cao F. The Tumor-Suppressive Human Circular RNA CircITCH Sponges miR-330-5p to Ameliorate Doxorubicin-Induced Cardiotoxicity Through Upregulating SIRT6, Survivin, and SERCA2a. Circ. Res. 2020;127:e108–e125. doi: 10.1161/CIRCRESAHA.119.316061. [DOI] [PubMed] [Google Scholar]
- 36.Jiang Q., Liu C., Li C.P., Xu S.S., Yao M.D., Ge H.M., Sun Y.N., Li X.M., Zhang S.J., Shan K. Circular RNA-ZNF532 regulates diabetes-induced retinal pericyte degeneration and vascular dysfunction. J. Clin. Invest. 2020;130:3833–3847. doi: 10.1172/JCI123353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cristóbal I., Caramés C., Rubio J., Sanz-Alvarez M., Luque M., Madoz-Gúrpide J., Rojo F., García-Foncillas J. Functional and Clinical Impact of CircRNAs in Oral Cancer. Cancers (Basel) 2020;12:1041. doi: 10.3390/cancers12041041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J., Li D., Luo H., Zhu X. Circular RNAs: The star molecules in cancer. Mol. Aspects Med. 2019;70:141–152. doi: 10.1016/j.mam.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Garikipati V.N.S., Verma S.K., Cheng Z., Liang D., Truongcao M.M., Cimini M., Yue Y., Huang G., Wang C., Benedict C. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2019;10:4317. doi: 10.1038/s41467-019-11777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y.J., Zheng B., Luo G.J., Ma X.K., Lu X.Y., Lin X.M., Yang S., Zhao Q., Wu T., Li Z.X. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526–3540. doi: 10.7150/thno.32796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jie M., Wu Y., Gao M., Li X., Liu C., Ouyang Q., Tang Q., Shan C., Lv Y., Zhang K. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol. Cancer. 2020;19:56. doi: 10.1186/s12943-020-01160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Conn V.M., Hugouvieux V., Nayak A., Conos S.A., Capovilla G., Cildir G., Jourdain A., Tergaonkar V., Schmid M., Zubieta C., Conn S.J. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 44.Lei M., Zheng G., Ning Q., Zheng J., Dong D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang W.C., Wong C.W., Liang P.P., Shi M., Cao Y., Rao S.T., Tsui S.K., Waye M.M., Zhang Q., Fu W.M., Zhang J.F. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia X., Li X., Li F., Wu X., Zhang M., Zhou H., Huang N., Yang X., Xiao F., Liu D. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol. Cancer. 2019;18:131. doi: 10.1186/s12943-019-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J., Liu T., Wang X., He A. Circles reshaping the RNA world: from waste to treasure. Mol. Cancer. 2017;16:58. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Mo Y., Gong Z., Yang X., Yang M., Zhang S., Xiong F., Xiang B., Zhou M., Liao Q. Circular RNAs in human cancer. Mol. Cancer. 2017;16:25. doi: 10.1186/s12943-017-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardini B., Sabo A.A., Birolo G., Calin G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers (Basel) 2019;11:1170. doi: 10.3390/cancers11081170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaichian S., Shafabakhsh R., Mirhashemi S.M., Moazzami B., Asemi Z. Circular RNAs: A novel biomarker for cervical cancer. J. Cell. Physiol. 2020;235:718–724. doi: 10.1002/jcp.29009. [DOI] [PubMed] [Google Scholar]
- 51.Ng W.L., Mohd Mohidin T.B., Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15:995–1005. doi: 10.1080/15476286.2018.1486659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bach D.H., Lee S.K., Sood A.K. Circular RNAs in Cancer. Mol. Ther. Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong S., Ouyang Q., Zhu D., Huang Q., Zhao J., Fan M., Cai Y., Yang M. Hsa_circ_0088036 promotes the proliferation and migration of fibroblast-like synoviocytes by sponging miR-140-3p and upregulating SIRT 1 expression in rheumatoid arthritis. Mol. Immunol. 2020;125:131–139. doi: 10.1016/j.molimm.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Luo Q., Liu J., Fu B., Zhang L., Guo Y., Huang Z., Li J. Circular RNAs Hsa_circ_0002715 and Hsa_circ_0035197 in Peripheral Blood Are Novel Potential Biomarkers for New-Onset Rheumatoid Arthritis. Dis. Markers. 2019;2019:2073139. doi: 10.1155/2019/2073139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X., Li J., Wu Y., Ni B., Zhang B. Aberrant dysregulated circular RNAs in the peripheral blood mononuclear cells of patients with rheumatoid arthritis revealed by RNA sequencing: novel diagnostic markers for RA. Scand. J. Clin. Lab. Invest. 2019;79:551–559. doi: 10.1080/00365513.2019.1674004. [DOI] [PubMed] [Google Scholar]
- 56.Tang X., Wang J., Xia X., Tian J., Rui K., Xu H., Wang S. Elevated expression of ciRS-7 in peripheral blood mononuclear cells from rheumatoid arthritis patients. Diagn. Pathol. 2019;14:11. doi: 10.1186/s13000-019-0783-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo Q., Zhang L., Li X., Fu B., Deng Z., Qing C., Su R., Xu J., Guo Y., Huang Z., Li J. Identification of circular RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for rheumatoid arthritis. Clin. Exp. Immunol. 2018;194:118–124. doi: 10.1111/cei.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iparraguirre L., Muñoz-Culla M., Prada-Luengo I., Castillo-Triviño T., Olascoaga J., Otaegui D. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum. Mol. Genet. 2017;26:3564–3572. doi: 10.1093/hmg/ddx243. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Zhou Y., Zhao M., Zou J., Zhu Y., Yuan X., Liu Q., Cai H., Chu C.Q., Liu Y. Differential Profile of Plasma Circular RNAs in Type 1 Diabetes Mellitus. Diabetes Metab. J. 2020;44:854–865. doi: 10.4093/dmj.2019.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li F., Liu Z., Zhang B., Jiang S., Wang Q., Du L., Xue H., Zhang Y., Jin M., Zhu X. Circular RNA sequencing indicates circ-IQGAP2 and circ-ZC3H6 as noninvasive biomarkers of primary Sjögren’s syndrome. Rheumatology (Oxford) 2020;59:2603–2615. doi: 10.1093/rheumatology/keaa163. [DOI] [PubMed] [Google Scholar]
- 61.Su L.C., Xu W.D., Liu X.Y., Fu L., Huang A.F. Altered expression of circular RNA in primary Sjögren’s syndrome. Clin. Rheumatol. 2019;38:3425–3433. doi: 10.1007/s10067-019-04728-6. [DOI] [PubMed] [Google Scholar]
- 62.Zhou Z., Sun B., Huang S., Zhao L. Roles of circular RNAs in immune regulation and autoimmune diseases. Cell Death Dis. 2019;10:503. doi: 10.1038/s41419-019-1744-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie R., Zhang Y., Zhang J., Li J., Zhou X. The Role of Circular RNAs in Immune-Related Diseases. Front. Immunol. 2020;11:545. doi: 10.3389/fimmu.2020.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia X., Tang X., Wang S. Roles of CircRNAs in Autoimmune Diseases. Front. Immunol. 2019;10:639. doi: 10.3389/fimmu.2019.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J., Zhao X., Wang Y., Ren F., Sun D., Yan Y., Kong X., Bu J., Liu M., Xu S. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Q., Mang G., Wu J., Sun P., Li T., Zhang H., Wang N., Tong Z., Wang W., Zheng Y. Circular RNA circSnx5 Controls Immunogenicity of Dendritic Cells through the miR-544/SOCS1 Axis and PU.1 Activity Regulation. Mol. Ther. 2020;28:2503–2518. doi: 10.1016/j.ymthe.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y., Zhang G., Liu Y., Chen R., Zhao D., McAlister V., Mele T., Liu K., Zheng X. GDF15 Regulates Malat-1 Circular RNA and Inactivates NFκB Signaling Leading to Immune Tolerogenic DCs for Preventing Alloimmune Rejection in Heart Transplantation. Front. Immunol. 2018;9:2407. doi: 10.3389/fimmu.2018.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cortes R., Forner M.J. Circular RNAS: novel biomarkers of disease activity in systemic lupus erythematosus? Clin. Sci. (Lond.) 2019;133:1049–1052. doi: 10.1042/CS20180826. [DOI] [PubMed] [Google Scholar]
- 70.Luo Q., Zhang L., Li X., Fu B., Guo Y., Huang Z., Li J. Identification of circular RNAs hsa_circ_0044235 and hsa_circ_0068367 as novel biomarkers for systemic lupus erythematosus. Int. J. Mol. Med. 2019;44:1462–1472. doi: 10.3892/ijmm.2019.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miao Q., Zhong Z., Jiang Z., Lin Y., Ni B., Yang W., Tang J. RNA-seq of circular RNAs identified circPTPN22 as a potential new activity indicator in systemic lupus erythematosus. Lupus. 2019;28:520–528. doi: 10.1177/0961203319830493. [DOI] [PubMed] [Google Scholar]
- 72.Li S., Zhang J., Tan X., Deng J., Li Y., Piao Y., Li C., Yang W., Mo W., Sun J. Microarray expression profile of circular RNAs and mRNAs in children with systemic lupus erythematosus. Clin. Rheumatol. 2019;38:1339–1350. doi: 10.1007/s10067-018-4392-8. [DOI] [PubMed] [Google Scholar]
- 73.Zhang M.Y., Wang J.B., Zhu Z.W., Li L.J., Liu R.S., Yang X.K., Leng R.X., Li X.M., Pan H.F., Ye D.Q. Differentially expressed circular RNAs in systemic lupus erythematosus and their clinical significance. Biomed. Pharmacother. 2018;107:1720–1727. doi: 10.1016/j.biopha.2018.08.161. [DOI] [PubMed] [Google Scholar]
- 74.Ouyang Q., Huang Q., Jiang Z., Zhao J., Shi G.P., Yang M. Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol. Immunol. 2018;101:531–538. doi: 10.1016/j.molimm.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 75.Zhang C., Huang J., Chen Y., Shi W. Low Expression and Clinical Value of hsa_circ_0049224 and has_circ_0049220 in Systemic Lupus Erythematous Patients. Med. Sci. Monit. 2018;24:1930–1935. doi: 10.12659/MSM.906507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo G., Wang H., Ye L., Shi X., Yan K., Lin K., Huang Q., Li B., Lin Q., Zhu L. Hsa_circ_0000479 as a Novel Diagnostic Biomarker of Systemic Lupus Erythematosus. Front. Immunol. 2019;10:2281. doi: 10.3389/fimmu.2019.02281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo Q., Zhang L., Fang L., Fu B., Guo Y., Huang Z., Li J. Circular RNAs hsa_circ_0000479 in peripheral blood mononuclear cells as novel biomarkers for systemic lupus erythematosus. Autoimmunity. 2020;53:167–176. doi: 10.1080/08916934.2020.1728529. [DOI] [PubMed] [Google Scholar]
- 78.Li L.J., Zhu Z.W., Zhao W., Tao S.S., Li B.Z., Xu S.Z., Wang J.B., Zhang M.Y., Wu J., Leng R.X. Circular RNA expression profile and potential function of hsa_circ_0045272 in systemic lupus erythematosus. Immunology. 2018;155:137–149. doi: 10.1111/imm.12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X., Zhang C., Wu Z., Chen Y., Shi W. CircIBTK inhibits DNA demethylation and activation of AKT signaling pathway via miR-29b in peripheral blood mononuclear cells in systemic lupus erythematosus. Arthritis Res. Ther. 2018;20:118. doi: 10.1186/s13075-018-1618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C., Wang X., Chen Y., Wu Z., Zhang C., Shi W. The down-regulation of hsa_circ_0012919, the sponge for miR-125a-3p, contributes to DNA methylation of CD11a and CD70 in CD4+ T cells of systemic lupus erythematous. Clin. Sci. (Lond.) 2018;132:2285–2298. doi: 10.1042/CS20180403. [DOI] [PubMed] [Google Scholar]
- 81.Zhang C., Zhang C., Ji J., Xiong X., Lu Y. Hsa_circ_0012919 regulates expression of MDA5 by miR-125a-3p in CD4+ T cells of systemic lupus erythematous. Lupus. 2020;29:727–734. doi: 10.1177/0961203320920706. [DOI] [PubMed] [Google Scholar]
- 82.Kato Y., Park J., Takamatsu H., Konaka H., Aoki W., Aburaya S., Ueda M., Nishide M., Koyama S., Hayama Y. Apoptosis-derived membrane vesicles drive the cGAS-STING pathway and enhance type I IFN production in systemic lupus erythematosus. Ann. Rheum. Dis. 2018;77:1507–1515. doi: 10.1136/annrheumdis-2018-212988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arazi A., Rao D.A., Berthier C.C., Davidson A., Liu Y., Hoover P.J., Chicoine A., Eisenhaure T.M., Jonsson A.H., Li S., Accelerating Medicines Partnership in SLE network The immune cell landscape in kidneys of patients with lupus nephritis. Nat. Immunol. 2019;20:902–914. doi: 10.1038/s41590-019-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caielli S., Veiga D.T., Balasubramanian P., Athale S., Domic B., Murat E., Banchereau R., Xu Z., Chandra M., Chung C.H. A CD4+ T cell population expanded in lupus blood provides B cell help through interleukin-10 and succinate. Nat. Med. 2019;25:75–81. doi: 10.1038/s41591-018-0254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hamilton J.A., Hsu H.C., Mountz J.D. Autoreactive B cells in SLE, villains or innocent bystanders? Immunol. Rev. 2019;292:120–138. doi: 10.1111/imr.12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goleva E., Babineau D.C., Gill M.A., Jackson L.P., Shao B., Hu Z., Liu A.H., Visness C.M., Sorkness C.A., Leung D.Y.M. Expression of corticosteroid-regulated genes by PBMCs in children with asthma. J. Allergy Clin. Immunol. 2019;143:940–947.e6. doi: 10.1016/j.jaci.2018.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bakhtiari T., Ghaderi A., Safaee Nodehi S.R., Aghazadeh Z., Tofighi Zavareh F., Jafarnezhad-Ansariha F., Barati A., Mirshafiey A. An in vitro assessment for evaluating the efficiency of β-d-mannuronic acid (M2000) in myelodysplastic syndrome. J. Cell. Physiol. 2019;234:12971–12977. doi: 10.1002/jcp.27966. [DOI] [PubMed] [Google Scholar]
- 88.Catalina M.D., Owen K.A., Labonte A.C., Grammer A.C., Lipsky P.E. The pathogenesis of systemic lupus erythematosus: Harnessing big data to understand the molecular basis of lupus. J. Autoimmun. 2020;110:102359. doi: 10.1016/j.jaut.2019.102359. [DOI] [PubMed] [Google Scholar]
- 89.Wu H., Chen Y., Zhu H., Zhao M., Lu Q. The Pathogenic Role of Dysregulated Epigenetic Modifications in Autoimmune Diseases. Front. Immunol. 2019;10:2305. doi: 10.3389/fimmu.2019.02305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karagianni P., Tzioufas A.G. Epigenetic perspectives on systemic autoimmune disease. J. Autoimmun. 2019;104:102315. doi: 10.1016/j.jaut.2019.102315. [DOI] [PubMed] [Google Scholar]
- 91.Surace A.E.A., Hedrich C.M. The Role of Epigenetics in Autoimmune/Inflammatory Disease. Front. Immunol. 2019;10:1525. doi: 10.3389/fimmu.2019.01525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mazzone R., Zwergel C., Artico M., Taurone S., Ralli M., Greco A., Mai A. The emerging role of epigenetics in human autoimmune disorders. Clin. Epigenetics. 2019;11:34. doi: 10.1186/s13148-019-0632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang X., Zhao C., Zhang C., Mei X., Song J., Sun Y., Wu Z., Shi W. Increased HERV-E clone 4-1 expression contributes to DNA hypomethylation and IL-17 release from CD4+ T cells via miR-302d/MBD2 in systemic lupus erythematosus. Cell Commun. Signal. 2019;17:94. doi: 10.1186/s12964-019-0416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alunno A., Padjen I., Fanouriakis A., Boumpas D.T. Pathogenic and Therapeutic Relevance of JAK/STAT Signaling in Systemic Lupus Erythematosus: Integration of Distinct Inflammatory Pathways and the Prospect of Their Inhibition with an Oral Agent. Cells. 2019;8:898. doi: 10.3390/cells8080898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jensen M.A., Niewold T.B. Interferon regulatory factors: critical mediators of human lupus. Transl. Res. 2015;165:283–295. doi: 10.1016/j.trsl.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hofmann S.R., Mäbert K., Kapplusch F., Russ S., Northey S., Beresford M.W., Tsokos G.C., Hedrich C.M. cAMP Response Element Modulator α Induces Dual Specificity Protein Phosphatase 4 to Promote Effector T Cells in Juvenile-Onset Lupus. J. Immunol. 2019;203:2807–2816. doi: 10.4049/jimmunol.1900760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Katsuyama T., Martin-Delgado I.J., Krishfield S.M., Kyttaris V.C., Moulton V.R. Splicing factor SRSF1 controls T cell homeostasis and its decreased levels are linked to lymphopenia in systemic lupus erythematosus. Rheumatology (Oxford) 2020;59:2146–2155. doi: 10.1093/rheumatology/keaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Katsuyama T., Li H., Comte D., Tsokos G.C., Moulton V.R. Splicing factor SRSF1 controls T cell hyperactivity and systemic autoimmunity. J. Clin. Invest. 2019;129:5411–5423. doi: 10.1172/JCI127949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matta B., Barnes B.J. Coordination between innate immune cells, type I IFNs and IRF5 drives SLE pathogenesis. Cytokine. 2020;132:154731. doi: 10.1016/j.cyto.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ko K., Koldobskaya Y., Rosenzweig E., Niewold T.B. Activation of the Interferon Pathway is Dependent Upon Autoantibodies in African-American SLE Patients, but Not in European-American SLE Patients. Front. Immunol. 2013;4:309. doi: 10.3389/fimmu.2013.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Niewold T.B., Kelly J.A., Kariuki S.N., Franek B.S., Kumar A.A., Kaufman K.M., Thomas K., Walker D., Kamp S., Frost J.M. IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Ann. Rheum. Dis. 2012;71:463–468. doi: 10.1136/annrheumdis-2011-200463. [DOI] [PMC free article] [PubMed] [Google Scholar]