Abstract
Long non-coding RNAs (lncRNAs) regulate the development of follicles and reproductive diseases, but the mechanisms by which lncRNAs regulate ovarian functions and fertility remain elusive. We profiled the expression of lncRNAs in ovarian tissues of Hu sheep with different prolificacy and identified 21,327 lncRNAs. Many of the lncRNAs were differentially expressed in different groups. We further characterized an lncRNA that was predominantly expressed in the ovaries of the low prolificacy FecB+ (LPB+) group and mainly present in granulosa cells (GCs), and the expression of this lncRNA decreased during follicular development, which we named follicular development-associated lncRNA (FDNCR). Next, we found that FDNCR directly binds miR-543-3p, and decorin (DCN) was identified as a target of miR-543-3p. FDNCR overexpression promoted GC apoptosis through increased expression of DCN, which could be attenuated by miR-543-3p. Furthermore, miR-543-3p increased and FDNCR reduced the expression of transforming growth factor-β (TGF-β) pathway-related genes, including TGF-β1 and inhibin beta A (INHBA), which were upregulated upon DCN silencing. Our results demonstrated that FDNCR sponges miR-543-3p in GCs and prevents miR-543-3p from binding to the DCN 3′ UTR, resulting in DCN transactivation and TGF-β pathway inhibition and promotion of GC apoptosis in Hu sheep. These findings provide insights into the mechanisms underlying prolificacy in sheep.
Keywords: FDNCR, miR-543-3p, granulosa cells, apoptosis, Hu sheep
Graphical abstract
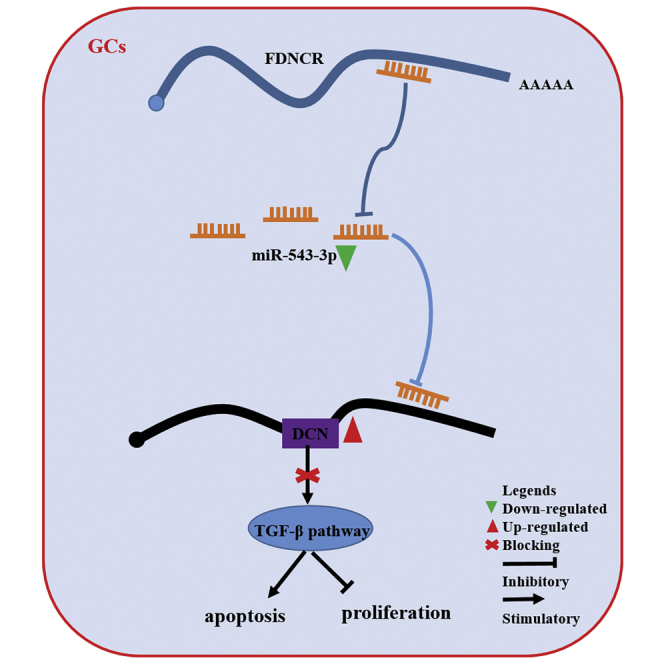
lncRNA FDNCR sponges miR-543-3p in GCs of Hu sheep, and it prevents miR-543-3p from binding to the 3′ UTR of DCN, resulting in the transactivation of DCN and inhibition of the TGF-β signaling pathway, ultimately promoting GC apoptosis. These findings provide insights into the mechanisms underlying prolificacy in Hu sheep.
Introduction
Low reproductive efficiency (i.e., litter size) limits the development of the sheep industry. Most sheep species exhibit seasonal estrus and produce one lamb per pregnancy. Thus, good breed selection is important for crossbreeding. Hu sheep are an excellent local breed in China and are famous for their precocious puberty, year-round estrus, and high fecundity (average litter size 2.06).1 Granulosa cells (GCs) play an essential role in the recruitment, selection, ovulation, and atresia of follicles.2 Therefore, elucidation of GC function is important for understanding follicular development, increasing the ovulation rate, and female fertility. This will provide useful information for the breeding and trait selection of Hu sheep.
FecB (BMPR1B mutation) is a fundamental fecundity gene in Hu sheep.2 Sheep with the homozygous mutation (FecBB [BB]) exhibit higher ovulation rates than do heterozygous (FecB+ [B+]) or wild-type (++) sheep.3, 4, 5 However, under similar conditions, some Hu sheep carrying the BB mutation gave birth to single lambs, although the underlying molecular mechanisms remain unknown.
Long non-coding RNAs (lncRNAs) with length >200 nt regulate genes at the transcriptional and post-transcription levels. At the post-transcriptional level, lncRNA can serve as efficient microRNA (miRNA) sponges—termed competing endogenous RNAs (ceRNAs)—that interact with miRNA to regulate gene expression.6 Recently, numerous lncRNAs have been identified in humans and other organisms that play diverse roles in regulating various phenomenon, such as cell proliferation and apoptosis,7,8 cell development,9 cell differentiation,10 and development of certain diseases.11,12 Specifically, lncRNAs are involved in regulating spermatogenesis,13,14 steroidogenesis,15 embryo implantation16 and development,17 follicular development,18 oocyte maturation,19 GC apoptosis and proliferation,20,21 and reproductive diseases,22,23 suggesting their importance in reproduction.24 However, only a few lncRNAs have been identified in domestic animals. Although lncRNA expression profiles in the ovary, uterus, and pituitary glands of sheep25, 26, 27, 28, 29, 30, 31 (Small Tail Han sheep, Dorset sheep, or Hu sheep), and goats32 (Anhui white goats) with different prolificacy indicate their importance in determining female fecundity, the mechanisms underlying the influence of lncRNA with respect to determining fecundity in Hu sheep remain elusive.
GC apoptosis can result in follicular atresia, a key process in follicle selection and development.33, 34, 35 Only 1% of the follicles eventually mature and ovulate, and >99% undergo atresia and degeneration in mammals.36 Although GC apoptosis is regulated by lncRNAs such as PVT1 and NORFA,22,37 the number of lncRNAs reported to be involved in GC apoptosis and follicular development remains limited.
In this study, we hypothesized that the differences in fecundity and FecB genotype could be correlated with different lncRNA expression profiles in the ovarian cells of Hu sheep, providing a target for treating ovulation failure. Hence, we investigated the differentially expressed (DE) lncRNAs in the ovaries of Hu sheep with different prolificacy (high prolificacy [HP], litter size = 3; low prolificacy [LP], litter size = 1) and FecB genotypes (BB and B+) using RNA sequencing (RNA-seq). We further characterized that a follicular development-associated lncRNA (FDNCR) acts as a ceRNA for miR-543-3p to augment decorin (DCN) expression and inhibit the transforming growth factor-β (TGF-β) signaling pathway, thereby promoting GC apoptosis in Hu sheep. The findings of our study would aid in extensively improving sheep breeding in China and would provide insights into the regulatory mechanisms responsible for determining the fecundity of Hu sheep. Additionally, this study also may provide a basis for identifying new therapeutic strategies for reproductive diseases, such as polycystic ovarian syndrome (PCOS), which leads to ovulation failure.
Results
Identification of DE lncRNA in the ovaries of Hu sheep
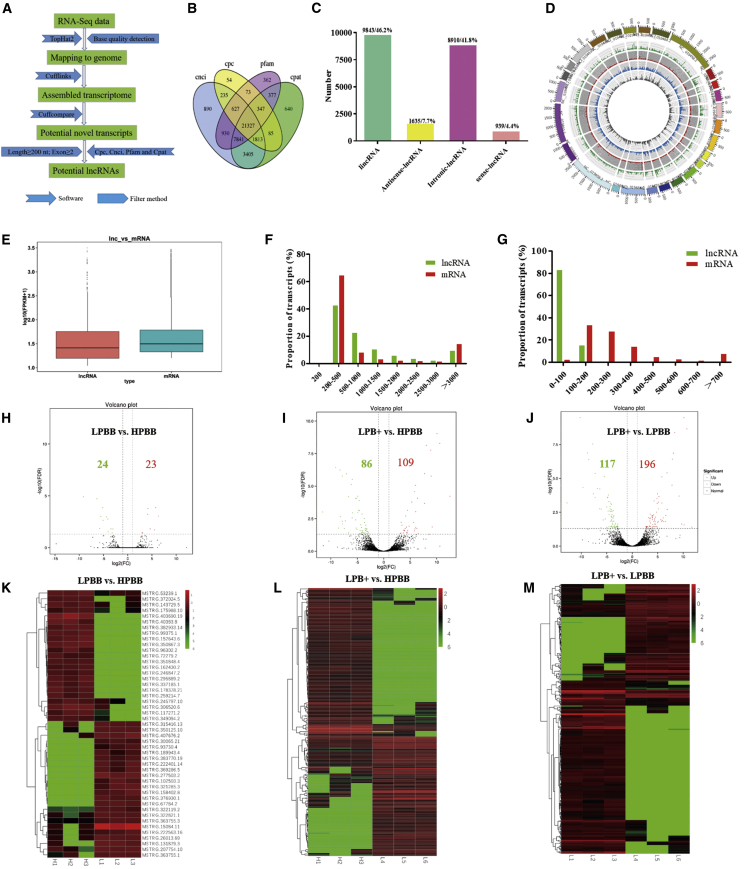
To identify putative transcripts in sheep ovaries, nine ovarian samples were obtained from different groups (HPBB, LPBB and LPB+). We acquired 59–83, 58–71, and 54–66 million unique mapped clean reads in LPB+, LPBB, and HPBB libraries, respectively (Table S1). Many lncRNAs were identified in Hu sheep ovaries according to the steps of the workflow shown in Figure 1A. In total, 21,327 lncRNAs were identified in Hu sheep ovaries (Figure 1B), including 9,843 long intergenic non-coding RNA (lincRNAs), 8,910 intronic lncRNAs, 1,635 anti-sense lncRNAs, and 939 sense lncRNAs (Figure 1C). Despite the non-uniform distribution of the lncRNA-coding sequences among the chromosomes, the number of reads mapped to the chromosome increased with increasing chromosome length (Figure 1D). The lncRNA expression level was lower than that of mRNA in Hu sheep ovaries (Figure 1E). lncRNA distribution transcripts were mainly located from 200 to 1,000 bp (Figure 1F). The lengths of the open reading frame coding for the lncRNAs and mRNA transcripts were in the range 0–100 bp and 100–300 bp, respectively (Figure 1G).
Figure 1.
Profile of lncRNA expression in the ovaries of Hu sheep and identification of DE lncRNAs
(A) Workflow for the preparation and analysis of lncRNA libraries. (B) Identification of lncRNAs in the ovarian tissue of Hu sheep. (C) Classification of lncRNAs. (D) Circos plot showing the distribution of lncRNAs in different chromosomes. The outermost ring represents different chromosomes. From the outside toward the inside: sense-lncRNA (green), lincRNA (red), intronic-lncRNA (blue), and antisense-lncRNA (gray). (E) Boxplots showing the expression levels of lncRNAs and mRNAs. FPKM, fragments per kilobase of transcript per million mapped reads. (F) Length of lncRNAs and mRNAs. (G) Lengths of open readings frames of lncRNAs and mRNAs. (H–J) Volcano plot of DE lncRNAs in each group. FDR, false discovery rate; FC, fold change. Red indicates upregulated and green indicates downregulated. (K–M) Hierarchical clustering of DE lncRNAs in each group.
There were 47 (23 upregulated and 24 downregulated), 195 (109 upregulated and 86 downregulated), and 313 (196 upregulated and 117 downregulated) DE lncRNAs identified in the LPBB versus HPBB (Figure 1H; Table S2), LPB+ versus HPBB (Figure 1I; Table S3), and LPB+ versus LPBB groups (Figure 1J; Table S4), respectively. Hierarchical clustering of the DE lncRNAs (Figures 1K–1M) revealed the expression patterns of the individuals for the same three comparisons. Additionally, we found non-uniform distribution of DE lncRNAs across the chromosomes (Figure S1).
To further evaluate RNA-seq reliability, seven DE lncRNA transcripts were randomly selected for quantitative reverse transcriptase PCR (qRT-PCR). The results obtained were consistent with those of RNA-seq, indicating the reliability of the RNA-seq data (Figure S2).
Functional enrichment and construction of lncRNA-mRNA interaction network
The roles of DE lncRNAs could be revealed by enrichment analyses of their target genes through Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The female reproduction-associated 4, 26, and 44 DE lncRNAs were screened in the LPBB versus HPBB (Table S5), LPB+ versus HPBB (Table S6), and LPB+ versus LPBB (Table S7) groups, respectively. Notably, DE lncRNA targets were most commonly enriched in reproductive signaling pathways, including TGF-β, prolactin, and insulin in the LPB+ versus HPBB and LPB+ versus LPBB groups. Moreover, certain DE lncRNAs (1, 2, and 2, respectively) were specifically expressed in the HPBB (Table S8), LPBB (Table S9) and LPB+ (Table S10) groups, indicating the potential role of these DE lncRNAs in Hu sheep prolificacy. Notably, MSTRG.98424.7 was predominantly expressed in the ovaries of the LPB+ group and had two nearly coding genes (DCN and LUM), wherein DCN is an intermediary of the TGF-β signaling pathway. Furthermore, a co-expression network was constructed for female reproduction-associated DE lncRNAs (Figure S3), which provided valuable information regarding the potential function of the analyzed lncRNAs with respect to regulating the expression of their target genes.
Identification and characterization of candidate lncRNAs
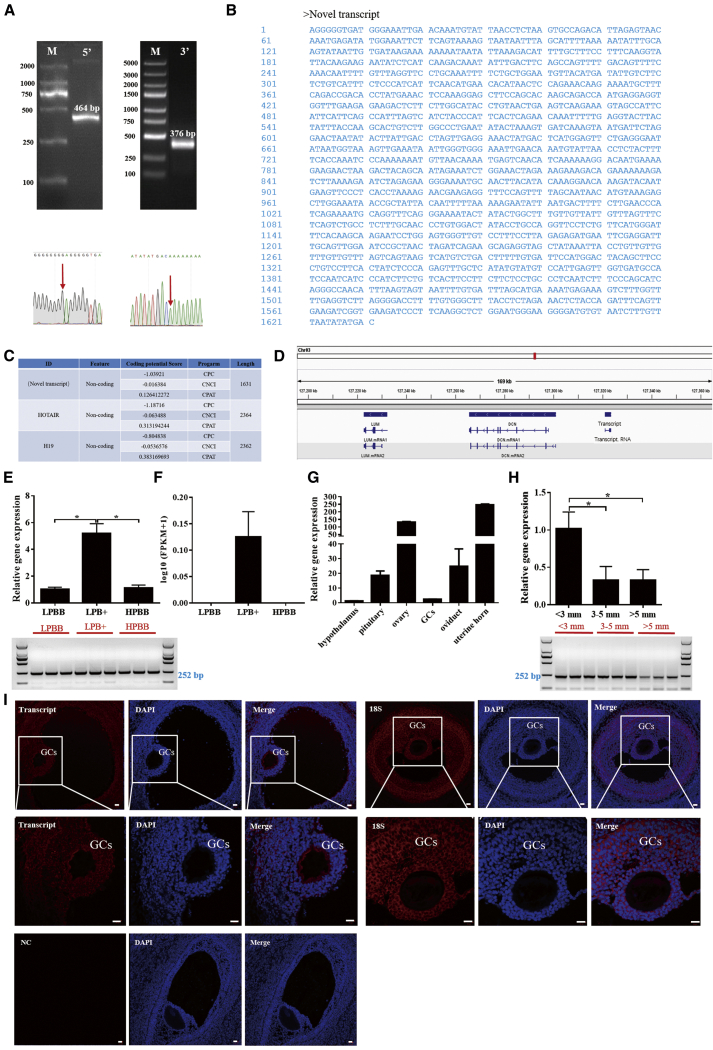
The 1,631-nt full-length sequence of the MSTRG.98424.7 transcript was generated using 5′ and 3′ rapid amplification of cloned cDNA ends (RACE; Figures 2A and 2B). The non-coding nature of this transcript was confirmed by coding-potential analysis, similar to other well-characterized lncRNAs, HOTAIR and H19 (Figure 2C; Figure S4). Furthermore, the transcript was located upstream of DCN and LUM on sheep chromosome 3 and consisted of two exons (Figure 2D). Moreover, compared with FDNCR, a region exhibiting high homology (65.54%) with LOC105607925 (known lncRNA in Ovis aries genome) (Figure S5A), annotated upstream of DCN and LUM, was observed at sheep chromosome 3 and consisted of five exons (Figure S5B).
Figure 2.
Identification and characterization of a candidate transcript in Hu sheep
(A) Representative images from RACE (5′ RACE and 3′ RACE) and Sanger DNA sequencing. (B) The full-length RNA sequence of this transcript. (C) The coding potential of this transcript and other RNAs was predicted using three computational approaches (CPC, coding potential calculator; CNCI, coding-non-coding index; CPAT, coding potential assessment tool). (D) Schematic view of the chromosomal location of this transcript. (E and F) Expression levels of this transcript in each group using qRT-PCR (E) and RNA-seq (F). (G and H) Expression level of this transcript in different tissues of the LPBB group (G) and healthy follicles of various sizes (H) from Hu sheep. (I) Localization of the transcript in the ovaries was detected by FISH. GC, granulosa cell. Scale bars, 20 μm. RNA-seq data are presented as the log10(FPKM+1) of each transcript. Values represent means ± SEM for three individuals. ∗p < 0.05.
Compared with either LPBB or HPBB group, both the expression level (Figure 2E) and RNA-seq (Figure 2F) of this transcript were significantly higher in the LPB+ group, indicating the high accuracy of the RNA-seq data. Notably, the transcript was highly expressed in the uterine horn and ovary, from the LPBB group of Hu sheep, with low expression in the hypothalamus (Figure 2G). Moreover, the transcript expression was reduced during follicular development, as determined by the follicle diameter (Figure 2H). Fluorescence in situ hybridization (FISH) indicated that the transcript was mainly expressed in the GCs and theca cells of the LPBB group (Figure 2I). Hence, the candidate transcript was termed FDNCR based on these characteristics.
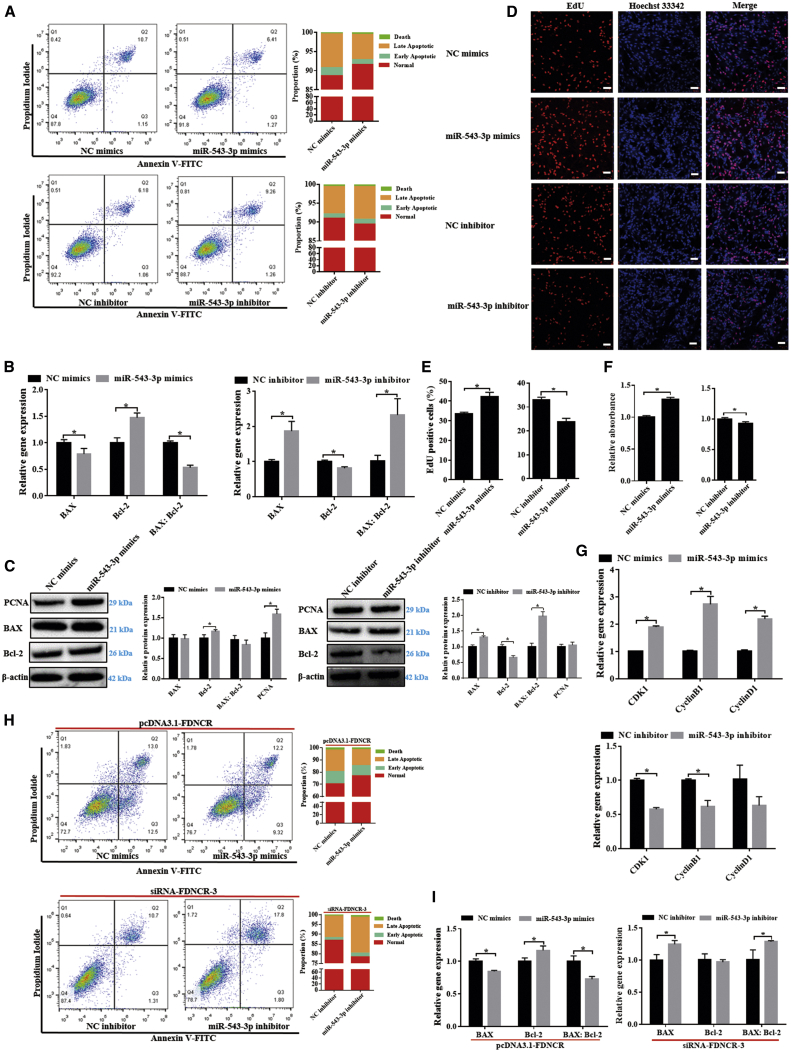
FDNCR is involved in GC apoptosis and proliferation
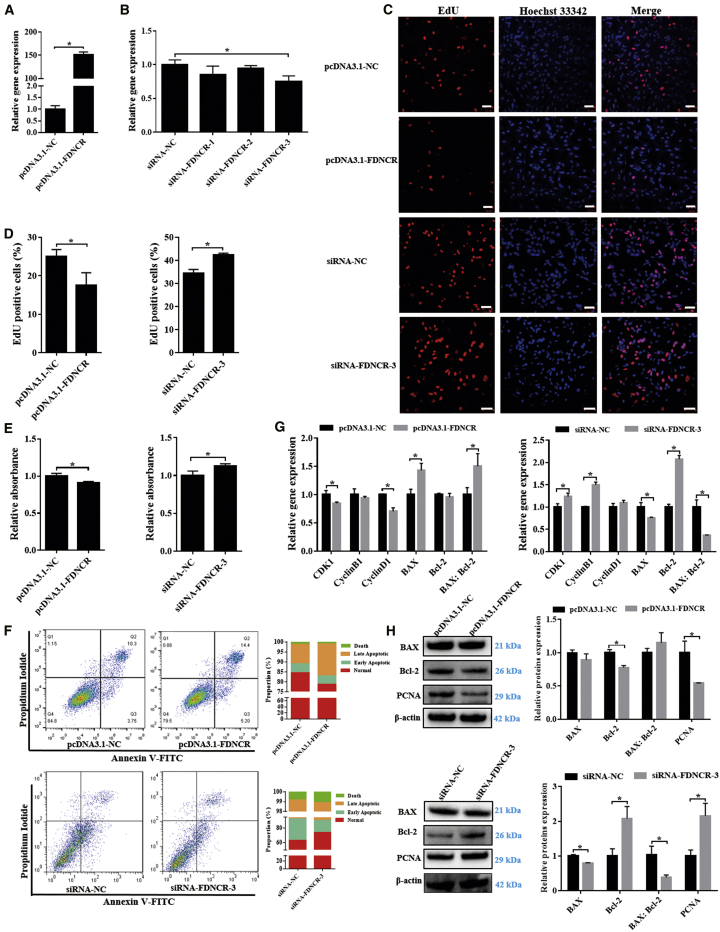
To determine the function of FDNCR in follicular development, we overexpressed and knocked down FDNCR in GCs using small interfering RNA (siRNA) and found that the transcript levels of FDNCR were significantly increased and decreased, respectively (Figures 3A and 3B). The 5-ethynyl-2′-deoxyuridine (EdU) assay indicated that overexpression or knockdown of FDNCR significantly inhibited and promoted cell proliferation, respectively (Figures 3C and 3D), which was consistent with the results obtained using the Cell Counting Kit-8 (CCK8) assay (Figure 3E). FDNCR overexpression and knockdown promoted (14.08% ± 0.51% versus 20.31% ± 0.35%) and inhibited (36.27% ± 2.81% versus 25.13% ± 3.36%) GC apoptosis, respectively (Figure 3F). Moreover, FDNCR overexpression significantly downregulated CDK1 and CyclinD1 and upregulated BAX and the the BAX/Bcl-2 ratio although not CyclinB1 and Bcl-2 mRNA expression; FDNCR knockdown dramatically upregulated CDK1, CyclinB1, and Bcl-2, although not CyclinD1 mRNA expression, and it downregulated BAX mRNA expression and the BAX/Bcl-2 ratio (Figure 3G). Similarly, PCNA and Bcl-2 protein expression levels were significantly downregulated or upregulated upon FDNCR overexpression and knockdown, respectively (Figure 3H). Furthermore, BAX protein abundance and the BAX/Bcl-2 ratio were significantly decreased upon FDNCR knockdown, whereas no change was observed upon FDNCR overexpression (Figure 3H). Overall, our data indicated that FDNCR plays vital roles in follicular development by promoting GC apoptosis.
Figure 3.
FDNCR promotes Hu sheep GC apoptosis
(A and B) mRNA expression of FDNCR in GCs in each group. (C–F) GC proliferation and apoptosis were detected using EdU (C and D), CCK8 (E), and flow cytometry (F). Scale bars, 50 μm. (G and H) mRNA (G) and protein (H) expression of cell cycle- and/or apoptosis-related genes in GCs in each group. Values represent means ± SEM for three individuals. ∗p < 0.05.
FDNCR acts as a ceRNA for miR-543-3p
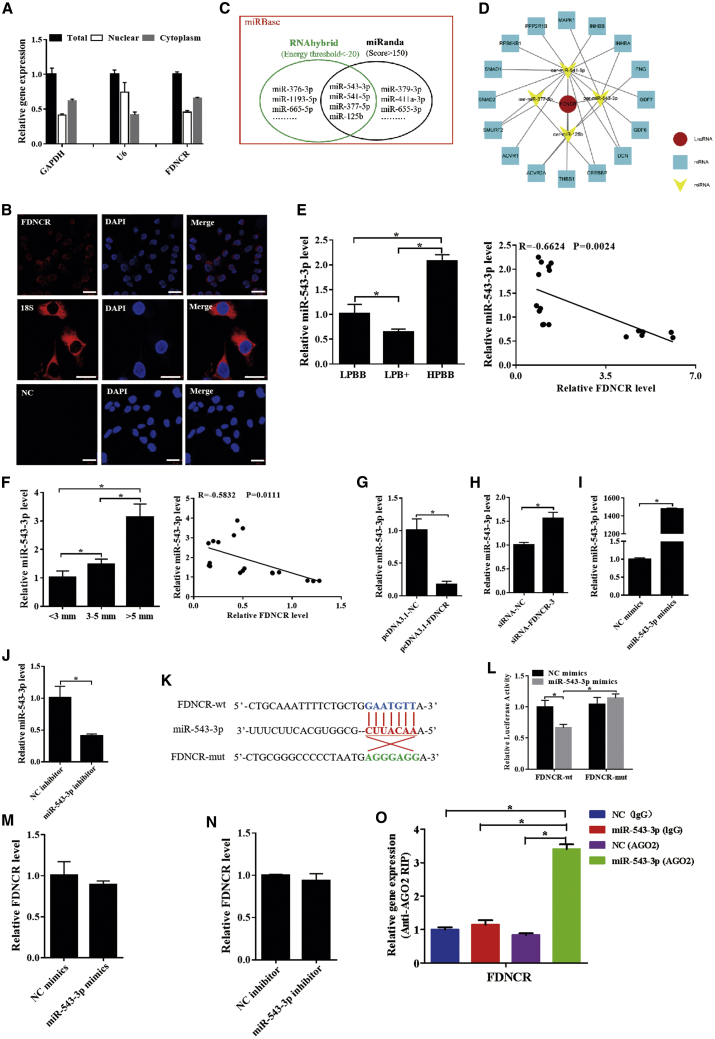
Because subcellular localization (nuclear or cytoplasmic) of lncRNAs may influence its functions, subcellular fractionation analyses were performed, which revealed that >65% of FDNCR was localized to the cytoplasm of GCs (Figure 4A); this was further confirmed using FISH (Figure 4B). Furthermore, the FDNCR-binding miRNAs miR-543-3p, miR-541-5p, miR-377-5p, and miR-125b were predicted as candidates using RNAhybrid and miRanda software (Figure 4C). A ceRNA (FDNCR-miRNA-mRNA) network with four miRNAs and 16 mRNAs was constructed in Hu sheep and belongs to the TGF-β signaling pathway (Figure 4D).
Figure 4.
FDNCR acts as a ceRNA and sponges miR-543-3p in the GCs of Hu sheep
(A and B) Subcellular localization of FDNCR was determined using qRT-PCR (A) and FISH (B). NC, negative control. Scale bars, 20 μm. (C) RNAhybrid and miRanda predicted that miRNA targets FDNCR. (D) Construction of the ceRNA network. (E and F) Expression levels of miR-543-3p in each group. Pearson’s correlation was determined between FDNCR and miR-543-3p. (G–J) Expression of miR-543-3p in the GCs in each group. (K) Schematic depicting the interactions of miR-543-3p with wild-type FDNCR (blue) and mutant FDNCR (green). Red nucleotides indicate the seed sequence of miR-543-3p. (L) The regulatory relationship between FDNCR and miR-543-3p was assessed using a dual-luciferase reporter gene assay. (M and N) Expression of FDNCR in the GCs in each group. (O) Association of FDNCR and miR-543-3p with AGO2 was investigated using the RIP assay. FDNCR and miR-543-3p expression was quantified by qRT-PCR. Values represent means ± SEM for three individuals. ∗p < 0.05.
The highest and lowest expression levels of miR-543-3p were in the HPBB group with >5-mm healthy follicles and the LPB+ group with <3-mm healthy follicles, respectively; both groups were negatively correlated with FDNCR expression (Figures 4E and 4F). Although miR-541-5p expression in the ovaries from LPBB was significantly higher than that in the LPB+ and HPBB groups, a orrelation with FDNCR expression was not apparent (Figure S6A). Moreover, miR-541-5p expression increased with increasing follicle diameter and was negatively correlated with FDNCR expression (Figure S6B). Thus, miR-543-3p and miR-541-5p may be involved in follicular development.
To further investigate the miRNA directly binding to FDNCR, qRT-PCR was performed. miR-543-3p was significantly downregulated and upregulated upon FDNCR overexpression and silencing in GCs, respectively (Figures 4G and 4H). Moreover, miR-543-3p levels increased and decreased after treatment with miR-543-3p mimics and inhibitors, respectively (Figures 4I and 4J). Dual-luciferase reporter constructs containing the miRNA response element (MRE; wild-type [WT]) and mutant (mut) plasmid were co-transfected with miR-543-3p mimics into GCs (Figure 4K). Luciferase activity of the FDNCR-MT construct was dramatically decreased, although that for the FDNCR-mut construct did not change (Figure 4L). Meanwhile, miR-543-3p overexpression or knockdown had no effect on FDNCR expression in GCs from Hu sheep (Figures 4M and 4N). Moreover, the RNA-binding protein immunoprecipitation (RIP) assay was performed using AGO2 antibody, followed by qRT-PCR, confirming the interaction between FDNCR and miR-543-3p (Figure 4O). These data indicated that FDNCR may act as a ceRNA to sponge miR-543-3p.
Next, we isolated and characterized the gene encoding miR-543-3p, which is 123 bp in length and conserved among sheep and other mammals such as goats and cows (Figure S7A). The mature sequences of miR-543-3p and the seed region of miR-543-3p are also highly conserved in other species (Figure S7B). Therefore, these results clearly demonstrated that miR-543-3p is evolutionarily conserved in mammals.
FDNCR inhibits the proliferation of GCs by sponging miR-543-3p in Hu sheep
We further explored the critical functions of miR-543-3p in GCs, and the results showed that miR-543-3p mimics reduced apoptosis (10.91% ± 0.94% versus 7.76% ± 0.08%), BAX mRNA expression, and the BAX/Bcl-2 ratio, whereas it elevated the mRNA and/or protein expression of Bcl-2 and PCNA (Figures 5A–5C). However, miR-543-3p overexpression had no effect on BAX protein levels or the BAX/Bcl-2 protein ratio (Figure 5C). In contrast, miR-543-3p inhibitor treatment increased apoptosis (8.43% ± 0.60% versus 9.95% ± 0.39%), BAX mRNA and protein expression, and the BAX/Bcl-2 ratio, and it reduced Bcl-2 mRNA and protein expression; however, miR-543-3p knockdown exerted no effect on PCNA protein levels (Figures 5A–5C). EdU and CCK8 assays showed that miR-543-3p mimics promoted GC proliferation, although cell growth was retarded upon miR-543-3p inhibitor treatment (Figures 5D–5F). miR-543-3p mimics significantly upregulated CDK1, CyclinD1, and CyclinB1 mRNA expression, whereas miR-543-3p inhibitor significantly downregulated CDK1 and CyclinB1 expression (Figure 5G). These data indicated that miR-543-3p functioned as an anti-apoptotic epigenetic regulator in GCs.
Figure 5.
miR-543-3p regulates GC proliferation and apoptosis and mediates FDNCR function
(A) GC apoptosis was detected using flow cytometry. (B and C) mRNA (B) and protein (C) expression of apoptosis-related genes in the GCs in each group. (D–F) GC proliferation was detected using EdU (D and E) and CCK8 (F). Scale bars, 50 μm. (G) Expression of cell cycle-related genes in the GCs in each group. (H and I) Apoptosis (H) and expression of apoptosis-related genes (I) were detected using flow cytometry and qRT-PCR, respectively. Values represent means ± SEM for three individuals. ∗p < 0.05.
Mediation of FDNCR functioning by miR-543-3p was further investigated. The results showed that overexpression of miR-543-3p rescued apoptosis (27.45% ± 2.60% versus 21.27% ± 1.65%) caused by FDNCR overexpression, FDNCR-induced BAX mRNA expression, and the BAX/Bcl-2 ratio, and it reduced Bcl-2 mRNA expression; conversely, inhibition of miR-543-3p prevented FDNCR-specific siRNA reduced apoptosis (12.19% ± 0.57% versus 19.17% ± 1.10%), and FDNCR-siRNA reduced BAX mRNA expression and the BAX/Bcl-2 ratio (Figures 5H and 5I). Thus, FDNCR promoted apoptosis by sponging miR-543-3p in GCs from Hu sheep.
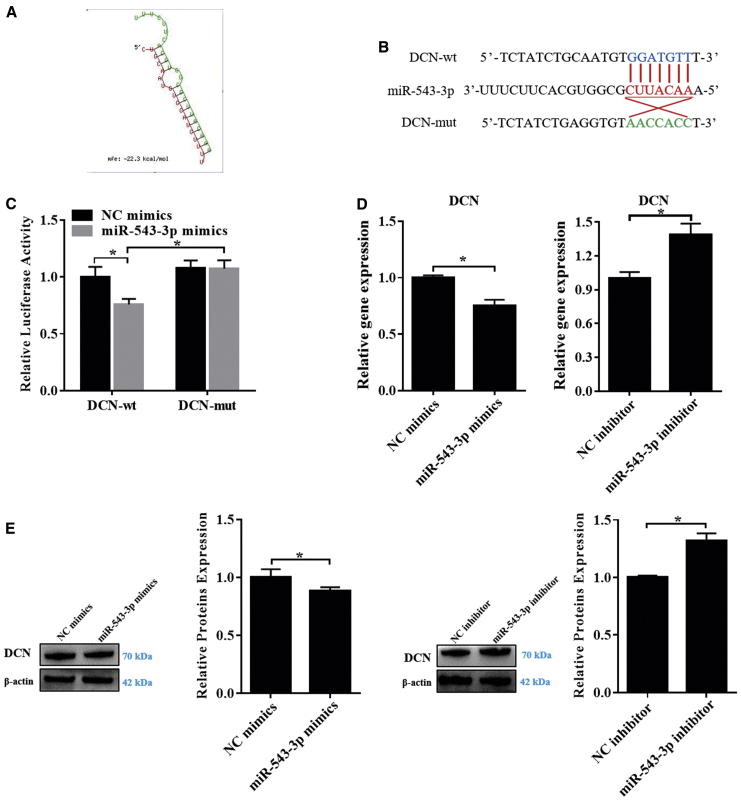
miR-543-3p regulates GC proliferation by targeting DCN
Based on the above ceRNA (FDNCR-miR-543-3p-mRNA) network, several genes (e.g., DCN and ACVR2A) were predicted to be miR-543-3p target genes. Software analysis showed that the miR-543-3p seed sequence was completely paired with the DCN 3′ UTR (Figure 6A). Luciferase reporter plasmids containing the DCN-MT MRE motif or the mutated versions were constructed (Figure 6B) and co-transfected with miR-543-3p mimics into GCs. miR-543-3p significantly reduced luciferase activity of the reporter containing the MRE-MT motif, with no change in reporter activity observed with the mutant MRE motif (Figure 6C), indicating that DCN is a direct target of miR-543-3p. Furthermore, miR-543-3p mimics and inhibitor markedly downregulated and upregulated DCN mRNA and protein expression levels in GCs, respectively (Figures 6D and 6E). Collectively, these results implied that DCN is a functional target of miR-543-3p in GCs of Hu sheep. DCN was predominantly localized in GCs in all follicle types and predominantly localized in the cytoplasm of cultured GCs (Figures S8A and SB). Additionally, the highest expression of DCN was noted in the uterine horn and ovary (Figure S8C), and expression increased with follicle diameter, although no significant difference was observed (Figure S8D), suggesting that DCN is important in follicular development.
Figure 6.
DCN is a direct target of miR-543-3p in the GCs of Hu sheep
(A) miRNA-response elements (MREs) within the 3′ UTR of sheep DCN that enable the binding of miR-543-3p were predicted using RNAhybrid. mfe, minimum free energy. (B) Schematic depicting the interaction of miR-543-3p with wild-type (blue) and mutant DCN (green). Red nucleotides indicate the seed sequence of miR-543-3p. (C) The regulatory relationship between miR-543-3p and DCN was assessed using a dual-luciferase reporter gene assay. (D and E) Expression of DCN mRNA (D) and protein (E) was detected in each group. Values represent means ± SEM for three individuals. ∗p < 0.05.
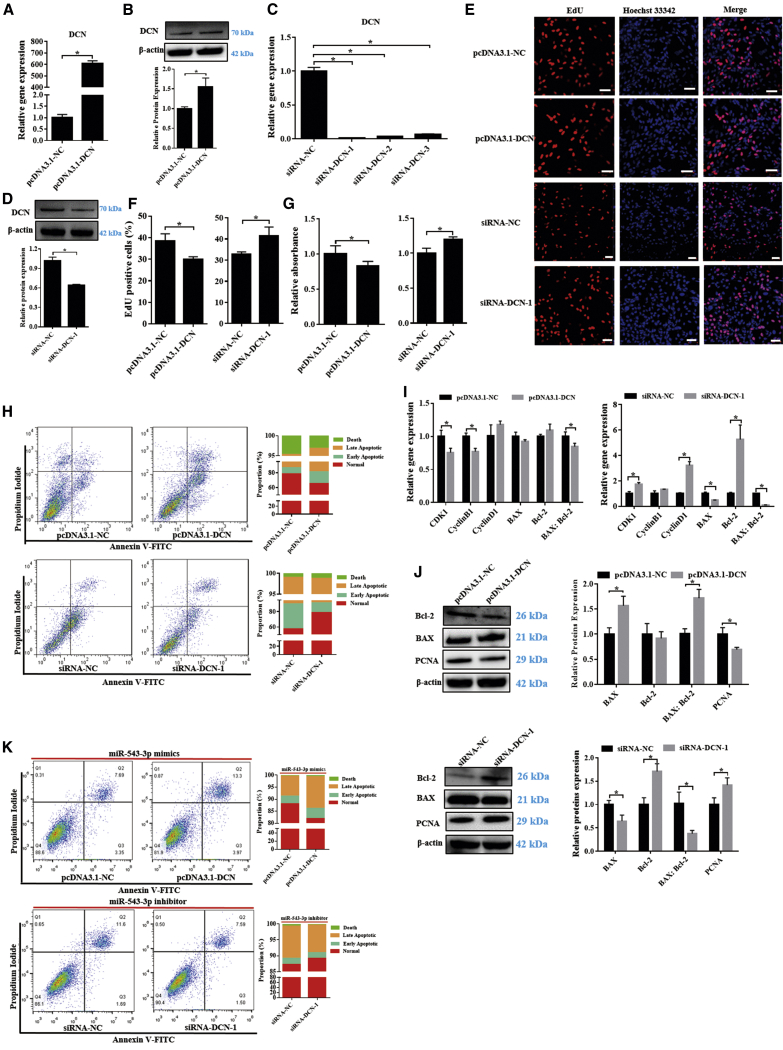
We investigated the potential functions of DCN in GCs from Hu sheep. The synthesized expression vector pcDNA3.1-DCN and DCN-specific siRNA (siRNA-DCN-1) increased and decreased DCN mRNA and protein levels, respectively (Figures 7A–7D). Notably, DCN overexpression by pcDNA3.1-DCN transfection and DCN knockdown by siRNA-DCN-1 inhibited and promoted GC proliferation, respectively (Figures 7E–7G). DCN overexpression and knockdown significantly promoted (17.13% ± 3.32% versus 31.20% ± 2.25%) and inhibited (41.38% ± 3.28% versus 19.91% ± 3.33%) GC apoptosis, respectively (Figure 7H). Moreover, DCN overexpression significantly downregulated CDK1 and CyclinB1 mRNA expression, as well as the BAX/Bcl-2 ratio; however, DCN overexpression had no effect on CyclinD1, BAX, and Bcl-2 mRNA levels (Figure 7I). Furthermore, DCN knockdown significantly upregulated CDK1, CyclinD1 and Bcl-2 mRNA levels and downregulated BAX mRNA expression and the BAX/Bcl-2 ratio; however, CyclinB1 mRNA expression was not significantly affected (Figure 7I). Furthermore, DCN overexpression significantly upregulated BAX protein expression and the BAX/Bcl-2 ratio, and it downregulated PCNA protein levels, whereas the opposite trend was observed in the DCN knockdown group (Figure 7J). Overall, these data suggest that DCN is a pro-apoptotic regulator in GCs from Hu sheep.
Figure 7.
miR-543-3p enhances GC proliferation by targeting DCN
(A–D) DCN expression at mRNA and protein levels in the GCs in each group. (E–H) GC proliferation and apoptosis were detected using EdU (E and F), CCK8 (G), and flow cytometry (H). Scale bars, 50 μm. (I and J) mRNA (I) and protein (J) expression of cell cycle- and/or apoptosis-related genes in the GCs in each group. (K) GC apoptosis in each group was detected using flow cytometry. Values represent means ± SEM for three individuals. ∗p < 0.05.
We further investigated the regulation of GC apoptosis by DCN-mediated miR-543-3p. miR-543-3p mimic or inhibitor and pcDNA3.1-DCN or siRNA-DCN-1 were co-transfected into GCs. Overexpression of DCN reduced the anti-apoptotic effect of miR-543-3p mimics (11.37% ± 0.22% versus 17.43% ± 2.69%), whereas apoptosis (11.92% ± 0.79% versus 10.36% ± 0.75%) induced by miR-543-3p inhibitor was recovered when DCN was knocked down (Figure 7K). Overall, these data strongly suggest that miR-543-3p regulates GC apoptosis by targeting DCN.
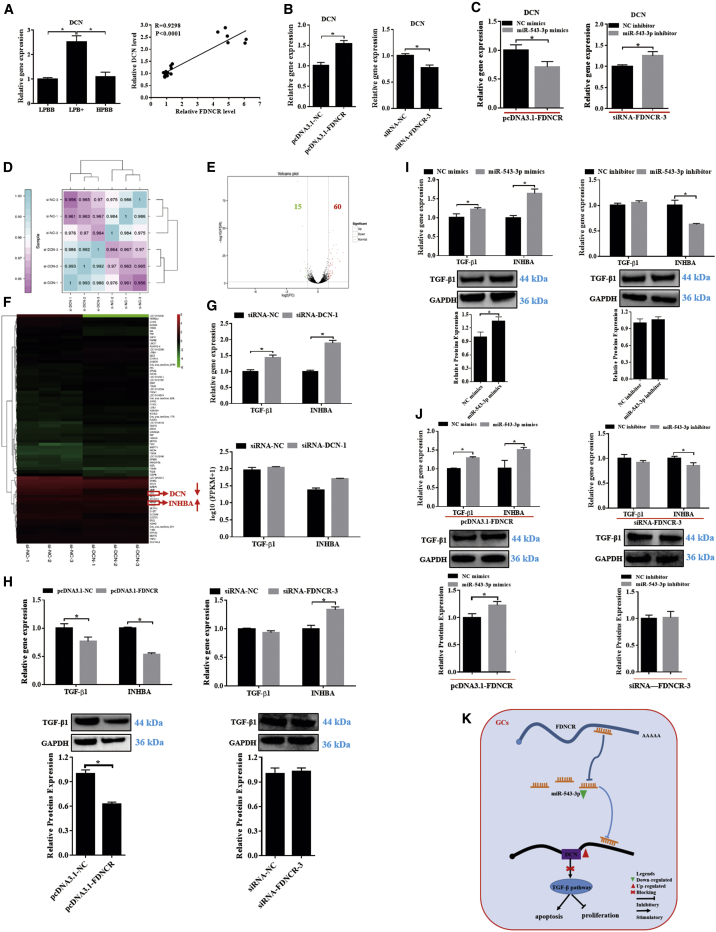
FDNCR regulates the TGF-β pathway via miR-543-3p-DCN axis
We investigated the mechanism through which FDNCR acted as an upstream regulator of miR-543-3p to affect DCN expression. Notably, ovarian FDNCR expression in Hu sheep with different prolificacy was positively correlated with DCN mRNA levels (Figure 8A). DCN mRNA levels were significantly upregulated and downregulated in GCs after transfection with pcDNA3.1-FDNCR and siRNA-FDNCR-3, respectively (Figure 8B), which indicates that FDNCR is a regulator of DCN. Moreover, miR-543-3p mimics inhibited FDNCR-induced DCN mRNA expression, and miR-543-3p inhibitor rescued siRNA-FDNCR-3-reduced DCN expression (Figure 8C). These data indicated that miR-543-3p mediated FDNCR-induced DCN expression in GCs of Hu sheep.
Figure 8.
FDNCR regulates DCN and TGF-β signaling pathways by affecting the levels of miR-543-3p in the GCs of Hu sheep
(A) DCN expression in the ovaries in each group. Pearson’s correlation between FDNCR and miR-543-3p. (B and C) Expression of DCN in the GCs in each group. (D) Correlation coefficient between siRNA-NC and siRNA-DCN. (E) Volcano plot showing the DE genes between the siRNA-NC and siRNA-DCN groups. Red indicates upregulated, green indicates downregulated genes. (F) Hierarchical clustering showing DE genes between the siRNA-NC group and siRNA-DCN group. (G) Validation of TGF-β1 and INHBA mRNA expression in different groups. (H–J) mRNA or protein expression of TGF-β1 and INHBA in the GCs in each group. (K) Proposed model of FDNCR regulation of GCs state in Hu sheep. RNA-seq data are presented as the log10(FPKM+1) of each transcript. Values represent means ± SEM for three individuals. ∗p < 0.05.
DCN inhibits TGF-β by binding to its receptors, which leads to interference with the Smad and non-Smad pathways downstream of the TGF-β receptors.38, 39, 40 Therefore, RNA-seq was performed to elucidate genes downstream of DCN after transfection with siRNA-DCN-1 in GCs, and that of the TGF-β signaling pathway was noted. RNA-seq data quality was verified by correlation coefficient trends; hence, further analyses were performed (Figure 8D). 15 downregulated and 60 upregulated DE genes were identified (Figure 8E; Table S11), and hierarchical clustering thereof revealed expression profiles involving the siRNA-negative control (NC) and siRNA-DCN groups (Figure 8F). TGF-β signaling pathway-related genes, including TGF-β1 and inhibin beta A (INHBA), were upregulated upon DCN silencing and exhibited similar expression patterns (Figure 8G).
Furthermore, we investigated the interaction between the FDNCR-miR-543-3p axis and TGF-β1 and INHBA. FDNCR overexpression downregulated TGF-β1 and INHBA mRNA or protein expression, and the mRNA or protein levels of INHBA, although not TGF-β1, were significantly increased after FDNCR knockdown (Figure 8H). However, miR-543-3p mimics increased TGF-β1 and INHBA mRNA or protein levels, whereas expression of INHBA, although not TGF-β1, mRNA or protein was downregulated upon miR-543-3p knockdown (Figure 8I). Moreover, miR-543-3p increased FDNCR-reduced TGF-β1 and INHBA mRNA or protein expression. Conversely, miR-543-3p inhibitor decreased FDNCR-induced INHBA, although not TGF-β1, mRNA or protein levels (Figure 8J). Collectively, these results indicated that the miR-543-3p-DCN axis could have mediated FDNCR function in the TGF-β pathway in GCs from Hu sheep (Figure 8K).
Discussion
RNA-seq data revealed that most lncRNAs, which may be byproducts of mRNA, showed low expression levels in ovarian tissue from Hu sheep. However, several abundant DE lncRNAs were identified in Hu sheep with different prolificacy. Moreover, the targets of DE lncRNAs were enriched in certain signaling pathways involved in female reproduction and GC functions, such as the prolactin,41 insulin,42 ovarian steroidogenesis,43 and TGF-β signaling pathways,44 suggesting that DE lncRNAs have specific roles in regulating fecundity in Hu sheep. Therefore, consistent with previous reports, lncRNAs might be associated with fecundity in sheep or goats.25, 26, 27, 28, 29,31,32 Moreover, FDNCR was highly expressed in the uterine horn and ovary, and expression levels decreased with increasing follicle diameters, suggesting that FDNCR is important for follicular development.
In this study, we demonstrated that FDNCR controls follicular development by regulating GC apoptosis and proliferation. Follicular atresia decreases the number of ovulations and restricts reproductive potential.45 FDNCR was predominantly expressed in the ovaries of the LPB+ group, indicating that FDNCR was involved in fecundity by promoting follicular atresia. FDNCR, which is highly expressed in the ovaries of the LPB+ group, may provide insight into the mechanisms through which lncRNA regulates fecundity in sheep. Further studies to investigate other functions of FDNCR in regulating GCs (e.g., steroidogenesis), follicular development, or ovulation in vivo and reproductive performance such as litter size in Hu sheep are required.
lncRNAs are known to modulate post-transcriptional regulation by acting as miRNA sponges to decrease their target mRNAs.46,47 NORFA reportedly regulates porcine GC apoptosis by sponging miR-126.37 lncRNAs PTV1 and PCAT6 act as ceRNAs to sponge miR-543 to regulate SERPINI1 and ZEB1 expression, respectively.48,49 In the present study, we found that FDNCR sponged miR-543-3p to negatively regulate miR-543-3p function in GC proliferation and apoptosis. Alternatively, miR-543-3p promoted GC proliferation, which was consistent with previous studies.50, 51, 52 This effect could be eliminated by FDNCR overexpression, suggesting that FDNCR regulates GC apoptosis or proliferation by sponging miR-543-3p. These results further demonstrated that FDNCR could serve as a regulator of follicular development.
Notably, our results showed that miR-543-3p targeted DCN. Several studies have indicated that DCN is widely expressed in reproductive tissues, including the ovary and uterus of goats, mice, sheep, and monkeys.53, 54, 55, 56 Consistently, DCN was highly expressed in the ovary, particularly in GCs and the uterine horn from the LPBB group of Hu sheep, suggesting its involvement in the regulation of follicular development. Moreover, miR-543-3p and DCN in GCs produced opposing effects, whereby DCN promoted GC apoptosis in Hu sheep. Consistently, DCN overexpression can induce GC apoptosis or suppress cell proliferation in goat and hamster ovaries.53,57 These observations indicate that miR-543-3p regulates GC apoptosis or proliferation by targeting DCN. Furthermore, FDNCR overexpression increased DCN expression, whereas this effect was eliminated by miR-543-3p. This result further demonstrated that binding of FDNCR to miR-543-3p regulates GC state and function by targeting DCN. Similarly, LSAMP-AS1 binds to miR-183-5p to suppress prostate cancer progression by upregulating DCN gene expression.58
DCN can inhibit TGF-β by binding to its receptors, leading to interference with the Smad and non-Smad pathways downstream of TGF-β receptors.38, 39, 40 In the present study, TGF-β1 and INHBA genes were upregulated during DCN silencing. Increasing evidence has demonstrated that lncRNA or miRNA directly interacts with the TGF-β pathway to regulate the state and function of GCs.37,59,60 Notably, this study demonstrated that miR-543-3p restored TGF-β1 and INHBA expression upon FDNCR overexpression. Hence, those results confirmed that the FDNCR-miR-543-3p axis interacts with the TGF-β pathway.
In conclusion, numerous lncRNAs with different abundance in Hu sheep ovaries with different prolificacy were annotated. We further identified FDNCR, which is predominantly expressed in the ovaries of the LPB+ group and involved in follicular development. Our findings confirm the mechanism whereby FDNCR sponges miR-543-3p in GCs and prevents miR-543-3p binding to the DCN 3′ UTR, which subsequently releases DCN and blocks the TGF-β pathway to promote GC apoptosis in Hu sheep. Ovulation is the culmination of female reproductive activity and is essential for successful pregnancy; hence, most female reproductive diseases, such as PCOS, are caused by ovulation defects. Recently, a number of lncRNAs implicated in PCOS, such as lncRNA PVT1 and HOTAIR, have been identified.22,23 Therefore, this study identified a candidate lncRNA (FDNCR) involved in Hu sheep fecundity, providing insights into the regulatory mechanisms underlying follicular development, which will provide a basis for new therapeutic strategies of reproductive diseases.
Materials and methods
Animals and sample collection
Hu sheep were raised at the Taizhou Sheep Farm (Jiangsu, China) under similar conditions, with free access to food and water and under natural lighting. Twenty non-pregnant healthy ewes (2∼3 years age) with identical litter size numbers of three records were randomly selected and divided into HP (each records of litter size=3; n = 4) and LP (each records of litter size=1; n = 16) groups.3,61 Finally, nine ewes were selected and divided into HPBB (n = 3), LPBB (n = 3), and LPB+ groups (n = 3). Synchronous estrus was established as described previously.26 The estrus status of the ewes was checked daily. Ewes were slaughtered within 12 h during the estrus stage, and ipsilateral ovary and female reproductive organs (oviduct, uterine horn, hypothalamus, and pituitary) were immediately stored at −80°C. The other ovary was fixed in 4% formaldehyde for 24 h and embedded in paraffin for further analysis. BMPRIB polymorphism genotyping and ovarian morphological characteristic results are shown in Figure S9. All experimental procedures, including animal care, were approved by the Institutional Animal Care and Use Committee of Nanjing Agricultural University (SYXK 2011-0036).
RNA-Seq and bioinformatics analysis
Total RNA was extracted from ovarian tissue of nine ewes, assessed by electrophoresis, and quantified using a NanoDrop 2000 (NanoDrop Technologies, Wilmington, DE, USA). Library preparation and Illumina sequencing analysis were performed as previously described.29 Unknown transcripts were used to screen putative lncRNAs, as described previously.26 To investigate interactions between lncRNAs and mRNAs, we constructed a complementary pair network comprising mRNA and lncRNA using Cytoscape 3.7.1 (https://cytoscape.org/).62 The coding potential of FDNCR was analyzed by PhyloCSF and open reading frame (ORF) Finder (https://www.ncbi.nlm.nih.gov/orffinder/). Venn diagram indicating the intersected genes was generated using a draw Venn diagram online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). Heatmaps were generated by using the R package (heatmap). The raw sequencing dataset supporting the results of this study have been submitted to NCBI Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/) under accession no. SUB8643608, BioProject: PRJNA681969.
5′ and 3′ RACE
The full-length sequence of FDNCR was obtained using a 5′-RACE/3′-RACE kit (Thermo Fisher Scientific, MA, USA), according to the manufacturer’s instructions. 5′-RACE and 3′-RACE were performed by nested PCR for each reaction. The abridged anchor and abridged universal anchor primers supplied with the kit were used. Specific primers are listed in Tables S12.
miRNA prediction for targeting FDNCR and DCN
DCN 3′ UTR and FDNCR sequences were obtained from NCBI (XM_012159321.3) and this study, respectively. Interactions involving miRNA-mRNA and FDNCR-miRNA were predicted based on the mature miRNA sequence of sheep using RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) and miRanda (https://www.miranda-ng.org/en/). Additionally, mature miRNA sequences from different species were obtained from miRBase (http://www.mirbase.org).
Vector construction
The full-length FDNCR and coding DNA sequence region (GenBank: NM_001009218.1) of DCN were cloned into a pcDNA3.1 overexpression vector. FDNCR or DCN 3′ UTR fragments containing potential miR-543-3p MRE motifs were isolated and inserted into pmirGLO. siRNAs (siRNA-FDNCR and siRNA-DCN) and miR-543-3p mimics and inhibitor, in addition to the corresponding NC, were synthesized by GenePharma (Shanghai, China; Table S13).
Ovarian GC culture treatments and follicle size classification
Hu sheep ovaries were collected from a local abattoir (Taicang, Jiangsu, China; 121°10ʹE, 31°45ʹN) during the breeding season (October to January). GCs were isolated from healthy follicles (2–5 mm) and cultured as previously described.63 Briefly, GCs were seeded into different plates (6 wells, 1 × 106 cells/well; 12 wells, 5 × 105 cells/well; 24 wells, 1 × 105 cells/well; 96 wells, 1 × 104 cells/well) in culture medium (Dulbecco’s modified Eagle’s medium/nutrient mixture F-12 [DMEM/F12] supplemented with 10% fetal bovine serum [FBS], 2 mM l-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin) at 37°C in a humidified atmosphere containing 5% CO2. After overnight incubation (at 60%∼70% confluence), transfection or co-transfection were performed using Lipofectamine 3000 (Invitrogen, Shanghai, China) for 48 h. 100 nM/mL siRNA-FDNCR, siRNA-DCN, and miR-543-3p mimics and inhibitor and 1.25 μg/mL pcDNA3.1-FDNCR and pcDNA3.1-DCN plasmid were used in this study. RNA-seq was performed to identify genes downstream of DCN by silencing DCN in GCs from Hu sheep. The full datasets have been submitted to NCBI SRA (accession no. SUB8657413, BioProject: PRJNA681969). All visible antral follicles were dissected, measured with a caliper, and classified into three size classes (<3, 3–5, and >5 mm) of approximately 30 ovaries. In total, 18 large follicles (>5 mm), 36 medium follicles (3–5 mm), and 80 small follicles (<3 mm) were collected. Subsequently, three follicles were randomly selected for DCN mRNA level analysis from the large category, while three pooled samples were formed from all collected follicles for the medium and small categories. All reagents used in this study were purchased from Life Technologies (Pleasanton, CA, USA), unless otherwise mentioned.
Cell proliferation analysis
Cell proliferation was analyzed using CCK8 (KeyGen, Nanjing, China) and EdU incorporation assays (KeyGen, Nanjing, China), as described previously.64,65 Nuclei were stained with Hoechst 33342.
Cell apoptosis analysis
Cell apoptosis was analyzed by flow cytometry (BD Biosciences, NJ, USA) using an annexin V-fluorescein isothiocyanate/propidium iodide apoptosis detection kit (Vazyme, Nanjing, China). All data were analyzed using FlowJo software.
Immunohistochemistry and immunofluorescence
Immunohistochemistry was performed following our previously described method.66 Rabbit anti-DCN and goat anti-rabbit immunoglobulin G (IgG) were the primary and secondary antibodies, respectively. The negative controls were incubated with Tris-buffered saline instead of primary antibodies. All sections were stained with diaminobenzidine (DAB; Boster, Wuhan, China) and examined with a light microscope (Nikon, Tokyo, Japan). All antibodies were purchased from commercial suppliers (Table S14).
Immunofluorescence analysis was performed as described previously.67 Rabbit anti-DCN and CoraLite 594-conjugated goat anti-rabbit IgG were used as primary and secondary antibodies, respectively. Nuclei were stained with DAPI and examined with an LSM 710 laser scanning confocal microscope (Carl Zeiss, Oberkochen, Germany).
FISH analysis
RNA FISH analysis was performed to assess FDNCR localization in GCs using a FISH kit (GenePharma, Shanghai, China) and an FDNCR FISH probe mix (Cy3 labeled), following the manufacturer’s instructions. 18S was used as a positive control for cytoplasmic fractions. Nuclei were stained with DAPI. Images were captured using an LSM 710 laser scanning confocal microscope.
qRT-PCR analysis
Total RNA extraction and cDNA synthesis were performed as described previously.68,69 All qRT-PCR reactions were performed in an ABI 7500 real-time PCR system (Applied Biosystems) using SYBR Green master mix (Roche, Mannheim, Germany) following the manufacturer’s instructions. Briefly, the reaction was performed at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and a dissociation step consisting of 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s. miR-16b (for miRNA) and GAPDH were used as internal controls. Primer sequences are listed in Tables S15 and S16. The 2–ΔΔCT method was used to analyze relative expression levels.
RIP assay
A RIP assay was performed using a RIP assay kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. Briefly, GCs were collected and lysed using RIP lysis buffer. Then, the cell lysates were incubated with magnetic beads conjugated with anti-Ago2 antibody (Abcam, Cambridge, UK). Thereafter, the RNA-protein complex was extracted, and the abundance of FDNCR and miR-543-3p in bound fractions was evaluated using qRT-PCR analysis.
Subcellular localization
For nuclear and cytoplasmic RNA separation, 1 × 106 cells were collected and extracted using a Paris kit (Life Technologies, Pleasanton, CA, USA), according to the manufacturer’s instructions. U6 and GAPDH were used as positive controls for the nucleus and cytoplasm, respectively.
Western blot analysis
Western blotting was performed according to our previously described methods with slight modification.66 Briefly, rabbit anti-DCN, anti-BAX, anti-Bcl-2, anti-PCNA, anti-β-actin, anti-TGF-β1, and mouse anti-GAPDH were used as primary antibodies, while goat anti-rabbit IgG and goat anti-mouse IgG were used as the secondary antibodies. Immunoreactions were visualized using an enhanced chemiluminescence detection system (Fujifilm, Tokyo, Japan). The protein band intensity was quantified using ImageJ software.
Dual-luciferase reporter assays
After 48 h of transfection, GCs were collected and luciferase activity was determined using the Dual-Luciferase reporter assay system (Vazyme, Nanjing, China), according to the manufacturer’s protocol.
Statistical analysis
All experiments were performed at least three times. Data are presented as means ± standard error of the mean based on three independent experiments. All data were normally distributed, and variance was similar between the statistically compared groups. Statistical analyses were performed using GraphPad Prism 6 (GraphPad, San Diego, CA, USA). A Student’s t test (two-tailed) or one-way analysis of variance (ANOVA) was performed followed by the Student-Newman-Keuls (SNK) method for multiple comparisons. p values <0.05 were considered statistically significant.
Acknowledgments
This work was supported by the earmarked funds for the National Natural Science Foundation of China (31872357 and 32002174) and by the Fundamental Research Funds for the Central Universities (KYYJ202001). We sincerely thank Shijia Ying and Rihong Guo from the Jiangsu Academy of Agricultural Sciences for their full support.
Author contributions
F.W. and X.Y. designed the study, and X.Y., X.G., and Y.B. performed the experiments and drafted the manuscript. J.Y., X.L., and Z.W. assisted in conducting the experiments and analyzed the data. X.Y., X.G., and Z. W. collected tissue samples. M.A.E.-S., G.Z., Y.Z., F.W., and W.L. assisted in revision of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.02.030.
Supplemental information
References
- 1.Yue G.H. Reproductive characteristics of Chinese Hu sheep. Anim. Reprod. Sci. 1996;44:223–230. [Google Scholar]
- 2.McGee E.A., Hsueh A.J.W. Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 2000;21:200–214. doi: 10.1210/edrv.21.2.0394. [DOI] [PubMed] [Google Scholar]
- 3.Chu M.X., Liu Z.H., Jiao C.L., He Y.Q., Fang L., Ye S.C., Chen G.H., Wang J.Y. Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep (Ovis aries) J. Anim. Sci. 2007;85:598–603. doi: 10.2527/jas.2006-324. [DOI] [PubMed] [Google Scholar]
- 4.Fabre S., Pierre A., Mulsant P., Bodin L., Di Pasquale E., Persani L., Monget P., Monniaux D. Regulation of ovulation rate in mammals: contribution of sheep genetic models. Reprod. Biol. Endocrinol. 2006;4:20. doi: 10.1186/1477-7827-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gootwine E., Reicher S., Rozov A. Prolificacy and lamb survival at birth in Awassi and Assaf sheep carrying the FecB (Booroola) mutation. Anim. Reprod. Sci. 2008;108:402–411. doi: 10.1016/j.anireprosci.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q.Y., Yang K., Liu F.G., Sun X.G., Chen L., Xiu H., Liu X.S. Long noncoding RNA CASC2c inhibited cell proliferation in hepatocellular carcinoma by inactivated ERK1/2 and Wnt/β-catenin signaling pathway. Clin. Transl. Oncol. 2020;22:302–310. doi: 10.1007/s12094-019-02223-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhou X.B., Lai L.F., Xie G.B., Ding C., Xu X., Wang Y. lncRNA GAS5 sponges miRNA-221 to promote neurons apoptosis by up-regulated PUMA under hypoxia condition. Neurol. Res. 2020;42:8–16. doi: 10.1080/01616412.2019.1672382. [DOI] [PubMed] [Google Scholar]
- 9.Kang X., Zhao Y., Van Arsdell G., Nelson S.F., Touma M. Ppp1r1b-lncRNA inhibits PRC2 at myogenic regulatory genes to promote cardiac and skeletal muscle development in mouse and human. RNA. 2020;26:481–491. doi: 10.1261/rna.073692.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu T., Wang S., Wang L., Zhang W., Chen W., Lv X., Li Y., Hussain Z., Sun W. Long noncoding RNA (lncRNA) CTTN-IT1 elevates skeletal muscle satellite cell proliferation and differentiation by acting as ceRNA for YAP1 through absorbing miR-29a in Hu sheep. Front. Genet. 2020;11:843. doi: 10.3389/fgene.2020.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dasgupta P., Kulkarni P., Majid S., Hashimoto Y., Shiina M., Shahryari V., Bhat N.S., Tabatabai L., Yamamura S., Saini S. lncRNA CDKN2B-AS1/miR-141/cyclin D network regulates tumor progression and metastasis of renal cell carcinoma. Cell Death Dis. 2020;11:660. doi: 10.1038/s41419-020-02877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Li D., Lu J., Chen L., Zhang S., Qi W., Li W., Xu H. Long noncoding RNA TTN-AS1 facilitates tumorigenesis and metastasis by maintaining TTN expression in skin cutaneous melanoma. Cell Death Dis. 2020;11:664. doi: 10.1038/s41419-020-02895-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trovero M.F., Rodríguez-Casuriaga R., Romeo C., Santiñaque F.F., François M., Folle G.A., Benavente R., Sotelo-Silveira J.R., Geisinger A. Revealing stage-specific expression patterns of long noncoding RNAs along mouse spermatogenesis. RNA Biol. 2020;17:350–365. doi: 10.1080/15476286.2019.1700332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rolland A.D., Evrard B., Darde T.A., Le Béguec C., Le Bras Y., Bensalah K., Lavoué S., Jost B., Primig M., Dejucq-Rainsford N. RNA profiling of human testicular cells identifies syntenic lncRNAs associated with spermatogenesis. Hum. Reprod. 2019;34:1278–1290. doi: 10.1093/humrep/dez063. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y., Wang J., Fan Y., Qin C., Xia X., Johnson J., Kallen A.N. Absence of the long noncoding RNA H19 results in aberrant ovarian STAR and progesterone production. Mol. Cell. Endocrinol. 2019;490:15–20. doi: 10.1016/j.mce.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D., Jiang W., Jiang Y., Wang S., Fang J., Zhu L., Zhu Y., Yan G., Sun H., Chen L., Zhang N. Preliminary functional inquiry of lncRNA ENST00000433673 in embryo implantation using bioinformatics analysis. Syst Biol Reprod Med. 2019;65:164–173. doi: 10.1080/19396368.2018.1563844. [DOI] [PubMed] [Google Scholar]
- 17.Wu M., Zhang S., Chen X., Xu H., Li X. Expression and function of lncRNA MALAT-1 in the embryonic development of zebrafish. Gene. 2019;680:65–71. doi: 10.1016/j.gene.2018.09.037. [DOI] [PubMed] [Google Scholar]
- 18.Ernst E.H., Nielsen J., Ipsen M.B., Villesen P., Lykke-Hartmann K. Transcriptome analysis of long non-coding RNAs and genes encoding paraspeckle proteins during human ovarian follicle development. Front. Cell Dev. Biol. 2018;6:78. doi: 10.3389/fcell.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X., Pan J., Wu B., Teng X. Construction and analysis of a lncRNA (PWRN2)-mediated ceRNA network reveal its potential roles in oocyte nuclear maturation of patients with PCOS. Reprod. Biol. Endocrinol. 2018;16:73. doi: 10.1186/s12958-018-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong Y., Yao G., He J., Yang G., Kong D., Sun Y. lncRNA LNC-GULP1-2:1 is involved in human granulosa cell proliferation by regulating COL3A1 gene. Fertil. Steril. 2018;110(4, Suppl):E320. [Google Scholar]
- 21.Zhao J., Xu J., Wang W., Zhao H., Liu H., Liu X., Liu J., Sun Y., Dunaif A., Du Y., Chen Z.J. Long non-coding RNA LINC-01572:28 inhibits granulosa cell growth via a decrease in p27 (Kip1) degradation in patients with polycystic ovary syndrome. EBioMedicine. 2018;36:526–538. doi: 10.1016/j.ebiom.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu G., Liu S., Xing G., Wang F. lncRNA PVT1/microRNA-17-5p/PTEN axis regulates secretion of E2 and P4, proliferation, and apoptosis of ovarian granulosa cells in PCOS. Mol. Ther. Nucleic Acids. 2020;20:205–216. doi: 10.1016/j.omtn.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Jiang B., Xue M., Xu D., Song J., Zhu S. Down-regulated lncRNA HOTAIR alleviates polycystic ovaries syndrome in rats by reducing expression of insulin-like growth factor 1 via microRNA-130a. J. Cell. Mol. Med. 2020;24:451–464. doi: 10.1111/jcmm.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor D.H., Chu E.T.J., Spektor R., Soloway P.D. Long non-coding RNA regulation of reproduction and development. Mol. Reprod. Dev. 2015;82:932–956. doi: 10.1002/mrd.22581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao X., Luo Q., Zhao H., Qin X. Ovarian transcriptomic study reveals the differential regulation of miRNAs and lncRNAs related to fecundity in different sheep. Sci. Rep. 2016;6:35299. doi: 10.1038/srep35299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng X., Li F., Wang F., Zhang G., Pang J., Ren C., Zhang T., Yang H., Wang Z., Zhang Y. Genome-wide differential expression profiling of mRNAs and lncRNAs associated with prolificacy in Hu sheep. Biosci. Rep. 2018;38 doi: 10.1042/BSR20171350. BSR20171350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao X., Luo Q., Zhao H., Qin X. Co-expression analysis and identification of fecundity-related long non-coding RNAs in sheep ovaries. Sci. Rep. 2016;6:39398. doi: 10.1038/srep39398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao X., Luo Q., Zhao H., Qin X. An integrated analysis of miRNAs and methylated genes encoding mRNAs and lncRNAs in sheep breeds with different fecundity. Front. Physiol. 2017;8:1049. doi: 10.3389/fphys.2017.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng J., Wang Z., Yang H., Yao X., Yang P., Ren C., Wang F., Zhang Y. Pituitary transcriptomic study reveals the differential regulation of lncRNAs and mRNAs related to prolificacy in different FecB genotyping sheep. Genes (Basel) 2019;10:157. doi: 10.3390/genes10020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao X., Yao X., Wang Z., Li X., Li X., An S., Wei Z.Y., Zhang G.M., Wang F. Long non-coding RNA366.2 controls endometrial epithelial cell proliferation and migration by upregulating WNT6 as a ceRNA of miR-1576 in sheep uterus. Biochim. Biophys. Acta Gene Regul. Mech. 2020;1863:194606. doi: 10.1016/j.bbagrm.2020.194606. [DOI] [PubMed] [Google Scholar]
- 31.La Y., Tang J., He X., Di R., Wang X., Liu Q., Zhang L., Zhang X., Zhang J., Hu W., Chu M. Identification and characterization of mRNAs and lncRNAs in the uterus of polytocous and monotocous Small Tail Han sheep (Ovis aries) PeerJ. 2019;7:e6938. doi: 10.7717/peerj.6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ling Y., Xu L., Zhu L., Sui M., Zheng Q., Li W., Liu Y., Fang F., Zhang X. Identification and analysis of differentially expressed long non-coding RNAs between multiparous and uniparous goat (Capra hircus) ovaries. PLoS ONE. 2017;12:e0183163. doi: 10.1371/journal.pone.0183163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuda F., Inoue N., Manabe N., Ohkura S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012;58:44–50. doi: 10.1262/jrd.2011-012. [DOI] [PubMed] [Google Scholar]
- 34.Lu C.L., Yang W., Hu Z.Y., Liu Y.X. Granulosa cell proliferation differentiation and its role in follicular development. Chin. Sci. Bull. 2005;50:2665–2671. [Google Scholar]
- 35.Zhao F., Zhao W., Ren S., Fu Y., Fang X., Wang X., Li B. Roles of SIRT1 in granulosa cell apoptosis during the process of follicular atresia in porcine ovary. Anim. Reprod. Sci. 2014;151:34–41. doi: 10.1016/j.anireprosci.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Asselin E., Xiao C.W., Wang Y.F., Tsang B.K. Mammalian follicular development and atresia: role of apoptosis. Biol. Signals Recept. 2000;9:87–95. doi: 10.1159/000014627. [DOI] [PubMed] [Google Scholar]
- 37.Du X., Liu L., Li Q., Zhang L., Pan Z., Li Q. NORFA, long intergenic noncoding RNA, maintains sow fertility by inhibiting granulosa cell death. Commun. Biol. 2020;3:131. doi: 10.1038/s42003-020-0864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hausser H., Gröning A., Hasilik A., Schönherr E., Kresse H. Selective inactivity of TGF-β/decorin complexes. FEBS Lett. 1994;353:243–245. doi: 10.1016/0014-5793(94)01044-7. [DOI] [PubMed] [Google Scholar]
- 39.Comalada M., Cardó M., Xaus J., Valledor A.F., Lloberas J., Ventura F., Celada A. Decorin reverses the repressive effect of autocrine-produced TGF-β on mouse macrophage activation. J. Immunol. 2003;170:4450–4456. doi: 10.4049/jimmunol.170.9.4450. [DOI] [PubMed] [Google Scholar]
- 40.Baghy K., Iozzo R.V., Kovalszky I. Decorin-TGFβ axis in hepatic fibrosis and cirrhosis. J. Histochem. Cytochem. 2012;60:262–268. doi: 10.1369/0022155412438104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perks C.M., Newcomb P.V., Grohmann M., Wright R.J., Mason H.D., Holly J.M.P. Prolactin acts as a potent survival factor against C2-ceramide-induced apoptosis in human granulosa cells. Hum. Reprod. 2003;18:2672–2677. doi: 10.1093/humrep/deg496. [DOI] [PubMed] [Google Scholar]
- 42.Diamanti-Kandarakis E., Argyrakopoulou G., Economou F., Kandaraki E., Koutsilieris M. Defects in insulin signaling pathways in ovarian steroidogenesis and other tissues in polycystic ovary syndrome (PCOS) J. Steroid Biochem. Mol. Biol. 2008;109:242–246. doi: 10.1016/j.jsbmb.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Jahromi M.S., Tehrani F.R., Noroozzadeh M., Zarkesh M., Ghasemi A., Zadeh-Vakili A. Elevated expression of steroidogenesis pathway genes; CYP17, GATA6 and StAR in prenatally androgenized rats. Gene. 2016;593:167–171. doi: 10.1016/j.gene.2016.07.067. [DOI] [PubMed] [Google Scholar]
- 44.Li X., Du X., Yao W., Pan Z., Li Q. TGF-β/SMAD4 signaling pathway activates the HAS2-HA system to regulate granulosa cell state. J. Cell. Physiol. 2020;235:2260–2272. doi: 10.1002/jcp.29134. [DOI] [PubMed] [Google Scholar]
- 45.Bhardwaj J.K., Sharma R.K. Scanning electron microscopic changes in granulosa cells during follicular atresia in Caprine ovary. Scanning. 2011;33:21–24. doi: 10.1002/sca.20217. [DOI] [PubMed] [Google Scholar]
- 46.Zhou X., Zhang W., Jin M., Chen J., Xu W., Kong X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017;8:e2929. doi: 10.1038/cddis.2017.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H., Yang J., Jiang R., Wei X., Song C., Huang Y., Lan X., Lei C., Ma Y., Hu L., Chen H. Long non-coding RNA profiling reveals an abundant MDNCR that promotes differentiation of myoblasts by sponging miR-133a. Mol. Ther. Nucleic Acids. 2018;12:610–625. doi: 10.1016/j.omtn.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu C., Dai C., Guo Y., Qin R., Liu J. Long non-coding RNA PVT1-mediated miR-543/SERPINI1 axis plays a key role in the regulatory mechanism of ovarian cancer. Biosci. Rep. 2020;40 doi: 10.1042/BSR20200800. BSR20200800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma Z., Gu G., Pan W., Chen X. lncRNA PCAT6 accelerates the progression and chemoresistance of cervical cancer through up-regulating ZEB1 by sponging miR-543. OncoTargets Ther. 2020;13:1159–1170. doi: 10.2147/OTT.S232354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang F., Ma J., Tang Q., Zhang W., Fu Q., Sun J., Wang H., Song B. MicroRNA-543 promotes the proliferation and invasion of clear cell renal cell carcinoma cells by targeting Krüppel-like factor 6. Biomed. Pharmacother. 2018;97:616–623. doi: 10.1016/j.biopha.2017.10.136. [DOI] [PubMed] [Google Scholar]
- 51.Chen Z.Y., Du Y., Wang L., Liu X.H., Guo J., Weng X.D. miR-543 promotes cell proliferation and metastasis of renal cell carcinoma by targeting Dickkopf 1 through the Wnt/β-catenin signaling pathway. J. Cancer. 2018;9:3660–3668. doi: 10.7150/jca.27124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du Y., Liu X.H., Zhu H.C., Wang L., Ning J.Z., Xiao C.C. miR-543 promotes proliferation and epithelial-mesenchymal transition in prostate cancer via targeting RKIP. Cell. Physiol. Biochem. 2017;41:1135–1146. doi: 10.1159/000464120. [DOI] [PubMed] [Google Scholar]
- 53.Peng J.Y., Gao K.X., Xin H.Y., Han P., Zhu G.Q., Cao B.Y. Molecular cloning, expression analysis, and function of decorin in goat ovarian granulosa cells. Domest. Anim. Endocrinol. 2016;57:108–116. doi: 10.1016/j.domaniend.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Salgado R.M., Favaro R.R., Zorn T.M.T. Modulation of small leucine-rich proteoglycans (SLRPs) expression in the mouse uterus by estradiol and progesterone. Reprod. Biol. Endocrinol. 2011;9:22. doi: 10.1186/1477-7827-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adam M., Saller S., Ströbl S., Hennebold J.D., Dissen G.A., Ojeda S.R., Stouffer R.L., Berg D., Berg U., Mayerhofer A. Decorin is a part of the ovarian extracellular matrix in primates and may act as a signaling molecule. Hum. Reprod. 2012;27:3249–3258. doi: 10.1093/humrep/des297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu W.X., Zhang Q., Unno N., Derks J.B., Nathanielsz P.W. Characterization of decorin mRNA in pregnant intrauterine tissues of the ewe and regulation by steroids. Am. J. Physiol. Cell Physiol. 2000;278:C199–C206. doi: 10.1152/ajpcell.2000.278.1.C199. [DOI] [PubMed] [Google Scholar]
- 57.Yamaguchi Y., Ruoslahti E. Expression of human proteoglycan in Chinese hamster ovary cells inhibits cell proliferation. Nature. 1988;336:244–246. doi: 10.1038/336244a0. [DOI] [PubMed] [Google Scholar]
- 58.Hua X., Liu Z., Zhou M., Tian Y., Zhao P.P., Pan W.H., Li C.X., Huang X.X., Liao Z.X., Xian Q. LSAMP-AS1 binds to microRNA-183-5p to suppress the progression of prostate cancer by up-regulating the tumor suppressor DCN. EBioMedicine. 2019;50:178–190. doi: 10.1016/j.ebiom.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Yao G., Yin M., Lian J., Tian H., Liu L., Li X., Sun F. MicroRNA-224 is involved in transforming growth factor-β-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Mol. Endocrinol. 2010;24:540–551. doi: 10.1210/me.2009-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang L., Du X., Liu L., Cao Q., Pan Z., Li Q. miR-1306 mediates the feedback regulation of the TGF-β/SMAD signaling pathway in granulosa cells. Cells. 2019;8:298. doi: 10.3390/cells8040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chu M., Jia L., Zhang Y., Jin M., Chen H., Fang L., Di R., Cao G., Feng T., Tang Q. Polymorphisms of coding region of BMPR-IB gene and their relationship with litter size in sheep. Mol. Biol. Rep. 2011;38:4071–4076. doi: 10.1007/s11033-010-0526-z. [DOI] [PubMed] [Google Scholar]
- 62.Saito R., Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Lotia S., Pico A.R., Bader G.D., Ideker T. A travel guide to Cytoscape plugins. Nat. Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang G.M., Deng M.T., Zhang Y.L., Fan Y.X., Wan Y.J., Nie H.T., Wang Z.Y., Wang F., Lei Z.H. Effect of PGC-1α overexpression or silencing on mitochondrial apoptosis of goat luteinized granulosa cells. J. Bioenerg. Biomembr. 2016;48:493–507. doi: 10.1007/s10863-016-9684-6. [DOI] [PubMed] [Google Scholar]
- 64.Li H., Yang J., Wei X., Song C., Dong D., Huang Y., Lan X., Plath M., Lei C., Ma Y. circFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J. Cell. Physiol. 2018;233:4643–4651. doi: 10.1002/jcp.26230. [DOI] [PubMed] [Google Scholar]
- 65.Yao X., Wang Z., El-Samahy M.A., Ren C., Liu Z., Wang F., You P. Roles of vitamin D and its receptor in the proliferation and apoptosis of luteinised granulosa cells in the goat. Reprod. Fertil. Dev. 2020;32:335–348. doi: 10.1071/RD18442. [DOI] [PubMed] [Google Scholar]
- 66.Yao X., Yang H., Zhang Y., Ren C., Nie H., Fan Y., Zhou W., Wang S., Feng X., Wang F. Characterization of GALNTL5 gene sequence and expression in ovine testes and sperm. Theriogenology. 2017;95:54–61. doi: 10.1016/j.theriogenology.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 67.Yao X., Ei-Samahy M.A., Fan L., Zheng L., Jin Y., Pang J., Zhang G., Liu Z., Wang F. In vitro influence of selenium on the proliferation of and steroidogenesis in goat luteinized granulosa cells. Theriogenology. 2018;114:70–80. doi: 10.1016/j.theriogenology.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 68.Yao X., Wang Z., Gao X., Li X., Yang H., Ei-Samahy M.A., Bao Y., Xiao S., Meng F., Wang F. Unconservative_15_2570409 suppresses progesterone receptor expression in the granulosa cells of Hu sheep. Theriogenology. 2020;157:303–313. doi: 10.1016/j.theriogenology.2020.08.011. [DOI] [PubMed] [Google Scholar]
- 69.Yao X., Zhang G., Guo Y., Ei-Samahy M., Wang S., Wan Y., Han L., Liu Z., Wang F., Zhang Y. Vitamin D receptor expression and potential role of vitamin D on cell proliferation and steroidogenesis in goat ovarian granulosa cells. Theriogenology. 2017;102:162–173. doi: 10.1016/j.theriogenology.2017.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.