Abstract
Despite significant advances in cystic fibrosis (CF) treatments, a one-time treatment for this life-shortening disease remains elusive. Stable complementation of the disease-causing mutation with a normal copy of the CF transmembrane conductance regulator (CFTR) gene fulfills that goal. Integrating lentiviral vectors are well suited for this purpose, but widespread airway transduction in humans is limited by achievable titers and delivery barriers. Since airway epithelial cells are interconnected through gap junctions, small numbers of cells expressing supraphysiologic levels of CFTR could support sufficient channel function to rescue CF phenotypes. Here, we investigated promoter choice and CFTR codon optimization (coCFTR) as strategies to regulate CFTR expression. We evaluated two promoters—phosphoglycerate kinase (PGK) and elongation factor 1-α (EF1α)—that have been safely used in clinical trials. We also compared the wild-type human CFTR sequence to three alternative coCFTR sequences generated by different algorithms. With the use of the CFTR-mediated anion current in primary human CF airway epithelia to quantify channel expression and function, we determined that EF1α produced greater currents than PGK and identified a coCFTR sequence that conferred significantly increased functional CFTR expression. Optimized promoter and CFTR sequences advance lentiviral vectors toward CF gene therapy clinical trials.
Keywords: Cystic fibrosis, lentiviral vectors, gene therapy, codon optimization, CFTR
Graphical abstract
Loza and colleagues identify a promoter and a codon-optimized CFTR sequence that, when delivered by a lentiviral vector, significantly increased the CFTR-mediated anion currents produced in primary human cystic fibrosis (CF) airway epithelia. These improvements help advance lentiviral vectors as a therapeutic strategy for CF.
Introduction
Since mutations in cystic fibrosis (CF) transmembrane conductance regulator (CFTR) were identified as the cause of CF,1 gene addition has held the promise of a one-time definitive treatment for this common life-shortening disease. Several CF gene therapy clinical trials using adenovirus,2,3 adeno-associated virus (AAV),4,5 or plasmid-based vectors6, 7, 8 have taken place, but none progressed to phase III. Lentiviral vectors are well suited for CF gene therapy because of their large packaging capacity, long-term expression, safety profile, and promising results in CF animal models.9, 10, 11 Despite these attractive features, clinical trials using lentiviral vectors have yet to be initiated.
The use of lentiviral vectors for the treatment of CF airway disease has been limited, in part, by the transduction efficiency of airway epithelial cells. Most lentiviral vectors used for clinical applications are pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G) because it produces high titer vectors with broad cell tropism.12 However, VSV-G receptors in airway epithelial cells are located predominantly on the basolateral surface13,14 and require additional steps, such as disruption of tight junctions,15, 16, 17 to achieve meaningful transduction. Alternative glycoproteins from filoviruses,18,19 baculovirus,14,20 influenza,21 and Sendai virus22, 23, 24 confer improved apical surface tropism. Compared to adenoviral or AAV vectors, however, the maximum achievable lentiviral vector titers with current manufacturing methods are lower. The achievable lentiviral titers, combined with the large surface area of human airway epithelium, are a driving force for alternative strategies to increase lentiviral-based CFTR expression.
The airway epithelium is a complex tissue consisting of a variety of cell types that are interconnected and electrochemically coupled through gap junctions.25, 26, 27, 28 Ciliated cells, one of the most abundant cell types, were previously thought to collectively produce the majority of CFTR function in surface airway epithelium, with low abundance per ciliated cell.29 The recent discovery of pulmonary ionocytes, however, revealed that this rare cell type, which comprises ∼1% of all proximal airway epithelial cells, produces ∼50% of all CFTR messenger RNA (mRNA) transcripts.30,31 Although the physiologic role and function of pulmonary ionocytes are under investigation, the natural occurrence of a rare cell type with high CFTR abundance supports the notion of vector-conferred gene addition as a therapeutic strategy. Additionally, a recent study provided detailed analysis of the regional distribution of FOXI1-expressing cells and also presented evidence that basal cells and secretory cells express CFTR transcripts.32 Thus, we reasoned that increasing CFTR expression in a small proportion of cells successfully transduced by lentiviral vectors could provide sufficient protein function to correct CF phenotypes. Gene expression can be modulated through promoter choice and codon usage. To develop a lentiviral vector that provides optimal CFTR expression, a direct comparison of these two vector components in a relevant cell culture model is needed.
Promoter strength varies among different species and cell types.33,34 Overexpression of CFTR with a strong constitutive promoter, such as cytomegalovirus (CMV), can cause protein mis-sorting to the basolateral membrane.35 Alternative promoters include human phosphoglycerate kinase (PGK) and elongation factor 1-α (EF1α), which are associated with reduced rates of gene silencing and low genotoxic risk when used with integrating vectors.36, 37, 38 In addition, they were used in clinical trials with no adverse effects reported to date,39, 40, 41 making them ideal candidates for cell-based pre-clinical studies.
Codon optimization is another strategy to increase transgene expression in clinical trials for genetic diseases.41, 42, 43, 44, 45, 46, 47 The genetic code is degenerate; therefore, multiple nucleotide sequences can yield the same final amino acid sequence. Codon optimization utilizes this feature of the genetic code to improve gene expression. Several factors related to nucleotide sequence influence gene expression, including tRNA abundance,48 total GC content,49 GC content at the third nucleotide position of a codon (GC3),49, 50, 51 and CpG content.52 A combination of these and other factors, such as predicted secondary structure, translation speed, and cryptic splice sites, are used by proprietary and publicly available algorithms to codon optimize genes. In addition, most codon optimization algorithms increase the codon adaptive index (CAI), a measure of how closely the codons in a gene match those of highly expressed genes in a particular species.53 Because each algorithm uses different parameters and produces a unique final sequence, we compared three different codon-optimized sequences for efficacy in a model cell line. We then validated the best performing sequence in primary human airway epithelial cells derived from CF donors.
Results
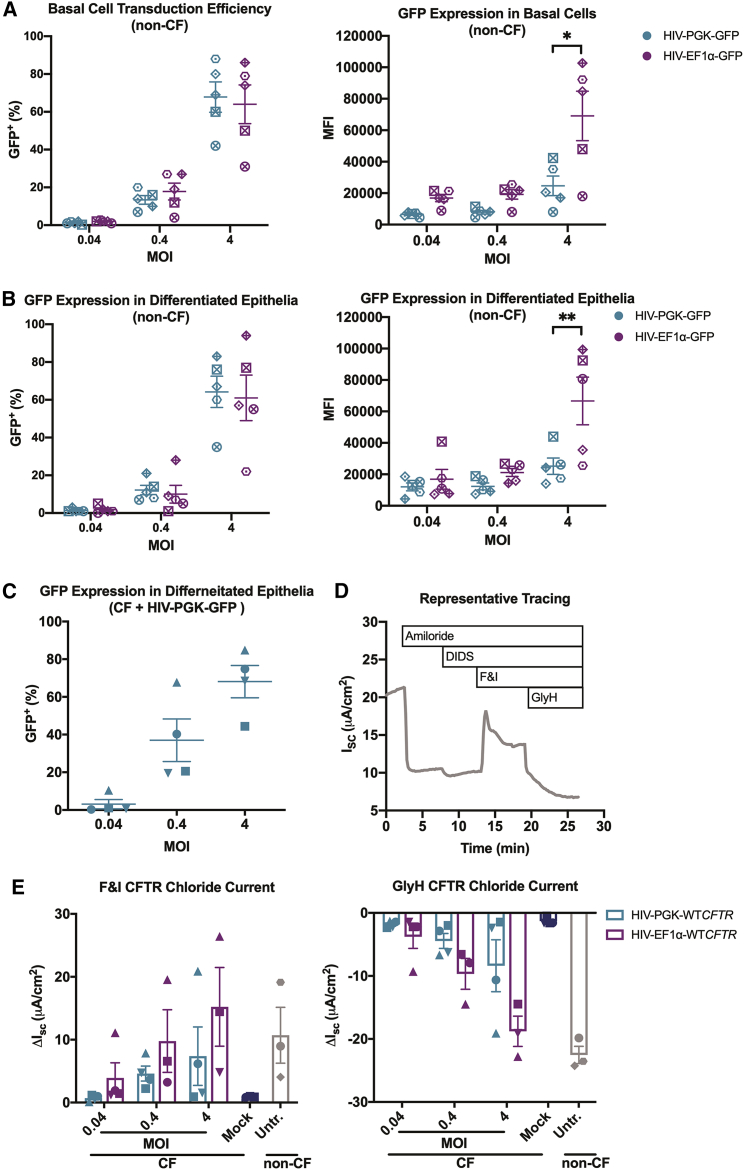
To compare the amount of protein produced by vectors with expression driven by the PGK or EF1α promoters, we cloned GFP into HIV-based lentiviral vectors that differed only in the promoter (HIV-PGK-GFP and HIV-EF1α-GFP) (Figure 1) and produced VSV-G-pseudotyped vectors. Basal cells, a progenitor cell type of the conducting airways, from human non-CF donors were transduced with these vectors at MOIs of 0.04, 0.4, and 4 and analyzed by flow cytometry. Similar numbers of GFP+ cells were observed with both vectors, ranging from <1% to >80%, among the different MOIs, 3−5 days post-transduction (Figure 2A), and after 4 weeks of differentiation in air-liquid interface culture conditions (Figure 2B). Cells transduced with HIV-EF1α-GFP, however, showed higher mean fluorescence intensity (MFI) at every dose tested compared to cells transduced with HIV-PGK-GFP, and this difference was statistically significant at MOI 4 (Figures 2A, p < 0.0006, and 2B, p < 0.002).
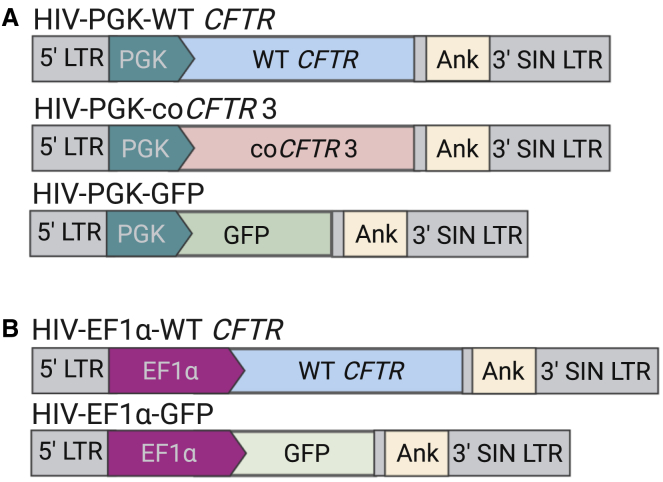
Figure 1.
Schematic of the vectors used in this study
The integrated forms of the HIV-based lentiviral vectors are shown. Two versions of vectors expressing wild-type (WT) cystic fibrosis (CF) transmembrane conductance regulator (CFTR), codon-optimized CFTR version 3 (coCFTR3), or GFP were compared. (A and B) Only the promoters used to drive transgene expression differ between the two versions, either (A) human phosphoglycerate kinase (PGK), or (B) human elongation factor 1-α (EF1α). A human ankyrin 1 (Ank) insulator element in the reverse orientation is present within the self-inactivating (SIN) 3′ LTR. Vectors shown are not to scale.
Figure 2.
EF1α drives higher transgene expression in primary human airway epithelial cells compared to PGK
(A and B) Basal progenitor cells from five human donors without CF (non-CF) donors were transduced with lentiviral vectors HIV-PGK-GFP or HIV-EF1α-GFP at MOI 0.04, 0.4, or 4. GFP+ cells and mean fluorescence intensity (MFI) were quantified by flow cytometry (A), 3−5 days post-transduction and (B) after 4 weeks of differentiation. No significant difference was observed in the number of GFP+ cells transduced by either vector at any dose. A significant increase in MFI was observed in cells transduced with HIV-EF1α-GFP compared to HIV-PGK-GFP at MOI 4 at both time points (∗p < 0.0006, ∗∗p < 0.002). Similarly, basal cells from four human CF donors were transduced with lentiviral vectors HIV-PGK-WTCFTR, HIV-EF1α-WTCFTR, or HIV-PGK-GFP. (C) After 4 weeks of differentiation under air-liquid interface conditions, the number of remaining transduced cells was estimated through quantification of GFP+ cells by flow cytometry. (D) Transepithelial Cl− current was measured in Ussing chambers. ENaC and non-CFTR chloride channels were inhibited with sequential addition of amiloride and 4,4′-diisothiocyano-2,2′-stilbenedisulfonic acid (DIDS), respectively, prior to activation of CFTR channels with cyclic AMP (cAMP) agonists forskolin and 3-isobutyl-1-methylxanthine (F&I). CFTR-specific current was verified by addition of CFTR inhibitor GlyH-101 (GlyH). (E) The short circuit current change (ΔISC) in response to F&I and GlyH was calculated. No significant difference between the two vectors was observed at any dose. Current measurements in cultures from one donor in two treatment groups (HIV-EF1α-WTCFTR MOI 0.04 and 4) could not be completed due to unsuccessful differentiation. Points on the graph represent the average of 1−3 epithelia. Each donor is represented by a unique symbol. Mean ± SE is shown.
We next transduced basal cells from CF donors with HIV-PGK-wild-type (WT)CFTR, HIV-EF1α-WTCFTR, or HIV-PGK-GFP. In these experiments, cells were seeded onto permeable filters at the time of transduction and allowed to differentiate at an air-liquid interface for a minimum of 4 weeks. The number of transduced cells present after differentiation was estimated by quantifying GFP+ cells by flow cytometry and ranged from <1% to 85% (Figure 2C). The CFTR-mediated chloride current was measured in Ussing chambers (Figure 2D). First, epithelial sodium channel (ENaC) and non-CFTR chloride channels were sequentially inhibited using amiloride and 4,4′-diisothiocyano-2,2′-stilbenedisulfonic acid (DIDS), respectively. CFTR was then stimulated with forskolin and 3-isobutyl-1-methylxanthine (F&I), and the change in short circuit current (ΔISC) was calculated. Finally, CFTR was inhibited with GlyH-101 (GlyH). At all MOIs tested, HIV-EF1α-WTCFTR produced more CFTR current, on average, compared to HIV-PGK-WTCFTR, but the difference was not statistically significant (Figure 2E). A condition using HIV-EF1α-GFP was not included in these experiments due to the limited number of primary cells available from each CF donor.
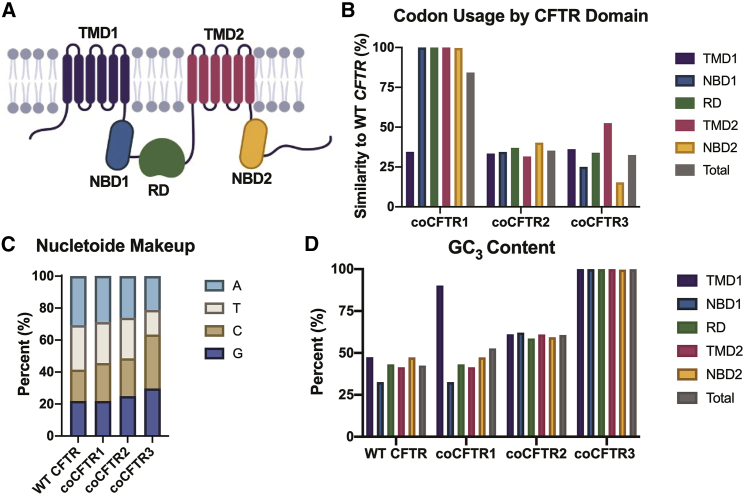
CFTR is composed of two sets of transmembrane domains (TMD1 and TMD2), two nucleotide-binding domains (NBD1 and NBD2), and a regulatory domain (RD) (Figure 3A). To investigate the efficacy of different codon-optimization strategies, we codon optimized CFTR (coCFTR) using three different methods. Previous studies suggest that codon usage affects CFTR co-translational folding, and these effects may be domain specific.54 Therefore, to determine the effect of codon optimization on a single domain, only the first domain, TMD1, was codon optimized in coCFTR1. The entire gene was optimized for coCFTR2 and coCFTR3 (Figure 3B). A proprietary algorithm (GenScript) was used for coCFTR2 and a publicly available algorithm (JCat)55 for coCFTR3. As expected, all strategies increased the CAI compared to WT CFTR, but the amino acid sequence remained the same (Table 1). The total GC content increased in all codon-optimized sequences compared to WT CFTR, but coCFTR3 had the highest increase (from 42% to 64%; Figure 3C). The GC3 content also increased compared to WT CFTR in the single codon-optimized domain of coCFTR1 and throughout all domains of coCFTR2 and coCFTR3, with coCFTR3 having a GC3 content of nearly 100% (Figure 3D).
Figure 3.
Comparison of three different coCFTR sequences
Different approaches to codon optimization were used to generate coCFTR1, coCFTR2, and coCFTR3. (A) CFTR is composed of five distinct protein domains, including two transmembrane domains (TMD1 and TMD2), two nucleotide-binding domains (NBD1 and NBD2), and a regulatory domain (RD). (B) The percent similarity of codons compared to WT CFTR is shown by protein domain. (C) The individual nucleotide content for each sequence was quantified. (D) The percentage of codons using a G or C at the third nucleotide position (GC3) for each sequence is shown by protein domain.
Table 1.
Codon usage of CFTR sequences
| WT CFTR | coCFTR1a | coCFTR2b | coCFTR3 | ||||
|---|---|---|---|---|---|---|---|
| Ala | G | C | G | 6 | 3 | 4 | 0 |
| A | 34 | 26 | 20 | 0 | |||
| T | 27 | 20 | 22 | 0 | |||
| C | 16 | 34 | 37 | 83 | |||
| Arg | G | C | G | 16 | 13 | 17 | 0 |
| A | 37 | 24 | 22 | 0 | |||
| C | G | G | 8 | 4 | 13 | 0 | |
| A | 9 | 33 | 7 | 0 | |||
| T | 3 | 2 | 7 | 0 | |||
| C | 5 | 2 | 12 | 78 | |||
| Asn | A | A | T | 25 | 21 | 24 | 0 |
| C | 29 | 33 | 30 | 54 | |||
| Asp | G | A | T | 38 | 32 | 26 | 0 |
| C | 20 | 26 | 32 | 58 | |||
| Cys | T | G | T | 8 | 6 | 10 | 0 |
| C | 10 | 12 | 8 | 18 | |||
| Gln | C | A | G | 31 | 37 | 47 | 67 |
| A | 36 | 30 | 20 | 0 | |||
| Glu | G | A | G | 30 | 45 | 61 | 93 |
| A | 63 | 48 | 32 | 0 | |||
| Gly | G | G | G | 16 | 10 | 21 | 0 |
| A | 38 | 30 | 18 | 0 | |||
| T | 15 | 14 | 13 | 0 | |||
| C | 15 | 30 | 32 | 84 | |||
| His | C | A | T | 11 | 9 | 7 | 0 |
| C | 14 | 16 | 18 | 25 | |||
| Ile | A | T | A | 33 | 26 | 14 | 0 |
| T | 50 | 39 | 46 | 0 | |||
| C | 36 | 54 | 59 | 119 | |||
| Leub | T | T | G | 34 | 27 | 29 | 0 |
| A | 37 | 29 | 13 | 0 | |||
| C | T | G | 37 | 26 | 72 | 183 | |
| A | 19 | 12 | 9 | 0 | |||
| T | 32 | 20 | 23 | 0 | |||
| C | 24 | 69 | 38 | 0 | |||
| Lys | A | A | G | 35 | 47 | 53 | 92 |
| A | 57 | 45 | 39 | 0 | |||
| Meta | A | T | G | 38 | 39 | 38 | 38 |
| Pheb | T | T | T | 47 | 35 | 37 | 0 |
| C | 38 | 50 | 47 | 85 | |||
| Pro | C | C | G | 1 | 0 | 5 | 0 |
| A | 13 | 21 | 12 | 0 | |||
| T | 20 | 15 | 13 | 0 | |||
| C | 11 | 9 | 15 | 45 | |||
| Ser | A | G | T | 19 | 15 | 18 | 0 |
| C | 24 | 20 | 28 | 123 | |||
| T | C | G | 4 | 3 | 9 | 0 | |
| A | 29 | 22 | 24 | 0 | |||
| T | 30 | 23 | 26 | 0 | |||
| C | 17 | 40 | 18 | 0 | |||
| Thr | A | C | G | 3 | 3 | 6 | 0 |
| A | 33 | 31 | 18 | 0 | |||
| T | 32 | 29 | 26 | 0 | |||
| C | 15 | 20 | 33 | 83 | |||
| Trp | T | G | G | 23 | 23 | 23 | 23 |
| Tyr | T | A | T | 22 | 15 | 15 | 0 |
| C | 18 | 25 | 25 | 40 | |||
| Vala | G | T | G | 36 | 46 | 48 | 89 |
| A | 12 | 11 | 9 | 0 | |||
| T | 23 | 17 | 12 | 0 | |||
| C | 18 | 14 | 20 | 0 | |||
| Stop | T | G | A | 0 | 0 | 0 | 0 |
| A | G | 1 | 1 | 1 | 0 | ||
| A | 0 | 0 | 0 | 1 | |||
| Total | 1481 | 1481 | 1481 | 1481 | |||
| CAI | 0.74 | 0.76 | 0.80 | 0.99 |
The codon adaptive index (CAI) is higher in all coCFTR sequences compared to WT CFTR. coCFTR1 and coCFTR2 were codon optimized with single amino acid changes that do not affect protein function.
coCFTR1 substitutes methionine for valine at position 1475.
coCFTR2 substitutes leucine for phenylalanine at position 833.
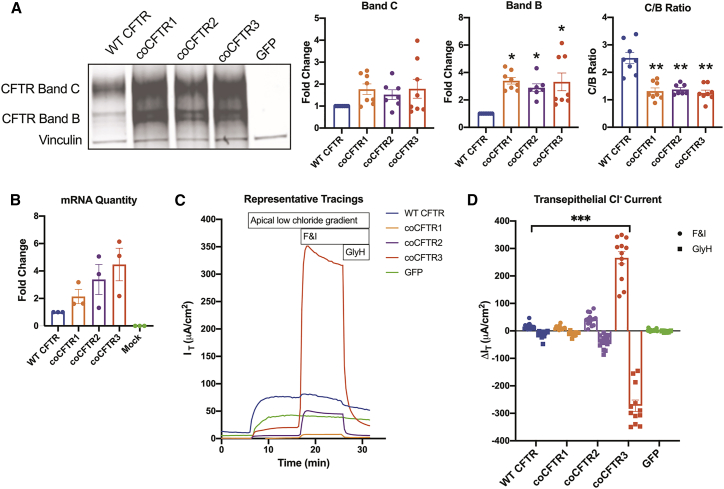
WT CFTR and all coCFTR cDNAs were each expressed from the mammalian expression vector pcDNA3.1(+) to compare their expression. Human embryonic kidney (HEK)293 cells were transfected, and 72 h later, total cellular mRNA and protein were collected. A GFP expression plasmid served as a control. Different forms of CFTR can be detected by western blot based on their glycosylation patterns, including bands B and C.56 Band C is the fully glycosylated form and was the most abundant form found in cells transfected with WT CFTR (Figure 4A). Bands B and C abundance increased in cells transfected with any of the coCFTR sequences, but only band B was significantly more abundant (p < 0.007), leading to a decrease in C/B ratio (p < 0.02). mRNA abundance was measured by qRT-PCR. Although all coCFTR sequences resulted in higher average mRNA content over WT CFTR, the fold increases were not statistically significant for any sequence (Figure 4B).
Figure 4.
coCFTR increases protein, mRNA, and transepithelial chloride current
293T human embryonic kidney cells (HEK293) cells were transfected with pcDNA3.1(+) plasmid expressing WT CFTR, coCFTR1, coCFTR2, coCFTR3, or a GFP control. Protein and mRNA were isolated from transfected cells. (A) CFTR abundance was quantified by western blot. Bands B and C glycosylation forms were detected, and vinculin was used as a loading control. Densitometry analysis demonstrated no significant increase for CFTR band C, a significant increase in CFTR band B (∗p ≤ 0.007), and a significant decrease in C/B ratio (∗∗p ≤ 0.02) with coCFTR1, coCFTR2, and coCFTR3 compared to WT CFTR. (B) CFTR mRNA quantification by qRT-PCR was performed using primers that target a portion of the polyadenylation sequence present in all plasmids, and GAPDH was used to normalize expression. No statistically significant increase in mRNA abundance compared to WT CFTR was observed with any of the codon-optimized sequences. To test protein function, Fischer rat thyroid (FRT) cells were electroporated with the same plasmids and grown at air-liquid interface culture conditions. 1 week later, transepithelial chloride current was measured in Ussing chambers. (C) Apical low chloride gradient was established, and once stable, CFTR channels were activated with cAMP agonists F&I. CFTR-specific current was verified by inhibiting CFTR with GlyH. (D) The change in current (ΔIT) in response to F&I and GlyH was measured. We observed a significant increase in CFTR-dependent current only with coCFTR3 compared to WT CFTR (∗∗∗p < 0.0001). Mean ± SE is shown. See also Figure S1.
To investigate the effect of codon optimization on protein function, a cell line that formed a polarized epithelium was required. Fischer rat thyroid (FRT) cells are a useful model for the study of CFTR function, because they express no endogenous CFTR and form a polarized epithelium when grown at air-liquid interface culture conditions.57 These epithelia are then used to measure transepithelial anion currents in Ussing chambers. FRT cells were electroporated with the pcDNA3.1(+) plasmids expressing WT CFTR, one of the coCFTR sequences, or a GFP control plasmid and seeded onto semipermeable membranes. 1 week later, transepithelial chloride current was measured under apical low chloride gradient conditions (Figure 4C). Compared to WT CFTR, coCFTR1 did not increase function, whereas coCFTR2 showed a modest increase, and coCFTR3 led to a significant increase (Figure 4D; p < 0.0001). Western blots performed in transfected FRT cells revealed a similar expression pattern as that observed in HEK293 cells, with all coCFTR sequences producing more CFTR compared to WT CFTR (Figure S1). Densitometry analysis, however, was not reliable due to the low abundance of WT CFTR.
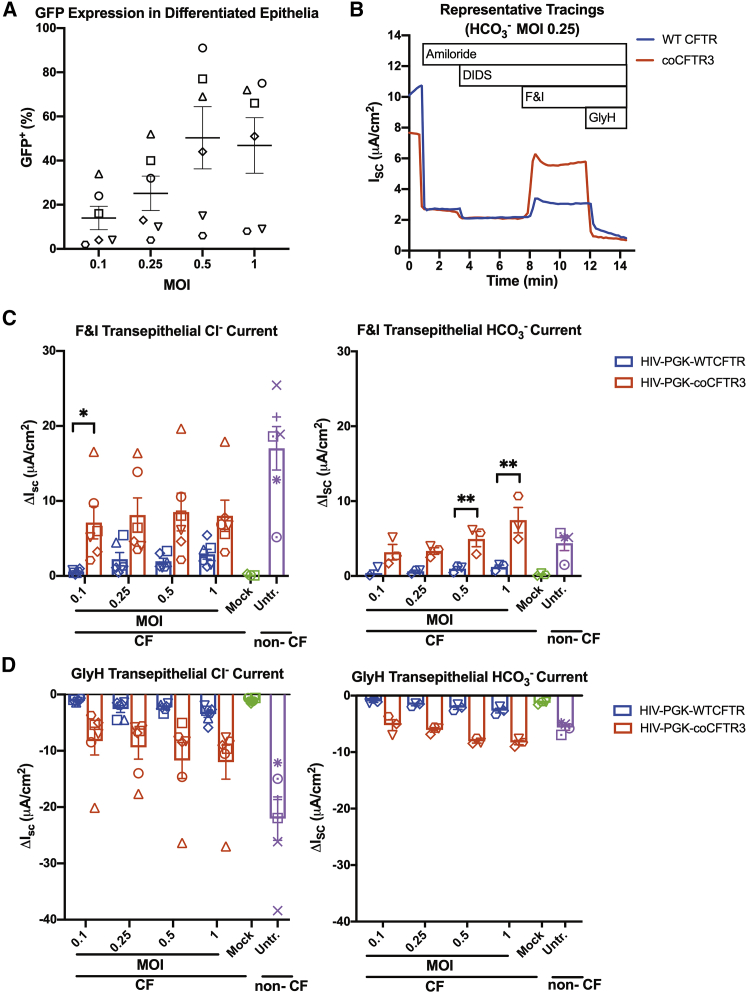
Since expression of coCFTR3 produced the greatest increase in protein function in the FRT cell assay, we next sought to determine if similar effects would be achieved in primary CF human airway epithelial cells. We introduced coCFTR3 into the same lentiviral vector backbone used in the promoter comparison experiments (Figure 1A). Given the significant increase in ISC observed with coCFTR3 compared to WT CFTR in FRT cells, PGK was chosen since it produced lower currents than EF1α, on average, and would allow us to more easily detect any effects of codon optimization. For the same reason, based on the ISC dose response observed in the promoter comparison experiments (Figure 2E), MOIs ≤ 1 were used. Basal cells from human CF donors were transduced with HIV-PGK-WTCFTR, HIV-PGK-coCFTR3, or HIV-PGK-GFP at four different MOIs (0.1, 0.25, 0.5, and 1). Epithelial cultures were transduced at the time of seeding and allowed to differentiate for four weeks (Figure S2).58 We predicted these doses would achieve a range of transduction efficiencies, allowing us to determine an optimal MOI to achieve non-CF levels of anion current. GFP+ cells were quantified by flow cytometry in epithelia transduced with the GFP vector to estimate the number of transduced cells present after differentiation. Donor-dependent differences in transduction permissiveness were observed, resulting in ∼2%–91% GFP+ cells at the doses tested (Figure 5A). Epithelia transduced with CFTR vectors were mounted in Ussing chambers and transepithelial anion currents measured (Figure 5B). HIV-PGK-coCFTR3 conferred significantly higher CFTR-dependent transepithelial chloride current at the lowest dose (MOI 0.1, p < 0.04) and higher bicarbonate currents at the highest doses (MOI 0.05 and 1, p < 0.02) compared to HIV-PGK-WTCFTR (Figures 5C and 5D).
Figure 5.
coCFTR increases transepithelial anion current in human CF primary airway epithelial cells
CF progenitor basal cells from six donors were transduced with lentiviral vectors HIV-PGK-WTCFTR, HIV-PGK-coCFTR3, or HIV-PGK-GFP at MOI 0.1, 0.25, 0.5, and 1 and seeded on polycarbonate membranes. Cells were allowed to differentiate for 4 weeks in air-liquid interface culture conditions. (A) Epithelia that received GFP were used to estimate the proportion of cells expressing the transgene after differentiation by flow cytometry. (B) CFTR transepithelial chloride and bicarbonate short circuit current was measured in Ussing chambers. Changes (ΔISC) in response to (C) F&I and (D) GlyH were quantified. HIV-PGK-coCFTR3 resulted in significantly higher chloride current at MOI 0.1 (∗p < 0.04) and higher bicarbonate current at MOI 0.5 and 1 (∗∗p < 0.02) compared to HIV-PGK-WTCFTR. Points on the graph represent the average of 2−3 epithelia. Each symbol represents cells from an individual donor. Mean ± SE is shown. See also Figure S2.
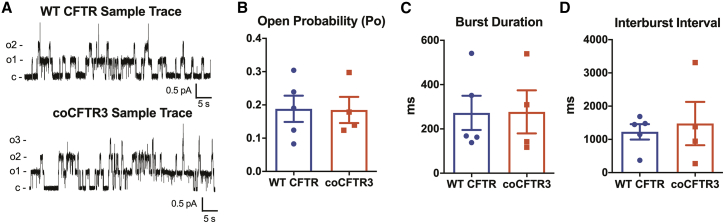
Previous studies demonstrate that synonymous codon substitutions may affect folding dynamics and could change protein function.59,60 The marked increase in chloride current observed with FRT cells (Figure 4C), combined with the significant increase in the immature (band B) form of CFTR (Figure 4A), and the increase in CFTR anion currents in primary airway epithelia observed with coCFTR3 (Figures 5C and 5D) raised the possibility that coCFTR3 could have channel properties different from WT CFTR. To study the single channel properties of WT CFTR and coCFTR3, we performed patch-clamp studies in HEK293 cells transfected with WT CFTR or coCFTR3 cDNA (Figure 6A). No significant differences were found in mean open state probability (Figure 6B), burst duration (BD) (Figure 6C), or interburst interval (IBI) (Figure 6D). These findings indicate that the CFTR channels produced by coCFTR3 have the same properties as those produced by WT CFTR. Furthermore, the increase in CFTR band B associated with coCFTR might be expected since protein processing mechanisms are downstream of those thought to be affected by codon optimization. Together, these results confirmed that coCFTR3 produces more functional CFTR in CF primary human airway epithelial cells, reaching the range of non-CF anion currents at MOIs < 1 without changing CFTR channel properties.
Figure 6.
Codon optimization of CFTR does not change CFTR single channel properties
Patch-clamp studies were performed using inside-out membrane patches from HEK293 transiently expressing WT CFTR or coCFTR3. (A) Representative tracings are shown. PKA catalytic subunit (22 nM) and ATP (1 mM) were present throughout. Holding voltage was −70 mV. For illustration purposes, traces were digitally low-pass filtered at 50 Hz. c, closed state; o, open state. (B−D) Mean channel open probability, burst duration, and interburst intervals were not different between WT CFTR and coCFTR3. Mean ± SE is shown.
Discussion
Several studies have attempted to approximate the proportion of airway epithelial cells that needs to express functional CFTR to correct CF phenotypes, with results ranging from 5% to 50%.35,61, 62, 63, 64 Achieving a transduction efficiency as high as 50% in intact airway epithelia with lentiviral vectors is challenging. In vivo studies in the CF pig model demonstrate that some phenotypic tissue correction is achievable, even if transgene expression was below detectable levels by mRNA quantification.10 Because airway epithelia are electrochemically coupled through gap junctions, we postulated that supra-physiologic levels of CFTR expression in a small number of cells could be therapeutic.
Promoter choice and codon optimization are two strategies that are often employed to increase transgene expression in gene therapy applications. We compared two constitutive promoters, PGK and EF1α, which are effective at driving expression of mammalian genes and are safely used in clinical trials.39, 40, 41 In CF airway epithelia, the CFTR currents achieved using the EF1α promoter trended higher than those attained with PGK, on average, but did not reach statistical significance. Of note, approximately 50% transduction efficiency was obtained with both vectors at MOI 4, and the CFTR-dependent currents at this dose reached non-CF levels. This is likely related to anion movement through gap junctions. Additionally, others have shown that the rate-limiting step for CFTR-dependent chloride secretion is the abundance of the NKCC1 cotransporter at the basolateral membrane.35,63,65 When quantifying GFP expression in non-CF cells, similar numbers of GFP+ cells were obtained with PGK or EF1α, and the MFI achieved with the EF1α promoter was higher on average at every dose, reaching statistical significance at the highest dose. The proportion of GFP+ non-CF cells, measured 3−5 days post-transduction, was within expected levels based on a similar transduction protocol.58 The proportion of GFP+ cells present in CF and non-CF cells after 4 weeks of differentiation, however, represents differences in individual cell division rates over that time, which may explain why some values exceed the theoretical maximum for a given MOI and why the number of transduced cells appears to change over time in some epithelia.
Codon optimization has been used to increase expression of other genes in several therapeutic applications66, 67, 68 and has been investigated as a strategy to improve non-viral approaches to CF gene therapy.69 Here, we compared different codon optimization strategies in cell line models and determined that each strategy conferred a unique increase in mRNA, protein, and protein function, evidenced by a CFTR-dependent transepithelial chloride current. Codon optimization of a single protein domain failed to improve expression over WT CFTR. Of two different strategies to codon optimize the entire gene, the one that resulted in higher GC and GC3 content, coCFTR3, also yielded the greatest increase in protein function, suggesting that these may be important characteristics for coCFTR.
When comparing WT CFTR and coCFTR3 in primary human CF airway epithelia transduced with a lentiviral vector, HIV-PGK-coCFTR3 significantly increased chloride currents at the lowest vector dose tested (MOI 0.1). As expected, we observed a dose-dependent increase in the chloride current with HIV-PGK-WTCFTR, but very similar chloride currents were observed at all doses tested with HIV-PGK-coCFTR3. This is consistent with previous studies indicating that chloride currents generated by epithelia composed entirely of non-CF cells can be achieved in epithelial sheets consisting of ∼50% non-CF and 50% CF cells and remain stable even when more non-CF cells are added.63 This suggests that the maximum chloride current for a given epithelium was achieved at MOI 0.1, explaining why no additional increase is seen at higher MOIs. Likewise, this could also explain why the chloride current difference between HIV-PGK-WTCFTR and HIV- EF1α-WTCFTR was not significant at any dose, but GFP expression was significantly higher with EF1α at the highest dose. That is, CFTR-mediated chloride current as a measure of CFTR expression has a lower maximum limit than MFI for GFP expression. CFTR bicarbonate currents, however, continued to increase with CFTR abundance,63 which is consistent with the dose-dependent increase observed with HIV-PGK-WTCFTR and HIV-PGK-coCFTR3, and the difference between the two was significant at the highest doses. Achieving transepithelial anion currents within the range of non-CF at MOI 0.1 with HIV-PGK-coCFTR3 is highly relevant for in vivo studies since we anticipate likely transducing less than 50% of surface epithelia.
Although EF1α conferred greater WT CFTR-mediated chloride currents and GFP expression, it is important to note that its sequence is over twice the length (∼1.1 kb) of PGK (∼0.5 kb). This is an important consideration when coupled with a large cDNA, such as CFTR (∼4.5 kb), since there is an inverse relationship between lentiviral vector titer and insert length.70,71 The extent to which the extra length of EF1α affects CFTR lentiviral vector titers remains to be determined and, if significant, weighed against the increase in CFTR production it provides.
One limitation of this work is that VSV-G-pseudotyped lentiviral vectors were used to transduce progenitor basal cells. It is reassuring that CF basal cells expressing exogenous CFTR under regulation of a constitutive promoter can differentiate, form epithelia, and support CFTR-mediated transepithelial anion movement, suggesting this could be an effective therapeutic strategy to achieve long-term CFTR expression. This strategy could also be used for cell-based therapies.72 The apical transduction efficiency of intact well-differentiated airway epithelia with VSV-G-pseudotyped lentiviral vectors is low.13,14 In future studies, lentiviral vectors pseudotyped with an envelope that allows more efficient apical transduction of differentiated epithelia will be required.
Having identified two independent methods to increase CFTR expression, it is tempting to suggest a combinational approach, using EF1α and coCFTR3 in the same vector, and we will test that strategy in future studies. However, we speculate that if the effects of each of these strategies are additive, then we may reach the counterproductive threshold. When CFTR was overexpressed with the CMV promoter in a large proportion of cells using an adenoviral vector, it led to aberrant localization to the basolateral membrane.35 If this happens in a small number of cells, then the anion concentration gradient would drive net movement in the desired direction. But, as the proportion of cells with both apical and basolateral CFTR expression increases, the driving force for anion secretion would dissipate.
We identified a promoter and coCFTR strategy that significantly increases CFTR-mediated anion transport. Questions remain regarding the effects of combining both features in a single vector and the effect of promoter length on vector titers. Ultimately, testing coCFTR3 in CF animal models will be needed to determine its efficacy at correcting CF phenotypes. We anticipate that incorporating EF1α, coCFTR3, or both could lower the proportion of cells that needs to be successfully targeted by a lentiviral vector and facilitate sufficient gene delivery to provide clinically relevant CF phenotype correction.
Materials and methods
Plasmids
WT CFTR and coCFTR1 (GenBank: MW222115) in the pcDNA3.1(+) vector were kind gifts from Drs. Jae Seok Yoon and William R. Skach. Only the first 356 amino acids of coCFTR1 were codon optimized. coCFTR1 contains a valine-for-methionine substitution at position 1475, which does not affect protein function.73 Codon optimization and synthesis of coCFTR2 (GenBank: MW116826) were commercially performed by GenScript in the Bluescript plasmid. coCFTR2 contains the leucine-for-phenylalanine substitution at position 833 present in the originally reported CFTR sequence,1 which increases protein solubility but does not affect protein structure.74 Codon optimization of coCFTR3 (GenBank: MW116827) was performed using JCat55 and was commercially synthesized by GenScript in the HIV-PGK-coCFTR3 vector. coCFTR2 and coCFTR3 were subcloned into pcDNA3.1(+). HIV-PGK-WTCFTR and HIV-EF1α-GFP were synthesized by GenScript and used to clone HIV-PGK-GFP, HIV-PGK-coCFTR3, and HIV-EF1α-WTCFTR. The HIV-based vectors used are second generation, self-inactivating (SIN), and contain a human ankyrin 1 element in the reverse orientation within the 3′ long terminal repeat (LTR) that improves long-term expression by avoiding silencing.75,76
Codon-optimized sequence comparisons
CFTR domains were defined by the following amino acid positions: TMD1 (1−356), NBD1 (357−678), RD (679−840), TMD2 (841−1,156), and NBD2 (1,157−1,481). Nucleotide content was determined using SeqBuilder Pro 17 (DNASTAR; Lasergene, Madison, WI, USA). Codon usage and GC3 content were determined using Sequence Manipulation Suite (bioinformatics.org).
Viral vector production
All viral vectors were produced by The University of Iowa Viral Vector Core. Briefly, a triple transfection (psPAX2, VSV-G, and HIV vector) was performed in 293FT (Thermo Fisher Scientific/Gibco, Waltham, MA, USA) cells using the TransIT-Lenti Reagent (Mirus, Madison, WI, USA). Cells were cultured in DMEM, supplemented with 5% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Vector-containing supernatant was collected at 48 and 72 h. After filtration (0.2 μm), vector was concentrated 750-fold by overnight centrifugation (7,000 rpm, 4°C). The vector pellet was resuspended in 4% D-lactose and stored at −80°C. Vectors were titered in HT1080 cells by flow cytometry for GFP vectors or by TaqMan qPCR/digital droplet PCR for CFTR vectors using primers: 5′-CGACTGGTGAGTACGCCAAA-3′ and 5′-CGCACCCATCTCTCTCCTTCT-3′ and probe 5′-ATTTTGACTAGCGGAGGC-3′.
FRT electroporation and epithelia formation
FRT cells were cultured in Ham F-12 Coon’s modified medium, supplemented with 5% FBS and 1% penicillin/streptomycin at 37°C in a 5% CO2 humidified environment. 1.2 × 106 FRT cells were electroporated with 4 μg of pcDNA3.1(+) plasmid (Amaxa Nucleofector 2b, Nucleofector Kit L; Lonza, Basel, Switzerland). The GFP plasmid included in the electroporation kit was used to verify successful electroporation, and these cells were used as negative controls in Ussing chamber experiments. Cells were resuspended in a total volume of 700 μL, and 200 μL was seeded in each of three, collagen-coated polycarbonate membrane tissue-culture inserts (6.5 mm, 0.4 μm pore; Corning Transwell, Corning, NY, USA) with 400 μL media on the basolateral compartment. The next day, apical media were removed, and the basolateral media were replaced. After 7 days of culture in air-liquid interface conditions, cells were mounted in Ussing chambers for electrophysiology measurements.
HEK293 cell transfection
HEK293 cells were cultured in 5% FBS and 1% penicillin/streptomycin at 37°C in a 5% CO2 humidified environment. 1 × 105 cells per well were seeded on a 6-well plate. The next day, 2 μg of pcDNA3.1(+) plasmid was transfected following the manufacturer’s instructions for Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). When confluent, cells were harvested for mRNA or western blot analysis. A GFP control plasmid was used to verify successful transfection and to serve as a negative control in mRNA and western blot analyses.
Transduction and differentiation of primary human airway basal cells
All primary human airway epithelial cells were obtained from The University of Iowa In Vitro Models and Cell Culture Core under approval from The University of Iowa Institutional Review Board. De-identified cells isolated from donated non-CF lungs or discarded CF lungs after transplant were provided. 8 × 105 freshly isolated primary basal cells from CF donors were seeded on collagen-coated 10 cm plates in BronchiaLife Epithelial Basal Medium (Lifeline, Carlsbad, CA, USA). Once cells reached ∼80% confluency, cells were dissociated using TrypLE (Thermo Fisher Scientific/Gibco, Waltham, MA, USA), and 1 × 105 cells were seeded on collagen-coated polycarbonate membrane tissue-culture inserts (Corning Transwell; 6.5 mm, 0.4 μm pore), along with the viral vector and polybrene (2 μg/mL) in a final volume of 200 μL. 400 μL defined culture media (Ultroser G; Crescent Chemical, Islandia, NY, USA) were maintained in the basolateral compartment throughout the differentiation process.77 Cells were maintained under submerged culture conditions for 48−72 h. Apical media were then removed, and cultures were maintained at air-liquid interface conditions for a minimum of 4 weeks. Once differentiated, cells were either mounted directly in Ussing chambers for electrophysiology analysis or dissociated for flow cytometry analysis using Accumax (Sigma-Aldrich, St. Louis, MO, USA). The following mutations were represented in the CF samples used in these studies: ΔF508, R75X, and 3,849 + 10 kb C > T.
For non-CF cells, 2 × 105 cells were seeded onto collagen-coated 6-well plates, along with viral vector and polybrene (2 μg/mL) in BronchiaLife Epithelial Basal Medium (1 mL final volume). Media were replaced as needed until cells reached confluency 3−5 days post-transduction and were harvested using TypLE for flow cytometry analysis.
Western blot analysis
Adherent cells in a 6-well plate were washed with PBS, and 0.5 mL radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific/Gibco, Waltham, MA, USA) with protease inhibitor (cOmplete ULTRA Tablets, Mini, EDTA-free; Roche, Basel, Switzerland) was added to each well. The plate was placed at −80°C for 10 min and then allowed to thaw at room temperature. Cell lysate was collected, and cell debris was separated by centrifugation. Protein-containing supernatant was collected and stored at −20°C. A bicinchoninic acid (BCA) assay (Pierce BCA Protein Assay Kit; Thermo Fisher Scientific/Gibco, Waltham, MA, USA) was performed prior to each western blot to quantify protein. 20 μg of protein per sample was loaded in a 3%–8% Criterion XT Tris-Acetate Gel (Bio-Rad, Hercules, CA, USA) and ran at 125 V for 90 min in Tris/tricine/SDS Running Buffer (Bio-Rad, Hercules, CA, USA). Transfer to Immobilon-FL polyvinylidene fluoride (PVDF) membrane (Millipore, Burlington, MA, USA) was performed at 110 μA, 4°C, overnight in high glycine transfer buffer (120 g glycine, 6 g Tris-base in 2 L water). The membrane was blocked with 0.1% Hammarsten casein in PBS for 1 h. Primary antibodies (mouse anti-human CFTR UNC-596 and rabbit anti-human vinculin; Thermo Fisher Scientific/Gibco, Waltham, MA, USA) were diluted 1:2,000 and incubated at room temperature for 2 h. Secondary IRDye antibodies (donkey anti-rabbit 680RD and donkey anti-mouse 800CW; LI-COR Biosciences, Lincoln, NE, USA) were diluted 1:20,000 and incubated for 1 h at room temperature. Three washes with Tris-buffered saline with Tween 20 (TBST) for 15 min were performed between each step. Membrane was imaged and analyzed using the LI-COR Biosciences Odyssey system.
mRNA measurements
Adherent cells in a 6-well plate were washed with PBS. Cell contents of each well were collected in 0.5 mL TRIzol (Invitrogen, Carlsbad, CA, USA). RNA was isolated following the manufacturer’s instructions for Direct-zol RNA Kit (Zymo Research, Irvine, CA, USA). cDNA was generated using a High-Capacity-RNA-to-cDNA Kit (Applied Biosystems, Foster City, CA, USA), and qPCR (Applied Biosystems 7900HT, Foster City, CA, USA) was performed using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific/Gibco, Waltham, MA, USA) using primers directed against the polyadenylation sequence of pcDNA3.1(+) plasmids 5′-CTCGACTGTGCCTTCTAGTTG-3′ and 5′-GCACCTTCCAGGGTCAAG-3′. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalize gene expression using primers 5′-GGATTTGGTCGTATTGGG-3′ and 5′-GGATTTGGTCGTATTGGG-3′.
Flow cytometry
Cells were resuspended in 100 μL PBS containing 2% FBS with LIVE/DEAD Fixable Far Red Stain Kit (Invitrogen, Carlsbad, CA, USA) and incubated for 30 min protected from light. Three washes with 1 mL 2% FBS in PBS were performed before resuspending cells in a final volume of 500 μL for flow cytometry analysis in an Attune NxT Flow Cytometer (Invitrogen, Carlsbad, CA, USA). Doublets and dead cells were excluded from analysis.
Short circuit measurements
For transepithelial chloride current measurements, epithelial cultures were mounted in Ussing chambers; submerged in a solution containing 135 mM NaCl, 5 mM HEPES, 0.6 mM KH2PO4, 2.4 mM K2HPO4, 2.2 mM MgCl2, 1.2 mM CaCl2, and 5 mM dextrose; and bubbled with air. For bicarbonate measurements, cultures were submerged in a chloride-free solution composed of 118.9 mM Na gluconate, 25 mM NaHCO3, 5 mM Ca gluconate, 1 mM Mg gluconate, 2.4 mM K2HPO4, 0.6 mM KH2PO4, and 5 mM dextrose and bubbled with CO2. Amiloride, ENaC inhibitor, and DIDS, a Cl− exchanger inhibitor, were sequentially added apically to a final concentration of 100 μM. CFTR was then apically activated with the cyclic AMP agonists F&I at a final concentration of 10 μM and 100 μM, respectively. Finally, the CFTR inhibitor GlyH (Cystic Fibrosis Foundation) was added apically to a final concentration of 100 μM. ΔISC, in response to CFTR activation and inhibition, was calculated. For FRT transepithelial current measurements, a Cl− gradient was used. Prior to F&I stimulation, the apical solution was replaced with one in which NaCl was replaced with NaC6H11O7, and changes in current (ΔIT) were calculated.
Patch-clamp experiments
Experiments were performed as previously described.78 We used excised, inside-out membrane patches from HEK293T cells transiently expressing WT CFTR or coCFTR3 using pcDNA3.1(+) vectors. The pipette (extracellular) solution contained the following: 140 mM N-methyl-D-glucamine, 3 mM MgCl2, 5 mM CaCl2, 100 mM L-aspartic acid, and 10 mM tricine (pH 7.3) with HCl. The bath (intracellular) solution contained 140 mM N-methyl-D-glucamine, 3 mM MgCl2, 1 mM Cs ethylene glycol bis (2-aminoethyl ether)-N,N,N′,N′ tetraacetic acid (CsEGTA), and 10 mM tricine (pH 7.3) with HCl. Following patch excision, CFTR channels were activated with a 22-nM protein kinase A (PKA) catalytic subunit (from bovine heart; EMD Millipore, Billerica, MA, USA) and 1 mM ATP (magnesium salt; Sigma-Aldrich, St. Louis, MO, USA). The PKA catalytic subunit was present in all cytosolic solutions that contained ATP. Experiments were performed at room temperature (23°C −26°C). Recordings from patches containing 2−8 channels were digitized at 5 kHz and prior to analysis, low-pass filtered at 500 Hz using an 8-pole Bessel filter (Model 900; Frequency Devices, Haverhill, MA, USA). Single channel openings and closings were analyzed with a burst delimiter of 20 ms using Clampfit software (version 10.3; Molecular Devices, Sunnyvale, CA, USA).79 Events <4 ms duration were ignored. The mean IBI was calculated using the formula Po = (BD × Po,Burst)/(BD + IBI), where Po is the mean open state probability andPo,Burst is the Po within a burst.80
Statistics
All data were analyzed using GraphPad Prism 8 software (San Diego, CA, USA). Statistical significance was determined using one-way ANOVA with Dunn’s multiple comparison test, one-way ANOVA on ranks (Kruskal-Wallis test) with Dunn’s multiple comparisons test, or multiple t tests with Bonferroni-Dunn correction as appropriate. Error bars represent mean ± SE.
Acknowledgments
We thank Ian M. Thornell for his critical review of the manuscript. William R. Skach and Jae Seok Yoon provided materials and valuable insight. BioRender.com was used to create images included in this manuscript. This work was supported by the NIH (F31 HL152500, UG3 HL147366, P01 HL51670, P01 HL091842, P01 HL152960, R01DK107489, and T32 GM007337), Cystic Fibrosis Foundation, The University of Iowa Center for Gene Therapy (DK54759), and Roy J. Carver Chair in Pulmonary Research (to P.B.M.).
Author contributions
Conceptualization, L.I.M.L., A.L.C., P.L.S., S.R., and P.B.M.; investigation, L.I.M.L. and Q.D.; writing – original draft, L.I.M.L.; writing – review & editing, L.I.M.L, A.L.C., Q.D., C.O.R., S.R., P.L.S., and P.B.M.; supervision, C.O.R., P.L.S., and P.B.M.
Declaration of interests
P.B.M. is on the Scientific Advisory Board and receives support for sponsored research from Spirovant Sciences, Inc.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2021.02.020.
Supplemental information
References
- 1.Riordan J.R., Rommens J.M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J.L. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Zabner J., Couture L.A., Gregory R.J., Graham S.M., Smith A.E., Welsh M.J. Adenovirus-mediated gene transfer transiently corrects the chloride transport defect in nasal epithelia of patients with cystic fibrosis. Cell. 1993;75:207–216. doi: 10.1016/0092-8674(93)80063-k. [DOI] [PubMed] [Google Scholar]
- 3.Zabner J., Ramsey B.W., Meeker D.P., Aitken M.L., Balfour R.P., Gibson R.L., Launspach J., Moscicki R.A., Richards S.M., Standaert T.A. Repeat administration of an adenovirus vector encoding cystic fibrosis transmembrane conductance regulator to the nasal epithelium of patients with cystic fibrosis. J. Clin. Invest. 1996;97:1504–1511. doi: 10.1172/JCI118573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss R.B., Milla C., Colombo J., Accurso F., Zeitlin P.L., Clancy J.P., Spencer L.T., Pilewski J., Waltz D.A., Dorkin H.L. Repeated aerosolized AAV-CFTR for treatment of cystic fibrosis: a randomized placebo-controlled phase 2B trial. Hum. Gene Ther. 2007;18:726–732. doi: 10.1089/hum.2007.022. [DOI] [PubMed] [Google Scholar]
- 5.Wagner J.A., Reynolds T., Moran M.L., Moss R.B., Wine J.J., Flotte T.R., Gardner P. Efficient and persistent gene transfer of AAV-CFTR in maxillary sinus. Lancet. 1998;351:1702–1703. doi: 10.1016/S0140-6736(05)77740-0. [DOI] [PubMed] [Google Scholar]
- 6.Alton E.W.F.W., Armstrong D.K., Ashby D., Bayfield K.J., Bilton D., Bloomfield E.V., Boyd A.C., Brand J., Buchan R., Calcedo R., UK Cystic Fibrosis Gene Therapy Consortium Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: a randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2015;3:684–691. doi: 10.1016/S2213-2600(15)00245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies G., Davies J.C., Gill D.R., Hyde S.C., Boyd C., Innes J.A., Porteous D.J., Cheng S.H., Scheule R.K., Higgins T. T4: Safety and expression of a single dose of lipid-mediated CFTR gene therapy to the upper and lower airways of patients with Cystic Fibrosis. Thorax. 2011;66(Suppl 4):A2. [Google Scholar]
- 8.Hyde S.C., Southern K.W., Gileadi U., Fitzjohn E.M., Mofford K.A., Waddell B.E., Gooi H.C., Goddard C.A., Hannavy K., Smyth S.E. Repeat administration of DNA/liposomes to the nasal epithelium of patients with cystic fibrosis. Gene Ther. 2000;7:1156–1165. doi: 10.1038/sj.gt.3301212. [DOI] [PubMed] [Google Scholar]
- 9.Limberis M., Anson D.S., Fuller M., Parsons D.W. Recovery of airway cystic fibrosis transmembrane conductance regulator function in mice with cystic fibrosis after single-dose lentivirus-mediated gene transfer. Hum. Gene Ther. 2002;13:1961–1970. doi: 10.1089/10430340260355365. [DOI] [PubMed] [Google Scholar]
- 10.Cooney A.L., Abou Alaiwa M.H., Shah V.S., Bouzek D.C., Stroik M.R., Powers L.S., Gansemer N.D., Meyerholz D.K., Welsh M.J., Stoltz D.A. Lentiviral-mediated phenotypic correction of cystic fibrosis pigs. JCI Insight. 2016;1:e88730. doi: 10.1172/jci.insight.88730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrow N., Miller D., Cmielewski P., Donnelley M., Bright R., Parsons D.W. Airway gene transfer in a non-human primate: lentiviral gene expression in marmoset lungs. Sci. Rep. 2013;3:1287. doi: 10.1038/srep01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns J.C., Friedmann T., Driever W., Burrascano M., Yee J.K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borok Z., Harboe-Schmidt J.E., Brody S.L., You Y., Zhou B., Li X., Cannon P.M., Kim K.J., Crandall E.D., Kasahara N. Vesicular stomatitis virus G-pseudotyped lentivirus vectors mediate efficient apical transduction of polarized quiescent primary alveolar epithelial cells. J. Virol. 2001;75:11747–11754. doi: 10.1128/JVI.75.23.11747-11754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinn P.L., Cooney A.L., Oakland M., Dylla D.E., Wallen T.J., Pezzulo A.A., Chang E.H., McCray P.B., Jr. Lentiviral vector gene transfer to porcine airways. Mol. Ther. Nucleic Acids. 2012;1:e56. doi: 10.1038/mtna.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cmielewski P., Farrow N., Devereux S., Parsons D., Donnelley M. Gene therapy for Cystic Fibrosis: Improved delivery techniques and conditioning with lysophosphatidylcholine enhance lentiviral gene transfer in mouse lung airways. Exp. Lung Res. 2017;43:426–433. doi: 10.1080/01902148.2017.1395931. [DOI] [PubMed] [Google Scholar]
- 16.Johnson L.G., Olsen J.C., Naldini L., Boucher R.C. Pseudotyped human lentiviral vector-mediated gene transfer to airway epithelia in vivo. Gene Ther. 2000;7:568–574. doi: 10.1038/sj.gt.3301138. [DOI] [PubMed] [Google Scholar]
- 17.Wang G., Zabner J., Deering C., Launspach J., Shao J., Bodner M., Jolly D.J., Davidson B.L., McCray P.B., Jr. Increasing epithelial junction permeability enhances gene transfer to airway epithelia In vivo. Am. J. Respir. Cell Mol. Biol. 2000;22:129–138. doi: 10.1165/ajrcmb.22.2.3938. [DOI] [PubMed] [Google Scholar]
- 18.Kobinger G.P., Weiner D.J., Yu Q.C., Wilson J.M. Filovirus-pseudotyped lentiviral vector can efficiently and stably transduce airway epithelia in vivo. Nat. Biotechnol. 2001;19:225–230. doi: 10.1038/85664. [DOI] [PubMed] [Google Scholar]
- 19.Sinn P.L., Coffin J.E., Ayithan N., Holt K.H., Maury W. Lentiviral Vectors Pseudotyped with Filoviral Glycoproteins. Methods Mol. Biol. 2017;1628:65–78. doi: 10.1007/978-1-4939-7116-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinn P.L., Hwang B.Y., Li N., Ortiz J.L.S., Shirazi E., Parekh K.R., Cooney A.L., Schaffer D.V., McCray P.B., Jr. Novel GP64 envelope variants for improved delivery to human airway epithelial cells. Gene Ther. 2017;24:674–679. doi: 10.1038/gt.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel M., Giddings A.M., Sechelski J., Olsen J.C. High efficiency gene transfer to airways of mice using influenza hemagglutinin pseudotyped lentiviral vectors. J. Gene Med. 2013;15:51–62. doi: 10.1002/jgm.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griesenbach U., Inoue M., Meng C., Farley R., Chan M., Newman N.K., Brum A., You J., Kerton A., Shoemark A. Assessment of F/HN-pseudotyped lentivirus as a clinically relevant vector for lung gene therapy. Am. J. Respir. Crit. Care Med. 2012;186:846–856. doi: 10.1164/rccm.201206-1056OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitomo K., Griesenbach U., Inoue M., Somerton L., Meng C., Akiba E., Tabata T., Ueda Y., Frankel G.M., Farley R. Toward gene therapy for cystic fibrosis using a lentivirus pseudotyped with Sendai virus envelopes. Mol. Ther. 2010;18:1173–1182. doi: 10.1038/mt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi M., Iida A., Ueda Y., Hasegawa M. Pseudotyped lentivirus vectors derived from simian immunodeficiency virus SIVagm with envelope glycoproteins from paramyxovirus. J. Virol. 2003;77:2607–2614. doi: 10.1128/JVI.77.4.2607-2614.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bou Saab J., Losa D., Chanson M., Ruez R. Connexins in respiratory and gastrointestinal mucosal immunity. FEBS Lett. 2014;588:1288–1296. doi: 10.1016/j.febslet.2014.02.059. [DOI] [PubMed] [Google Scholar]
- 26.Chanson M., Kotsias B.A., Peracchia C., O’Grady S.M. Interactions of connexins with other membrane channels and transporters. Prog. Biophys. Mol. Biol. 2007;94:233–244. doi: 10.1016/j.pbiomolbio.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheckenbach K.E., Losa D., Dudez T., Bacchetta M., O’Grady S., Crespin S., Chanson M. Prostaglandin E2regulation of cystic fibrosis transmembrane conductance regulator activity and airway surface liquid volume requires gap junctional communication. Am. J. Respir. Cell Mol. Biol. 2011;44:74–82. doi: 10.1165/rcmb.2009-0361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanderson M.J., Chow I., Dirksen E.R. Intercellular communication between ciliated cells in culture. Am. J. Physiol. 1988;254:C63–C74. doi: 10.1152/ajpcell.1988.254.1.C63. [DOI] [PubMed] [Google Scholar]
- 29.Kreda S.M., Mall M., Mengos A., Rochelle L., Yankaskas J., Riordan J.R., Boucher R.C. Characterization of wild-type and deltaF508 cystic fibrosis transmembrane regulator in human respiratory epithelia. Mol. Biol. Cell. 2005;16:2154–2167. doi: 10.1091/mbc.E04-11-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montoro D.T., Haber A.L., Biton M., Vinarsky V., Lin B., Birket S.E., Yuan F., Chen S., Leung H.M., Villoria J. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plasschaert L.W., Žilionis R., Choo-Wing R., Savova V., Knehr J., Roma G., Klein A.M., Jaffe A.B. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560:377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okuda K., Dang H., Kobayashi Y., Carraro G., Nakano S., Chen G., Kato T., Asakura T., Gilmore R.C., Morton L.C. Secretory Cells Dominate Airway CFTR Expression and Function in Human Airway Superficial Epithelia. Am. J. Respir. Crit. Care Med. 2020 doi: 10.1164/rccm.202008-3198OC. Published online December 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin J.Y., Zhang L., Clift K.L., Hulur I., Xiang A.P., Ren B.Z., Lahn B.T. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS ONE. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng L., Ziegelhoffer P.R., Yang N.S. In vivo promoter activity and transgene expression in mammalian somatic tissues evaluated by using particle bombardment. Proc. Natl. Acad. Sci. USA. 1993;90:4455–4459. doi: 10.1073/pnas.90.10.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farmen S.L., Karp P.H., Ng P., Palmer D.J., Koehler D.R., Hu J., Beaudet A.L., Zabner J., Welsh M.J. Gene transfer of CFTR to airway epithelia: low levels of expression are sufficient to correct Cl- transport and overexpression can generate basolateral CFTR. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005;289:L1123–L1130. doi: 10.1152/ajplung.00049.2005. [DOI] [PubMed] [Google Scholar]
- 36.Zychlinski D., Schambach A., Modlich U., Maetzig T., Meyer J., Grassman E., Mishra A., Baum C. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol. Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- 37.Montini E., Cesana D., Schmidt M., Sanvito F., Ponzoni M., Bartholomae C., Sergi Sergi L., Benedicenti F., Ambrosi A., Di Serio C. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 38.Stein S., Ott M.G., Schultze-Strasser S., Jauch A., Burwinkel B., Kinner A., Schmidt M., Krämer A., Schwäble J., Glimm H. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 39.Río P., Navarro S., Wang W., Sánchez-Domínguez R., Pujol R.M., Segovia J.C., Bogliolo M., Merino E., Wu N., Salgado R. Successful engraftment of gene-corrected hematopoietic stem cells in non-conditioned patients with Fanconi anemia. Nat. Med. 2019;25:1396–1401. doi: 10.1038/s41591-019-0550-z. [DOI] [PubMed] [Google Scholar]
- 40.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 41.De Ravin S.S., Wu X., Moir S., Anaya-O’Brien S., Kwatemaa N., Littel P., Theobald N., Choi U., Su L., Marquesen M. Lentiviral hematopoietic stem cell gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 2016;8:335ra57. doi: 10.1126/scitranslmed.aad8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cehajic-Kapetanovic J., Xue K., Martinez-Fernandez de la Camara C., Nanda A., Davies A., Wood L.J., Salvetti A.P., Fischer M.D., Aylward J.W., Barnard A.R. Initial results from a first-in-human gene therapy trial on X-linked retinitis pigmentosa caused by mutations in RPGR. Nat. Med. 2020;26:354–359. doi: 10.1038/s41591-020-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer M.D., McClements M.E., Martinez-Fernandez de la Camara C., Bellingrath J.S., Dauletbekov D., Ramsden S.C., Hickey D.G., Barnard A.R., MacLaren R.E. Codon-Optimized RPGR Improves Stability and Efficacy of AAV8 Gene Therapy in Two Mouse Models of X-Linked Retinitis Pigmentosa. Mol. Ther. 2017;25:1854–1865. doi: 10.1016/j.ymthe.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohn D.B., Booth C., Kang E.M., Pai S.Y., Shaw K.L., Santilli G., Armant M., Buckland K.F., Choi U., De Ravin S.S., Net4CGD consortium Lentiviral gene therapy for X-linked chronic granulomatous disease. Nat. Med. 2020;26:200–206. doi: 10.1038/s41591-019-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rangarajan S., Walsh L., Lester W., Perry D., Madan B., Laffan M., Yu H., Vettermann C., Pierce G.F., Wong W.Y., Pasi K.J. AAV5-Factor VIII Gene Transfer in Severe Hemophilia A. N. Engl. J. Med. 2017;377:2519–2530. doi: 10.1056/NEJMoa1708483. [DOI] [PubMed] [Google Scholar]
- 48.Dittmar K.A., Goodenbour J.M., Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kudla G., Lipinski L., Caffin F., Helwak A., Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4:e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elhaik E., Tatarinova T. GC3 biology in eukaryotes and prokaryotes. In: Tatarinova T., editor. DNA Methylation - From Genomics to Technology. InTech; 2012. pp. 3–68. [Google Scholar]
- 51.Gebert D., Jehn J., Rosenkranz D. Widespread selection for extremely high and low levels of secondary structure in coding sequences across all domains of life. Open Biol. 2019;9:190020. doi: 10.1098/rsob.190020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bauer A.P., Leikam D., Krinner S., Notka F., Ludwig C., Längst G., Wagner R. The impact of intragenic CpG content on gene expression. Nucleic Acids Res. 2010;38:3891–3908. doi: 10.1093/nar/gkq115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharp P.M., Li W.H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S.J., Yoon J.S., Shishido H., Yang Z., Rooney L.A., Barral J.M., Skach W.R. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348:444–448. doi: 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- 55.Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng S.H., Gregory R.J., Marshall J., Paul S., Souza D.W., White G.A., O’Riordan C.R., Smith A.E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 57.Sheppard D.N., Carson M.R., Ostedgaard L.S., Denning G.M., Welsh M.J. Expression of cystic fibrosis transmembrane conductance regulator in a model epithelium. Am. J. Physiol. 1994;266:L405–L413. doi: 10.1152/ajplung.1994.266.4.L405. [DOI] [PubMed] [Google Scholar]
- 58.Baumlin-Schmid N., Salathe M., Fregien N.L. Optimal Lentivirus Production and Cell Culture Conditions Necessary to Successfully Transduce Primary Human Bronchial Epithelial Cells. J. Vis. Exp. 2016;(113):e54176. doi: 10.3791/54176. [DOI] [PubMed] [Google Scholar]
- 59.Bartoszewski R., Króliczewski J., Piotrowski A., Jasiecka A.J., Bartoszewska S., Vecchio-Pagan B., Fu L., Sobolewska A., Matalon S., Cutting G.R. Codon bias and the folding dynamics of the cystic fibrosis transmembrane conductance regulator. Cell. Mol. Biol. Lett. 2016;21:23. doi: 10.1186/s11658-016-0025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walsh I.M., Bowman M.A., Soto Santarriaga I.F., Rodriguez A., Clark P.L. Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. Proc. Natl. Acad. Sci. USA. 2020;117:3528–3534. doi: 10.1073/pnas.1907126117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldman M.J., Yang Y., Wilson J.M. Gene therapy in a xenograft model of cystic fibrosis lung corrects chloride transport more effectively than the sodium defect. Nat. Genet. 1995;9:126–131. doi: 10.1038/ng0295-126. [DOI] [PubMed] [Google Scholar]
- 62.Johnson L.G., Olsen J.C., Sarkadi B., Moore K.L., Swanstrom R., Boucher R.C. Efficiency of gene transfer for restoration of normal airway epithelial function in cystic fibrosis. Nat. Genet. 1992;2:21–25. doi: 10.1038/ng0992-21. [DOI] [PubMed] [Google Scholar]
- 63.Shah V.S., Ernst S., Tang X.X., Karp P.H., Parker C.P., Ostedgaard L.S., Welsh M.J. Relationships among CFTR expression, HCO3- secretion, and host defense may inform gene- and cell-based cystic fibrosis therapies. Proc. Natl. Acad. Sci. USA. 2016;113:5382–5387. doi: 10.1073/pnas.1604905113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang L., Button B., Gabriel S.E., Burkett S., Yan Y., Skiadopoulos M.H., Dang Y.L., Vogel L.N., McKay T., Mengos A. CFTR delivery to 25% of surface epithelial cells restores normal rates of mucus transport to human cystic fibrosis airway epithelium. PLoS Biol. 2009;7:e1000155. doi: 10.1371/journal.pbio.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frizzell R.A., Hanrahan J.W. Physiology of epithelial chloride and fluid secretion. Cold Spring Harb. Perspect. Med. 2012;2:a009563. doi: 10.1101/cshperspect.a009563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno-Carranza B., Gentsch M., Stein S., Schambach A., Santilli G., Rudolf E., Ryser M.F., Haria S., Thrasher A.J., Baum C. Transgene optimization significantly improves SIN vector titers, gp91phox expression and reconstitution of superoxide production in X-CGD cells. Gene Ther. 2009;16:111–118. doi: 10.1038/gt.2008.143. [DOI] [PubMed] [Google Scholar]
- 67.Ward N.J., Buckley S.M., Waddington S.N., Vandendriessche T., Chuah M.K., Nathwani A.C., McIntosh J., Tuddenham E.G., Kinnon C., Thrasher A.J., McVey J.H. Codon optimization of human factor VIII cDNAs leads to high-level expression. Blood. 2011;117:798–807. doi: 10.1182/blood-2010-05-282707. [DOI] [PubMed] [Google Scholar]
- 68.Zhang R., Wang Q., Zhang L., Chen S. Optimized human factor IX expression cassettes for hepatic-directed gene therapy of hemophilia B. Front. Med. 2015;9:90–99. doi: 10.1007/s11684-015-0390-2. [DOI] [PubMed] [Google Scholar]
- 69.Padegimas L., Kowalczyk T.H., Adams S., Gedeon C.R., Oette S.M., Dines K., Hyatt S.L., Sesenoglu-Laird O., Tyr O., Moen R.C., Cooper M.J. Optimization of hCFTR lung expression in mice using DNA nanoparticles. Mol. Ther. 2012;20:63–72. doi: 10.1038/mt.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.al Yacoub N., Romanowska M., Haritonova N., Foerster J. Optimized production and concentration of lentiviral vectors containing large inserts. J. Gene Med. 2007;9:579–584. doi: 10.1002/jgm.1052. [DOI] [PubMed] [Google Scholar]
- 71.Kumar M., Keller B., Makalou N., Sutton R.E. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 2001;12:1893–1905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- 72.Berical A., Lee R.E., Randell S.H., Hawkins F. Challenges Facing Airway Epithelial Cell-Based Therapy for Cystic Fibrosis. Front. Pharmacol. 2019;10:74. doi: 10.3389/fphar.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smit L.S., Wilkinson D.J., Mansoura M.K., Collins F.S., Dawson D.C. Functional roles of the nucleotide-binding folds in the activation of the cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. USA. 1993;90:9963–9967. doi: 10.1073/pnas.90.21.9963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baker J.M., Hudson R.P., Kanelis V., Choy W.Y., Thibodeau P.H., Thomas P.J., Forman-Kay J.D. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat. Struct. Mol. Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Romero Z., Campo-Fernandez B., Wherley J., Kaufman M.L., Urbinati F., Cooper A.R., Hoban M.D., Baldwin K.M., Lumaquin D., Wang X. The human ankyrin 1 promoter insulator sustains gene expression in a β-globin lentiviral vector in hematopoietic stem cells. Mol. Ther. Methods Clin. Dev. 2015;2:15012. doi: 10.1038/mtm.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Breda L., Casu C., Gardenghi S., Bianchi N., Cartegni L., Narla M., Yazdanbakhsh K., Musso M., Manwani D., Little J. Therapeutic hemoglobin levels after gene transfer in β-thalassemia mice and in hematopoietic cells of β-thalassemia and sickle cells disease patients. PLoS ONE. 2012;7:e32345. doi: 10.1371/journal.pone.0032345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karp P.H., Moninger T.O., Weber S.P., Nesselhauf T.S., Launspach J.L., Zabner J., Welsh M.J. An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol. Biol. 2002;188:115–137. doi: 10.1385/1-59259-185-X:115. [DOI] [PubMed] [Google Scholar]
- 78.Dong Q., Randak C.O., Welsh M.J. A mutation in CFTR modifies the effects of the adenylate kinase inhibitor Ap5A on channel gating. Biophys. J. 2008;95:5178–5185. doi: 10.1529/biophysj.108.140897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carson M.R., Travis S.M., Welsh M.J. The two nucleotide-binding domains of cystic fibrosis transmembrane conductance regulator (CFTR) have distinct functions in controlling channel activity. J. Biol. Chem. 1995;270:1711–1717. doi: 10.1074/jbc.270.4.1711. [DOI] [PubMed] [Google Scholar]
- 80.Cotten J.F., Welsh M.J. Covalent modification of the nucleotide binding domains of cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 1998;273:31873–31879. doi: 10.1074/jbc.273.48.31873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.