Abstract
Although thalidomide is highly teratogenic, it has been prescribed for treating multiple myeloma and Hansen's disease. However, its mechanism of action is not fully understood. Here, we employed a reverse transcription quantitative PCR array to measure the expression of 84 genes in human induced pluripotent stem cells (hiPSCs) and their mesodermal differentiation. Thalidomide altered the expression of undifferentiated marker genes in both cell types. Thalidomide affected more genes in the mesoderm than in the hiPSCs. Ectoderm genes were upregulated but mesendoderm genes were downregulated by thalidomide during mesoderm induction, suggesting that thalidomide altered mesoderm differentiation. We found that FABP7 (fatty acid binding protein 7) was dramatically downregulated in the hiPSCs. FABP is related to retinoic acid, which is important signaling for limb formation. Moreover, thalidomide altered the expression of the genes involved in TGF-β signaling, limb formation, and multiple myeloma, which are related to thalidomide-induced malformations and medication. In summary, iPSCs can serve as useful tools to elucidate the mechanisms underlying thalidomide malformations in vitro.
Keywords: Thalidomide, Human induced pluripotent cell, Mesodermal differentiation, FABP, Retinoic acid, Multiple myeloma
Highlights
-
•
Thalidomide downregulated FABP7, a fatty acid binding protein (FABP) cording gene.
-
•
FABP is related to retinoic acid, which is important signaling for limb formation.
-
•
Thalidomide treatment affected the expression of limb formation related genes.
-
•
Thalidomide treatment affected 5 genes related to multiple myeloma.
-
•
Thalidomide upregulated ectoderm but downregulated mesendoderm markers in mesoderm.
1. Introduction
Although thalidomide functioned as an excellent sedative, its use was discontinued in the 1960s because it is highly teratogenic [[1], [2], [3], [4]]. Recently, thalidomide has been prescribed for treating multiple myeloma and Hansen's disease [5,6]. Thus, thalidomide is an old but new medicine.
The primary target of thalidomide was identified as cereblon (CRBN) in 2010 [7]. However, the mechanism underlying its teratogenic effects remains unclear even after more than 50 years [8]. This is largely because thalidomide teratogenicity is not observed in experimental mammals, such as mice, but is only observed in other vertebrates, such as chicks and fish [9,10]. Recently, to test thalidomide teratogenicity in humans, many groups have used human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs), both of which have characteristics similar to those of epiblasts that give rise to the fetus [[11], [12], [13], [14], [15], [16], [17]].
Previously, we showed that thalidomide increased apoptosis in undifferentiated hiPSCs and early mesoderm [15,16]. Because the epiblast gives rise to the fetus and mesoderm give rise to bones and muscles, and given that thalidomide causes mild effects in the entire body and severe malformation in the limbs, hiPSCs could function as good in vitro models for studying the teratogenic effects of thalidomide.
In this study, we employed a reverse transcription quantitative polymerase chain reaction (RT-qPCR) array to study the expression of 84 genes (Supplementary Table 1), including pluripotent stem cell markers and early differentiation markers in hiPSCs and during the mesodermal differentiation of hiPSCs (Supplementary Table 1), to identify candidate genes responsible for thalidomide teratogenicity.
2. Materials and methods
2.1. hiPSC culture
The hiPSC cell line, 201B7 [18], was obtained from the RIKEN BRC Cell Bank (HPS0063, Tsukuba, Japan) via the National Bio-Resource Project for the Ministry of Education, Culture, Sports, Science, and Technology, Japan. hiPSCs were maintained as previously described [16,19,20]. Briefly, cells were maintained in knockout serum replacement (KSR)-based medium and cultured for at least two passages (0.5 U/mL dispase, Thermo Fisher Scientific, Waltham, Massachusetts, U.S.) under serum- and feeder-free conditions in the maintenance medium (hESF9a medium, Supplementary Table 2) containing 5 μM Rho-associated coil kinase inhibitor (ROCK inhibitor, Y-27632, FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) in 2 μg/cm2 fibronectin (from bovine blood plasma; Sigma-Aldrich, St. Louis, MO, USA) coated dishes [16,19,20].
2.2. Thalidomide treatment
Undifferentiated hiPSCs were dissociated into single cells by incubation in 0.02% (w/w) ethylenediaminetetraacetic acid (EDTA-4Na, FUJIFILM Wako) prepared in calcium- and magnesium-free phosphate-buffered saline (PBS) and were then re-plated at a density of 1 × 104 cells/cm2 (for undifferentiated cells) and 5 × 103 cells/cm2 (for mesodermal differentiation) on 2 μg/cm2 fibronectin-coated dishes in maintenance medium containing 5 μM Y-27632 (day-1). For undifferentiated hiPSCs, the medium was replaced with maintenance medium containing 5 μM thalidomide (+/-Thalidomide, 200–15131, FUJIFILM Wako) prepared in 0.1% dimethyl sulfoxide (DMSO) at day 0 (24 h after re-plating, Fig. 1A). Cells were harvested on day 4. For the mesoderm condition, the medium was replaced with hESF6 medium (Supplementary Table 2) containing 50 ng/mL activin, 3 μM CHIR99021 (R&D Systems, Minneapolis, MN), and Y-27632 at day 0 (24 h after re-plating, Fig. 2A). CHIR99021 was removed on day 1 and Y-27632 was removed on day 3, and cells were treated with thalidomide prepared in 0.1% dimethyl sulfoxide (DMSO) on day 0. Cells were harvested on day 5.
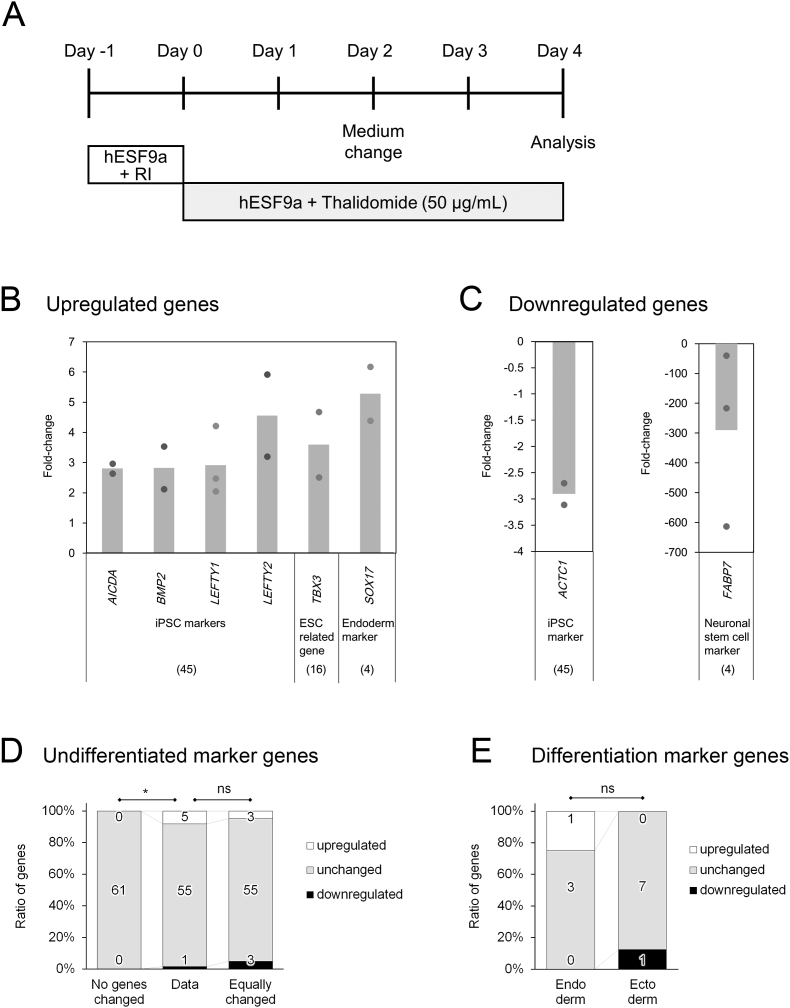
Fig. 1.
Effects of thalidomide on undifferentiated hiPSCs. A: Schematic of the experimental design. RI: Y-27632, hESF9a: Supplementary Table 2. BC: Genes upregulated (B) and downregulated (C) more than two-fold in response to thalidomide treatment. The circles show each experimental value, and the column shows the mean value. DE: Ratio of the numbers of upregulated genes, unchanged genes, and downregulated genes. D: Ratio of undifferentiated marker genes (data) compared to the hypothetical ratio with no gene changed (unchanged) and the hypothetical ratio with equal up- and downregulation (equally changed). E: Ratio of endoderm and ectoderm differentiation markers. The numbers in the bar represent the number of genes. *P < 0.05, and ns > 0.05 by Fisher's exact test.
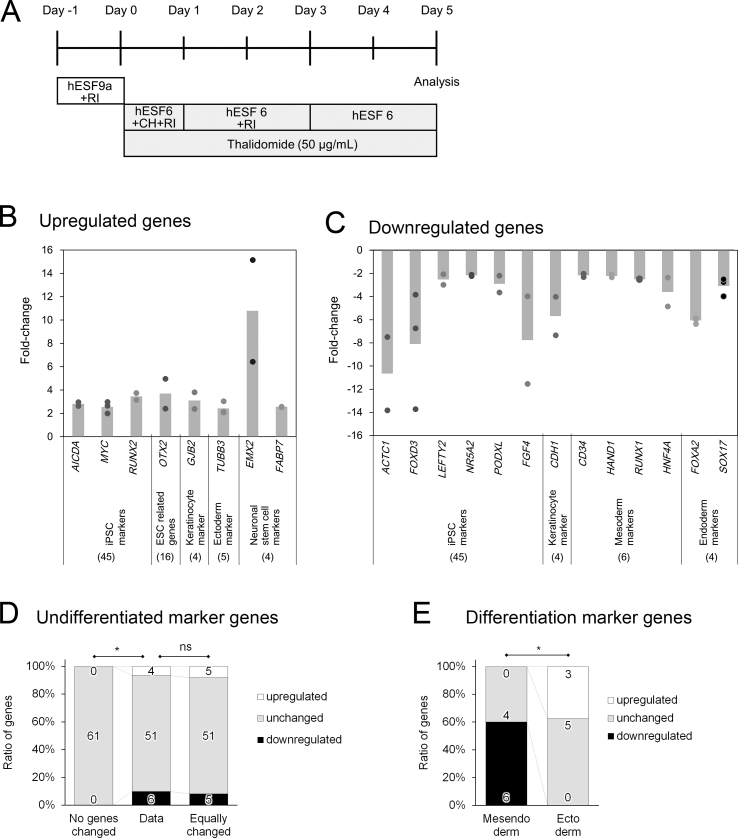
Fig. 2.
Effects of thalidomide on mesoderm differentiation. A: Schematic of the experimental design. RI: Y-27632, hESF9a and hESF6: Supplementary Table 2. BC: Genes upregulated (B) and downregulated (C) more than two-fold in response to thalidomide treatment. The circles show each experimental value, and the column shows the mean value. DE: Ratio of the numbers of upregulated genes, unchanged genes, and downregulated genes. D: Ratio of undifferentiated marker genes (data) compared to the hypothetical ratio with no gene changed (unchanged) and the hypothetical ratio with equal up- and downregulation (equally changed). E: Ratio of mesendoderm and ectoderm differentiation markers. The numbers in the bar represent the number of genes. *P < 0.05, and ns > 0.05 by Fisher's exact test.
2.3. RT-qPCR array analysis
RNA was extracted from the harvested cells using the RNeasy Mini Kit (74104, Qiagen, Hilden, Germany) and cDNA was synthesized using the RT2 First Strand Kit (330401, Qiagen). A mixture of cDNA and RT2 SYBR Green ROX qPCR Mastermix (330404, Qiagen) was added to the RT2 Profiler PCR Array Human Induced Pluripotent Stem Cells (PAHS-092ZE-4, Qiagen), which was placed in a 7900HT Fast Real-Time PCR machine (Applied Biosystems, CA, USA). An online analysis tool (https://www.qiagen.com/jp/shop/genes-and-pathways/data-analysis-center-overview-page/) was used for data analysis. Changes in gene expression levels were determined by comparing the normalized expression levels in cells treated with thalidomide with those in the control cells.
The RT-qPCR array targeted 84 genes, including 45 hiPSC genes, 16 ESC genes, four endoderm genes, six mesoderm genes, nine ectoderm (including neural stem cell) genes, and others (Supplementary Table 1).
2.4. Statistical analysis
Fisher's exact test (3 × 3 or 3 × 2 contingency table, two-tailed) was performed using R (http://www.R-project.org/). The three numbers shown in parentheses in each Fisher's exact test indicate the number of upregulated genes, the number of genes without change, and the number of downregulated genes.
3. Results
3.1. Effects of thalidomide on undifferentiated hiPSCs
We previously reported that thalidomide induced apoptosis in undifferentiated hiPSCs; however, the expression of SSEA-4, an undifferentiated cell marker, and SSEA-1, an early differentiation marker, did not appear to change based on immunofluorescence analysis [15]. Therefore, we used an RT-qPCR array to comprehensively determine the changes in gene expression patterns induced by thalidomide in undifferentiated hiPSCs (Fig. 1A).
First, we focused on the effects of thalidomide on the expression of 61 undifferentiated marker genes (45 hiPSC markers and 16 hESC-related genes). Thalidomide upregulated five genes (AICDA, BMP, LEFTY1, LEFTY2, and TBX3) and downregulated one gene (ACTC1) (Table 1, Fig. 1B and C). The ratio of regulated genes, genes without change, and downregulated genes (5, 55, 1) was significantly different from the hypothetical ratio without change (0, 61, 0) (Fig. 1D, Fisher's exact test, P = 0.027). However, the ratio (5, 55, 1) was not significantly different from the hypothetical ratio with equal up- and downregulation (3, 55, 3) (Fig. 1D, Fisher's exact test, P = 0.581). These results suggest that although thalidomide affects undifferentiated hiPSCs, it might not facilitate or inhibit the undifferentiated state. Notably, three transforming growth factor β (TGF-β) genes (BMP2, LEFTY1, and LEFTY2), which are related to mesoderm differentiation, including limb formation, were upregulated (Fig. 1B).
Table 1.
List of genes whose expression altered more than two-fold by thalidomide treatment.
| Gene | hiPSC | mesoderm | marker | Descriptions and [References] |
|---|---|---|---|---|
| ACTC1 | down | down | ES/iPSCs | Actin alpha cardiac muscle 1. Sarcomere gene |
| AICDA | up | up | ES/iPSCs (reprogramming-related) | Activation-induced cytidine deaminase. Genes associated with DNA demethylation |
| BMP2 | up | ES/iPSCs | Bone morphogenetic protein-2 encodes a TGF-β super family morphogen. Related to limb growth [38]. | |
| CD34 | down | mesoderm (blood stem cells) | Protein localized to hematopoietic progenitor cells and stem cells | |
| CDH1 | down | E-cadherin and keratinocyte | Intercellular adhesion factor. Association of E-cadherin expression | |
| EMX2 | up | ectoderm (neural stem cells) | Encodes empty spiracles homolog 2, which is associated with neural differentiation and cell proliferation in the brain, and limb formation [39] | |
| FABP7 | down | up | ectoderm (neural stem cells) | Encodes fatty acid binding protein 7, which localizes in the brain and is involved in neural development [21,22]. |
| FGF4 | down | ES/iPSCs (pluripotency markers) | Encodes fibroblast growth factor 4, which plays various roles in development, including limb bud development [38] | |
| FOXA2 | down | endoderm | Encodes forkhead box A2 | |
| FOXD3 | down | ES/iPSCs (pluripotency markers) | Encodes forkhead box protein D3, which acts as a transcriptional repressor. | |
| GJB2 | up | keratinocytes (CX26) | Encode the gap junction structural component connexin 26 (GJB2), which is a component of gap junctions and is related to hearing loss [42,47,48] | |
| HAND1 | down | mesoderm | essential gene for cardiovascular development and is involved in mammalian heart chamber formation [41]. | |
| HNF4A | down | mesoderm | Hepatocyte nuclear factor 4, alpha | |
| LEFTY1 | up | ES/iPSCs (pluripotency markers) | TGF-β super family related genes that determine the left-right asymmetry of the body | |
| LEFTY2 | up | down | ES/iPSCs (pluripotency markers) | TGF-β super family related genes that determine the left-right asymmetry of the body |
| MYC | up | ES/iPSCs (reprogramming factors) | oncogene. MYC can induce DNA breaks in vivo and in vitro, independent of reactive oxygen species. | |
| NR5A2 | down | ES/iPSCs (reprogramming factors) | Encodes nuclear receptor subfamily 5, group A, member 2, which is responsible for transcriptional activation of OCT3/4. | |
| OTX2 | up | ES/iPSCs | Encodes orthodenticle Homeobox 2, a marker gene for ectoderm and neural differentiation. | |
| PODXL | down | ES/iPSCs (pluripotency markers) | Encodes podocalyxin-like 1, which is related to cell adhesion. | |
| RUNX1 | down | mesoderm | Encodes Runt-related transcription factor 1 | |
| RUNX2 | up | ES/iPSC | Runt-related transcription factor 2 related to osteoblast differentiation. | |
| SOX17 | up | down | endoderm | Encodes sex-determining region Y-box 17 (SOX17). |
| TBX3 | up | ES/iPSCs (ESC related) | Encodes T-box transcription factor TBX3, which is essential for embryonic development and is involved in heart and angiogenesis. | |
| TUBB3 | up | ectoderm | Encodes tubulin beta 3 class III (TUB-β3), which is used for microtubule formation and is a neural differentiation marker. |
up: upregulated genes, down: downregulated genes.
We then focused on whether thalidomide affected spontaneous differentiation. With respect to marker genes of the three germ layers, although the expression of one of four endoderm genes (SOX17) was upregulated (1, 3, 0) and that of one of the eight ectoderm marker genes (FABP7) was downregulated (0, 7, 1), there was no significant difference between the ratios (Fig. 1E, Fisher's exact test, P = 0.58), suggesting that thalidomide did not induce endodermal differentiation nor did it suppress ectodermal differentiation. Surprisingly, FABP7 was dramatically (average of approximately 300-fold) downregulated by thalidomide (Fig. 1C). FABP7 is a member of the fatty acid binding protein family and is primarily expressed in the brain and is involved in neural development [21,22]. FABP7 was downregulated 40-600-fold in three experiments, suggesting that thalidomide strongly inhibits FABP7 expression in undifferentiated hiPSCs.
3.2. Effects of thalidomide on mesodermal differentiation
Next, we focused on the effects of thalidomide on mesodermal differentiation, which is responsible for the development of limbs. Given that the most common symptom of thalidomide teratogenicity is limb deformation, thalidomide may also affect mesodermal differentiation. We previously reported that, in response to thalidomide treatment, the number of apoptotic and dead mesoderm cells induced from hiPSCs increased on day 2, but the number of dead cells decreased on day 5, suggesting that mesoderm cells were damaged by thalidomide [16]. However, immunocytochemical analysis did not indicate any change in the expression of early mesoderm marker genes [16]. To gain further insights into the function of thalidomide, we performed a comprehensive gene expression analysis (Fig. 2A).
More than twice the number of genes were changed in mesoderm differentiation (21 genes) than in undifferentiated condition (8 genes), suggesting that mesoderm differentiation was more strongly affected by thalidomide compared to the undifferentiated state (Table 1). In both conditions, ACTC1 and AICDA were upregulated and downregulated, respectively; however, LEFTY2 and SOX17 were oppositely regulated between the undifferentiated and mesoderm differentiation conditions (Table 1).
With respect to the effects of thalidomide on the expression of 61 undifferentiated marker genes, four genes (AICDA, MYC, RUNX2, and OTX2) were upregulated (Fig. 2B) and six genes (ACTC1, FOXD3, LEFTY2, NR5A2, PODXL, and FGF4) were downregulated (Fig. 2C, Table 1). The ratio of upregulated genes, genes without change, and downregulated genes (4, 51, 6) were significantly different from the hypothetical ratio with no genes changed (0, 61, 0) (Fig. 2D, Fisher's exact test P = 0.005). However, the ratio was not significantly different from the hypothetical ratio with equal up- and downregulation (5, 51, 5) (Fig. 2D, Fisher's exact test, P = 1). These results suggest that although thalidomide affected the expression of undifferentiated marker genes during mesoderm differentiation, it might not be involved in the escape from the undifferentiated state.
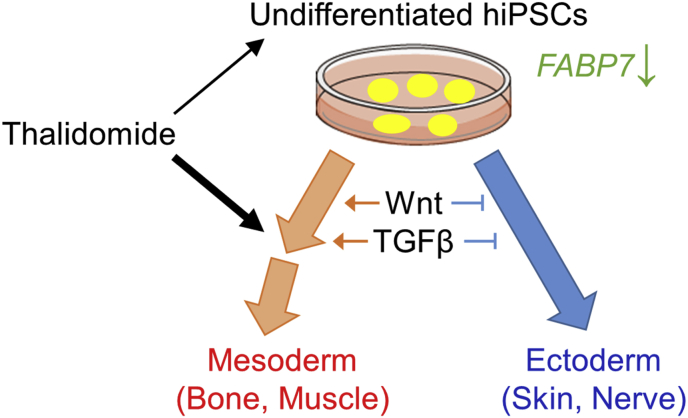
The effects of thalidomide on the three germ-layer marker genes were markedly different. Among the mesoderm and endoderm marker genes, although no genes were upregulated, four of the six mesodermal marker genes (CD34, HAND1, HNF4A, and RUNX1) were downregulated (0, 2, 4) (Fig. 2B), and twoof the four endodermal marker genes (FOXA2, and SOX17) were downregulated (0, 2, 2) (Fig. 2C). In contrast, although three of the eight ectodermal marker genes (TUBB3, EMX2, and FABP7) were upregulated (Fig. 2B), no ectodermal marker gene was downregulated (3, 5, 0) (Fig. 2C). Differentiation into the mesendoderm, the early stage of both the mesoderm and endoderm, and the ectoderm occurs with exclusivity; although mesendoderm differentiation is induced upon the activation of TGF-β and Wnt signals, ectoderm differentiation is induced upon the suppression of these signals (Fig. 3) [16,[23], [24], [25]]. Statistical analysis revealed significantly different gene expression results between the mesendoderm (0, 4, 6) and ectoderm (3, 5, 0) (Fig. 2E, Fisher's exact test, P = 0.0072). Thus, these results suggest that thalidomide slightly altered mesendoderm differentiation toward ectoderm by modulating TGF-β and Wnt signals (Fig. 3).
Fig. 3.
Schematic representation of the results. The thickness of the black arrows reflects the strength of the effects. The thick orange and blue arrows represent differentiation direction. The thin orange arrows represent activating signals, and thin blue blunt-ended lines represent inhibitory signals. The type of cells is shown in parentheses. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In the present study, we identified changes in gene expression in undifferentiated hiPSCs and during mesodermal differentiation of hiPSCs. In both conditions, although thalidomide treatment altered undifferentiated marker gene expression, it might not be involved in the escape from the undifferentiated state, which is consistent with the fact that thalidomide does not induce severe effects on embryos during the implantation period [26]. The effects of thalidomide are more prominent during mesoderm differentiation than in the undifferentiated state. In addition, ectoderm genes were upregulated, but mesendoderm genes were downregulated in mesodermal differentiation, in response to thalidomide (Fig. 3). Thus, our results do not contradict the previous reports on thalidomide-induced malformations.
We found that FABP7 was markedly downregulated in undifferentiated conditions and was upregulated during mesoderm differentiation (Fig. 3). FABPs are a family of proteins associated with intracellular fatty acid transport that regulate lipid metabolism, signal transduction, and gene expression, and FABP7 is particularly abundant in neurons [21,22]. Although there are no reports of a direct relationship between thalidomide and FABP7, there are many reports connecting them. FABPs are reported to be related to multiple myeloma, which is the main target of thalidomide as a medication [27]. FABPs crosstalk with peroxisome proliferator activated receptor (PPAR) and retinoic acid X receptor (RXR) to regulate retinoic acid (RA) signaling [28,29]. RA signaling regulates the development of many organs and tissues, including the body axis, spinal cord, heart, and limbs [30]. RA specifies the proximal region of the limb bud during development [31,32]. Moreover, RA is also known to directly activate SALL4 expression [33], which is a thalidomide-dependent neosubstrate in the CRBN pathway that may be involved in thalidomide embryopathy [[34], [35], [36]]. CRBN, which forms an E3 ubiquitin ligase complex, is the primary target of thalidomide [7]. Thalidomide has also been reported to inhibit hiPSC mesendoderm differentiation by modulating the CRBN-dependent degradation of SALL4 [17]. Thus, it is possible that FABP7 is related to thalidomide-induced limb malformations via RA and SALL4.
We also found that thalidomide alters the expression of many genes that might be related to thalidomide-induced malformations. The expression of TGF-β related genes (BMP2, Lefty1, and Lefty2) was altered, although BMP2, Lefty1, and Lefty2 were upregulated in undifferentiated conditions and Lefty2 was downregulated in the mesoderm. This difference may be due to the difference in TGF-β action on different cell types. For example, although a low dose (<10 ng/mL) of activin (a member of the TGF-β super family, whose action is related to the other TGF-β, such as BMP and Lefty) maintains the undifferentiated state of hiPSCs, a high dose of activin (>50 ng/mL) induces endodermal differentiation [20]. Thalidomide has been reported to induce the formation of limb deformities by perturbing the Bmp/Dkk1/Wnt signaling pathway in studies using chick and human embryonic fibroblasts [37]. Furthermore, thalidomide treatment altered the expression of BMP2, FGF-4, and EMX-2, which are related to limb formation [[38], [39], [40]]. In addition, thalidomide treatment during mesoderm differentiation altered the expression of HAND1, which is indispensable for heart formation, and GJB2 mutations, which cause hearing loss [41,42]. Thalidomide is also known to affect the heart and ear. Moreover, we found that the expression of BMP2, CD34, EMX2, MYC, and RUNX2, which are related to multiple myeloma, a disease medicinally targeted by thalidomide [[43], [44], [45], [46]]—was altered. These results suggest that these genes might be related to thalidomide-induced malformations and its mechanism of action as a medication.
In this study, we used hiPSCs, which are model cells that do not exist in vivo. Thus, the gene expression patterns that we observed were not the same as those observed during human embryogenesis. However, it has been reported that undifferentiated pluripotent stem cells can be used to predict the outcomes during the later stages of development [14]. Thus, although we used hiPSCs, it remains possible that our results are also relevant to thalidomide-induced malformations.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.100978.
Author contributions
Maho Shimizu: Validation, Formal analysis, Writing - Review & Editing, Visualization.
Saoko Tachikawa: Formal analysis, Investigation, Writing - Original Draft.
Nagatsuki Saitoha: Validation.
Kohei Nakazonoa: Validation.
Liu Yu-Jung: Investigation.
Mika Suga: Investigation.
Kiyoshi Ohnuma: Formal analysis, Writing - Review & Editing, Supervision, Funding acquisition.
Funding
This work was supported in part by AMED under Grant Number JP17bk0104011h0005 (to K.O.), and the funding bodies had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Franks M.E., Macpherson G.R., Figg W.D. Thalidomide. Lancet. 2004;363:1802–1811. doi: 10.1016/S0140-6736(04)16308-3. [DOI] [PubMed] [Google Scholar]
- 2.Knobloch J., Ruther U. Shedding light on an old mystery: thalidomide suppresses survival pathways to induce limb defects. Cell Cycle. 2008;7:1121–1127. doi: 10.4161/cc.7.9.5793. [DOI] [PubMed] [Google Scholar]
- 3.Melchert M., List A. The thalidomide saga. Int. J. Biochem. Cell Biol. 2007;39:1489–1499. doi: 10.1016/j.biocel.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Miller M.T., Stromland K. Teratogen update: thalidomide: a review, with a focus on ocular findings and new potential uses. Teratology. 1999;60:306–321. doi: 10.1002/(SICI)1096-9926(199911)60:5<306::AID-TERA11>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 5.Sheskin J. Thalidomide in the treatment of lepra reactions. Clin. Pharmacol. Ther. 1965;6:303–306. doi: 10.1002/cpt196563303. [DOI] [PubMed] [Google Scholar]
- 6.Singhal S., Mehta J., Desikan R., Ayers D., Roberson P., Eddlemon P., Munshi N., Anaissie E., Wilson C., Dhodapkar M., Zeddis J., Barlogie B. Antitumor activity of thalidomide in refractory multiple myeloma. N. Engl. J. Med. 1999;341:1565–1571. doi: 10.1056/NEJM199911183412102. [DOI] [PubMed] [Google Scholar]
- 7.Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y., Yamaguchi Y., Handa H. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 8.Vargesson N. The teratogenic effects of thalidomide on limbs. J. Hand Surg. Eur. 2019;44:88–95. doi: 10.1177/1753193418805249. [DOI] [PubMed] [Google Scholar]
- 9.Debock C.A., Peters A. Effect of thalidomide on the development of the chick embryo. Nature. 1963;199:1204–1206. doi: 10.1038/1991204a0. [DOI] [PubMed] [Google Scholar]
- 10.Evseenko D., Zhu Y., Schenke-Layland K., Kuo J., Latour B., Ge S., Scholes J., Dravid G., Li X., MacLellan W.R., Crooks G.M. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13742–13747. doi: 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 12.Xing J., Toh Y.C., Xu S., Yu H. A method for human teratogen detection by geometrically confined cell differentiation and migration. Sci. Rep. 2015;5:10038. doi: 10.1038/srep10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aikawa N., Kunisato A., Nagao K., Kusaka H., Takaba K., Ohgami K. Detection of thalidomide embryotoxicity by in vitro embryotoxicity testing based on human iPS cells. J. Pharmacol. Sci. 2014;124:201–207. doi: 10.1254/jphs.13162fp. [DOI] [PubMed] [Google Scholar]
- 14.Yamane J., Aburatani S., Imanishi S., Akanuma H., Nagano R., Kato T., Sone H., Ohsako S., Fujibuchi W. Prediction of developmental chemical toxicity based on gene networks of human embryonic stem cells. Nucleic Acids Res. 2016;44:5515–5528. doi: 10.1093/nar/gkw450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tachikawa S., Nishimura T., Nakauchi H., Ohnuma K. Vitro Cellular & Developmental Biology-Animal. vol. 53. 2017. Thalidomide induces apoptosis in undifferentiated human induced pluripotent stem cells; pp. 841–851. [DOI] [PubMed] [Google Scholar]
- 16.Tachikawa S., Shimizu M., Maruyama K., Ohnuma K. Thalidomide induces apoptosis during early mesodermal differentiation of human induced pluripotent stem cells. Vitro Anim. Cell Dev. Biol. 2018;54:231–240. doi: 10.1007/s11626-018-0234-x. [DOI] [PubMed] [Google Scholar]
- 17.Belair D.G., Lu G., Waller L.E., Gustin J.A., Collins N.D., Kolaja K.L. Thalidomide inhibits human iPSc mesendoderm differentiation by modulating CRBn-dependent degradation of SALL4. Sci. Rep. 2020;10:1–18. doi: 10.1038/s41598-020-59542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Suga M., Tachikawa S., Tateyama D., Ohnuma K., Furue M.K. Imaging-cytometry revealed spatial heterogeneities of marker expression in undifferentiated human pluripotent stem cells. In Vitro Cell Dev. Biol. Anim. 2017;53:83–91. doi: 10.1007/s11626-016-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ninomiya H., Mizuno K., Terada R., Miura T., Ohnuma K., Takahashi S., Asashima M., Michiue T. Improved efficiency of definitive endoderm induction from human induced pluripotent stem cells in feeder and serum-free culture system. In Vitro Cell. Dev. Biol. Anim. 2015;51:1–8. doi: 10.1007/s11626-014-9801-y. [DOI] [PubMed] [Google Scholar]
- 21.Storch J., Thumser A.E. Tissue-specific functions in the fatty acid-binding protein family. J. Biol. Chem. 2010;285:32679–32683. doi: 10.1074/jbc.R110.135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharifi K., Morihiro Y., Maekawa M., Yasumoto Y., Hoshi H., Adachi Y., Sawada T., Tokuda N., Kondo H., Yoshikawa T. FABP7 expression in normal and stab-injured brain cortex and its role in astrocyte proliferation. Histochem. Cell Biol. 2011;136:501. doi: 10.1007/s00418-011-0865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X., Wang L., Liu B., Xie W., Chen Y.-G. Activin/Smad2 and Wnt/β-catenin up-regulate HAS2 and ALDH3A2 to facilitate mesendoderm differentiation of human embryonic stem cells. J. Biol. Chem. 2018;293:18444–18453. doi: 10.1074/jbc.RA118.003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goulburn A.L., Stanley E.G., Elefanty A.G., Anderson S.A. Generating GABAergic cerebral cortical interneurons from mouse and human embryonic stem cells. Stem Cell Res. 2012;8:416–426. doi: 10.1016/j.scr.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Carlson B.M. Elsevier Health Sciences; 2018. Human Embryology and Developmental Biology. [Google Scholar]
- 26.Nowack E. Die sensible phase bei der thalidomid-embryopathie. Humangenetik. 1965:516–536. doi: 10.1007/BF00338341. 1965. [DOI] [PubMed] [Google Scholar]
- 27.Masarwi M., DeSchiffart A., Ham J., Reagan M.R. Multiple myeloma and fatty acid metabolism. JBMR Plus. 2019;3 doi: 10.1002/jbm4.10173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder F., Petrescu A.D., Huang H., Atshaves B.P., McIntosh A.L., Martin G.G., Hostetler H.A., Vespa A., Landrock D., Landrock K.K. Role of fatty acid binding proteins and long chain fatty acids in modulating nuclear receptors and gene transcription. Lipids. 2008;43:1–17. doi: 10.1007/s11745-007-3111-z. [DOI] [PubMed] [Google Scholar]
- 29.Liu R.Z., Graham K., Glubrecht D.D., Lai R., Mackey J.R., Godbout R. A fatty acid-binding protein 7/RXRβ pathway enhances survival and proliferation in triple-negative breast cancer. J. Pathol. 2012;228:310–321. doi: 10.1002/path.4001. [DOI] [PubMed] [Google Scholar]
- 30.Ghyselinck N.B., Duester G. Retinoic acid signaling pathways. Development. 2019;146:dev167502. doi: 10.1242/dev.167502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercader N., Leonardo E., Piedra M.E., Martinez-A C., Ros M., Torres M. Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development. 2000;127:3961–3970. doi: 10.1242/dev.127.18.3961. [DOI] [PubMed] [Google Scholar]
- 32.Zeller R., López-Ríos J., Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat. Rev. Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- 33.Gely-Pernot A., Raverdeau M., Teletin M., Vernet N., Féret B., Klopfenstein M., Dennefeld C., Davidson I., Benoit G., Mark M. Retinoic acid receptors control spermatogonia cell-fate and induce expression of the SALL4A transcription factor. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matyskiela M.E., Couto S., Zheng X., Lu G., Hui J., Stamp K., Drew C., Ren Y., Wang M., Carpenter A. SALL4 mediates teratogenicity as a thalidomide-dependent cereblon substrate. Nat. Chem. Biol. 2018;14:981–987. doi: 10.1038/s41589-018-0129-x. [DOI] [PubMed] [Google Scholar]
- 35.Donovan K.A., An J., Nowak R.P., Yuan J.C., Fink E.C., Berry B.C., Ebert B.L., Fischer E.S. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. Elife. 2018;7 doi: 10.7554/eLife.38430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asatsuma-Okumura T., Ando H., De Simone M., Yamamoto J., Sato T., Shimizu N., Asakawa K., Yamaguchi Y., Ito T., Guerrini L. p63 is a cereblon substrate involved in thalidomide teratogenicity. Nat. Chem. Biol. 2019;15:1077–1084. doi: 10.1038/s41589-019-0366-7. [DOI] [PubMed] [Google Scholar]
- 37.Knobloch J.r., Shaughnessy J.D., Jr., Rüther U. Thalidomide induces limb deformities by perturbing the Bmp/Dkk1/Wnt signaling pathway. Faseb. J. 2007;21:1410–1421. doi: 10.1096/fj.06-7603com. [DOI] [PubMed] [Google Scholar]
- 38.Niswander L., Martin G.R. FGF-4 and BMP-2 have opposite effects on limb growth. Nature. 1993;361:68–71. doi: 10.1038/361068a0. [DOI] [PubMed] [Google Scholar]
- 39.Beauchemin M., Del Rio-Tsonis K., Tsonis P.A., Tremblay M., Savard P. Graded expression of Emx-2 in the adult newt limb and its corresponding regeneration blastema. J. Mol. Biol. 1998;279:501–511. doi: 10.1006/jmbi.1998.1782. [DOI] [PubMed] [Google Scholar]
- 40.Capellini T.D., Vaccari G., Ferretti E., Fantini S., He M., Pellegrini M., Quintana L., Di Giacomo G., Sharpe J., Selleri L. Scapula development is governed by genetic interactions of Pbx1 with its family members and with Emx2 via their cooperative control of Alx1. Development. 2010;137:2559–2569. doi: 10.1242/dev.048819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reamon-Buettner S.M., Ciribilli Y., Inga A., Borlak J. A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Hum. Mol. Genet. 2008;17:1397–1405. doi: 10.1093/hmg/ddn027. [DOI] [PubMed] [Google Scholar]
- 42.Cohen-Salmon M., Ott T., Michel V., Hardelin J.-P., Perfettini I., Eybalin M., Wu T., Marcus D.C., Wangemann P., Willecke K. Targeted ablation of connexin26 in the inner ear epithelial gap junction network causes hearing impairment and cell death. Curr. Biol. 2002;12:1106–1111. doi: 10.1016/s0960-9822(02)00904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maes K., Nemeth E., Roodman G.D., Huston A., Esteve F., Freytes C., Callander N., Katodritou E., Tussing-Humphreys L., Rivera S. In anemia of multiple myeloma, hepcidin is induced by increased bone morphogenetic protein 2, Blood. J. Am. Soc. Hematol. 2010;116:3635–3644. doi: 10.1182/blood-2010-03-274571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei X.F., Chen Q.L., Fu Y., Zhang Q.K. Wnt and BMP signaling pathways co‐operatively induce the differentiation of multiple myeloma mesenchymal stem cells into osteoblasts by upregulating EMX2. J. Cell. Biochem. 2019;120:6515–6527. doi: 10.1002/jcb.27942. [DOI] [PubMed] [Google Scholar]
- 45.Anguiano A., Tuchman S.A., Acharya C., Salter K., Gasparetto C., Zhan F., Dhodapkar M., Nevins J., Barlogie B., Shaughnessy J.D., Jr. Gene expression profiles of tumor biology provide a novel approach to prognosis and may guide the selection of therapeutic targets in multiple myeloma. J. Clin. Oncol. 2009;27:4197. doi: 10.1200/JCO.2008.19.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu X., Zhang C., Trotter T.N., Gowda P.S., Lu Y., Ponnazhagan S., Javed A., Li J., Yang Y. Runx2 deficiency in osteoblasts promotes myeloma progression by altering the bone microenvironment at new bone sites. Canc. Res. 2020;80:1036–1048. doi: 10.1158/0008-5472.CAN-19-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snoeckx R.L., Huygen P.L., Feldmann D., Marlin S., Denoyelle F., Waligora J., Mueller-Malesinska M., Pollak A., Ploski R., Murgia A. GJB2 mutations and degree of hearing loss: a multicenter study. Am. J. Hum. Genet. 2005;77:945–957. doi: 10.1086/497996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takemori S., Tanaka Y., Suzuki J.-I. Thalidomide anomalies of the ear. Arch. Otolaryngol. 1976;102:425–427. doi: 10.1001/archotol.1976.00780120073010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.