Abstract
West Nile virus (WNV) is an important neurotropic flavivirus that is widely distributed globally. WNV strain XJ11129 was first isolated in Xinjiang, China, and its genetic and biological characteristics remain largely unknown. In this study, phylogenetic and sequence analyses revealed that XJ11129 belongs to lineage 1a and shares high genetic identity with the highly pathogenic strain NY99. Then, the full-length genomic cDNA of XJ11129 was amplified and assembled using a modified Gibson assembly (GA) method. The virus (named rXJ11129) was successfully rescued in days following this method. Compared with other wild-type WNV isolates, rXJ11129 exhibited virulence indistinguishable from that of the NY99 strain in vivo. In summary, the genomic and virulence phenotypes of rXJ11129 were characterized in vivo and in vitro, and these data will improve the understanding of the spread and pathogenesis of this reemerging virus.
Electronic supplementary material
The online version of this article (10.1007/s12250-020-00246-x) contains supplementary material, which is available to authorized users.
Keywords: WNV isolates from China, Phylogenetic analysis, GA method, Biological characteristic
Introduction
West Nile virus (WNV) is an important mosquito-borne flavivirus in the family Flaviviridae. It is mainly maintained in an enzootic transmission cycle between birds and mosquitoes and can infect humans and other vertebrates as final hosts (Komar 2003; van der Meulen et al. 2005). After infection with WNV, most humans are asymptomatic, although approximately 20% of infected individuals may develop symptoms of West Nile fever (WNF) (Jeha et al. 2003). WNF presents as a minor influenza-like illness accompanied by clinical signs and symptoms such as fever, headache, skin rash and vomiting, and it occasionally develops into meningoencephalitis episodes or flaccid paralysis; less frequently, death occurs (McDonald et al. 2019; Yu et al. 2020). To date, no licensed vaccines or therapeutics are available for human use (Donadieu et al. 2013; Lim and Shi 2013).
The WNV virion contains a positive-sense, single-stranded RNA genome containing approximately 11,000 nucleotides that comprises a single open reading frame flanked by two untranslated regions (UTRs) (Coia et al. 1988). The viral genomic RNA codes for a single polypeptide that is cleaved into three structural proteins (C, prM/M and E) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) by cellular and viral proteases (Wengler et al. 1985). The three structural proteins compose the WNV virion, and the seven NS proteins are multifunctional, playing critical roles in viral RNA synthesis and/or assembly (Donadieu et al. 2013).
WNV has evolved into up to 9 lineages (Chancey et al. 2015), among which lineage 1 has the largest geographic range and is generally the most pathogenic, causing severe central nervous system infection and death (Zehender et al. 2011). Strains of WNV in lineage 1 have been divided further into clades 1a, 1b and 1c (also named lineage 5). The most representative strain in clade 1a is NY99, which is highly pathogenic to humans. Since it was introduced into North America in 1999 (Lanciotti SR 1999; Nash et al. 2001), NY99 has infected 4 million humans in the US, causing more than 780,000 WNF cases, 16,000 cases of neurologic disease and over 1500 associated fatalities (Petersen et al. 2013).
The WNV epidemic is also a threat in China. Seropositivity for WNV has been reported in birds and humans (Li et al. 2013). Confirmed human cases of WNV in China were reported in 2013 in Xinjiang Province in northwestern China (Lu et al. 2014; Cao et al. 2017, 2019). Isolation of WNV has been attempted, and WNV has successfully been obtained from mosquitoes in Xinjiang, China (XJ11129, GenBank No. JX442279).
The biological characteristics and pathogenicity of XJ11129 are incompletely understood and need further elucidation. Herein, a phylogenetic tree was constructed and sequence alignment was conducted to analyze the relationship of XJ11129 with other strains of WNV. In addition, a modified Gibson assembly (GA) method that completely eliminates the need for both plasmid DNA production in bacteria and in vitro RNA transcription was employed to rescue XJ11129. Via this method, the virus was successfully rescued in days, and its biological features and pathogenicity were further examined both in vitro and in vivo.
Materials and Methods
Sequence Evolution Analysis
A total of 28 WNV strains, including XJ11129, were used for phylogenetic analysis in this study. The nucleic acid sequences of polyproteins were downloaded from GenBank, along with information such as the location and time of viral isolation (Supplementary Table S1). All sequences were aligned with MEGA5 software using ClustalW. A phylogenetic tree was then constructed using the neighbor joining (NJ) method with 1000 bootstrap replicates. The trees were rooted by using Japanese encephalitis virus (JEV) as the outgroup virus.
Eight strains were selected as representative strains of 8 lineages: AF196835 (lineage 1a), KT934797 (lineage 1b), GQ851605 (lineage 1c or 5), KP780840 (lineage 2), AY765264 (lineage 3), AY277251 (lineage 4), KY703855 (lineage 7), and KY703856 (lineage 8). Lineage 6 was excluded from our study because only a partial sequence was available. The complete polyprotein sequences and nucleic acid sequences of these strains were used for comparison of percent sequence identities with MEGA5 software using the ClustalW alignment program.
Cell Lines and Viruses
Vero (CCL-81), BHK-21 (CCL-10), C6/36 (CRL-1660) and DF-1 (CRL-141) were purchased from the American Type Culture Collection (ATCC) and maintained in our laboratory. BHK-21 and Vero cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS) (Lonza), 1 mmol/L sodium pyruvate, 100 U/mL penicillin and 50 µg/mL streptomycin and were maintained in an atmosphere containing 5% CO2 at 37 °C. C6/36 cells were grown in RPMI 1640 medium supplemented with 8% FBS (Lonza), 1 mmol/L sodium pyruvate, 100 U/mL penicillin and 50 µg/mL streptomycin and were maintained in an atmosphere containing 5% CO2 at 30 °C.
WNV strain NY99 was rescued from the two-plasmid infectious clone (Kinney et al. 2006). WNV strain XJ11129 was isolated from mosquito in Xinjiang, China, and stocked in our lab (Lu et al. 2014).The viruses were propagated in BHK-21 cells and titrated by the standard plaque assay as described below.
GA
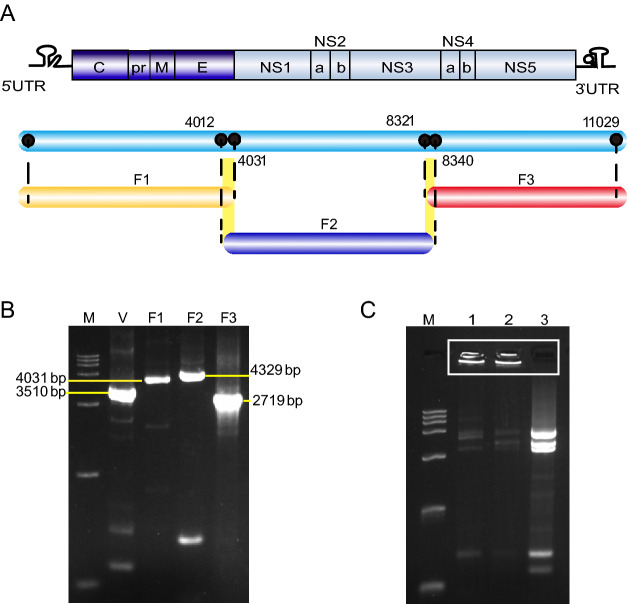
Viral cDNA was reverse transcribed using the SuperScript III reverse system (Promega) and served as the template for DNA fragment amplification. In our study, the whole genome of XJ11129 was divided into three fragments, as shown in Fig. 2. The first fragment contained nucleotides 1–4031; the second fragment, nucleotides 4012 to 8340; and the last fragment, nucleotides 8321–11,029. The DNA-lunched infectious clone of enterovirus 71 (EV71) was constructed in our lab using genetic engineering technique as previously described and used as the template for vector fragment amplification (Tan et al. 2016). This plasmid contained the CMV promoter placed immediately upstream of the first nucleotide of the EV71 sequence and the hepatitis delta virus ribozyme sequence located directly downstream of the last nucleotide of the EV71 sequence followed by the SV40 poly (A) signal. All the DNA fragments contained a 20-bp overlap at both the upstream and downstream ends. The sequences of all primers used for DNA fragment amplification are shown in Supplementary Table S2.
Fig. 2.
Strategy for rapid rescue of XJ11129 by GA. A Full-length preparation strategy of the XJ11129 genome by RT-PCR with overlapping primers. Full length of XJ11129 genome was divided into three fragments, named F1, F2 and F3. B PCR-amplified fragments. M: DNA Ladder maker DL10000 (TAKARA). V: vector fragment, which used pCMV/EV71 plasmid as the template and contained the CMV promoter, the hepatitis delta virus ribozyme sequence followed by the SV40 polyA signal. Length of vector fragment is 3510 bp. F1, F2, F3 are the WNV genome fragments. Length of the three fragments is 4031 bp, 4329 bp and 2719 bp, respectively. C GA reaction of the four fragments. M: DNA Ladder maker DL10000 (TAKARA). Lane 1, 2: GA reaction of the four fragments; Lane 3: Mixture of the four fragments without GA reaction.
The PCR products were column purified (Promega) and assembled for Gibson reactions (CWBIO) following the standard protocol. In brief, 2.5 µL of the four DNA fragments were mixed with 10 µL of 2 × GA master mix and incubated at 50 °C for 1 h. Approximately 20 µL of the reaction product was stored at −20 °C for further use.
Transfection and Virus Rescue
BHK-21 cells were seeded in 6-well culture plates. The GA products were directly transfected into the cells using Lipofectamine 3000 (Invitrogen) as indicated by the manufacturer. Three days post transfection, the recovered viruses were passaged in new BHK-21 cells. Once cytopathic effects (CPE) were observed, cell supernatants were harvested. Viral RNA was isolated using a PureLink RNA Mini Kit (Thermo Fisher Scientific), and cDNA was generated from 1 mg of RNA using SuperScript III (Thermo Fisher Scientific) according to the manufacturer’s instructions with random primers 9-mer (TAKARA). For determination of viral consensus sequences, PCR products were directly sequenced in both directions using virus-specific primers (Supplementary Table S3). Sequence fragments were assembled into a consensus sequence with DNA STAR software (version 7.0).
Rescued virus was propagated in BHK-21 cells, and the viral titer was determined by the standard plaque assay as described below.
Indirect Immunofluorescence Assay (IFA)
The IFA was performed on BHK-21 cells as previously described (Wang et al. 2014). In brief, BHK-21 cells were infected with each virus for 48 h and were then fixed with ice-cold acetone and incubated with the primary anti-flavivirus monoclonal antibody (mAb) 4G2. After three washes with PBS, cells were incubated with Alexa Fluor 488-conjugated secondary antibodies (ZSGB-BIO). Then, cells were incubated with DAPI to stain nuclei. Cells with positive staining were examined using a fluorescence microscope (Olympus).
Plaque Assay
The plaque assay on BHK-21 cells was conducted as follows. BHK-21 cells were seeded in 6-well or 12-well culture plates to form a monolayer and were infected with serial 10-fold dilutions of virus-containing supernatants for 1 h at 37 °C. After infection, the inoculum was removed and replaced with DMEM containing 1% low melting point agarose. After 3 days, cells were fixed by the addition of 5% formaldehyde, washed with water and stained with 1% crystal violet in 10% ethanol. After cells were rinsed with water, the plaques were counted, and the viral titers were calculated as plaque-forming units/mL (PFU/mL).
Growth Curve Analysis
Growth curve analysis was conducted on BHK-21, Vero, C6/36 and DF-1 cells. Cells were seeded in 24-well plates and infected with rXJ11129 at a multiplicity of infection (MOI) of 0.01. Culture supernatants were collected at successive 24 h intervals, and viral titers were then quantitated via a plaque assay with BHK-21 cells.
Ethics Statement
All animal experiments conducted in this study were approved and carried out in strict accordance with the guidelines of the Institutional Experimental Animal Welfare and Ethics Committee (IACUC-13-2016-001).
Virulence Assay in Mice
The BALB/c mice used in this study were purchased from Beijing Vital River Laboratory Animal Technology Co. and maintained under specific pathogen-free (SPF) conditions.
For the neuroinvasiveness assay, groups of 6-week-old BALB/c mice were inoculated intraperitoneally (i.p.) with the indicated doses of rXJ11129 or NY99. For the neurovirulence assay, groups of 6-week-old BALB/c mice were administered 10 PFU or 1 PFU of rXJ11129 or NY99 by the intracerebral (i.c.) route. Mice were then monitored for clinical symptoms and mortality for 21 days.
Statistical Analysis
Kaplan–Meier survival curves were analyzed with the log-rank test. Statistical analyses were performed with standard GraphPad Prism software (version 5.0).
Results
Phylogenetic Analysis of XJ11129
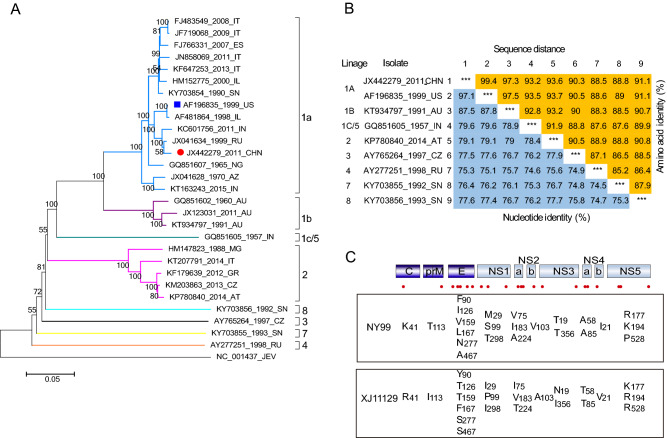
In order to understand the phylogenetic status of XJ11129, the complete sequence information of 28 WNV strains isolated at different times and locations was analyzed to construct a phylogenetic tree. These WNV strains are classified into eight major lineages. XJ11129 belongs to lineage 1 and is further clustered into clade 1a, which clusters most closely with viruses isolated from Russia and India, followed by strains isolated from the US (NY99) and Israel (Fig. 1A).
Fig. 1.
The phylogeny and genetic diversity of the West Nile virus lineages. A Phylogenetic tree of XJ11129 strains compared with reference strains. Each strain is listed by GenBank accession number, geographic origin, and collection date. Scale bar indicates substitutions per site. Abbreviations of geographic origin are list in Supplementary Table S1. XJ11129 was highlighted with a red solid circle and NY99 was highlighted with a blue solid box. B Pairwise percent identity between nucleotide (lower left) and amino acid (upper right) sequences of the polyprotein. Sequences are labeled in the following format: accession number, 2-letter country code, and year of isolation. C Amino acid differences between XJ11129 and NY99. A total of 23 amino acids were different between these two strains. The numeral subscript after amino acid is the location of each gene.
Further, the genetic identity of XJ11129 was compared at the gene and protein levels with that of strains representing 8 lineages. Since both XJ11129 and NY99 belong to lineage 1 and clade 1a, XJ11129 exhibited the highest nucleotide identity (97.1%) and amino acid identity (99.4%) with NY99. In addition, XJ11129 shared 75.3% –87.5% nucleotide similarity and 88.5% –97.3% amino acid similarity with the strains from the other lineages (Fig. 1B).
Next, the amino acid differences between XJ11129 and NY99 were compared. A total of 23 amino acids differed between the two strains (Fig. 1C). Six amino acid differences were located in the E protein: Y90F, T126I, T159V, F167L, S277N, and S467A. NS1, NS2A, and NS5 each contained three amino acid differences. NS3 and NS4A each contained two amino acid differences. C, M, NS2B and NS4B each had one amino acid difference.
The above results show that XJ11129 is classified in clade 1a of lineage 1 and showed high sequence identity with the pathogenic strain NY99. However, whether XJ11129 exhibits a high virulence level similar to that of NY99 needs further experimental verification.
Rapid Rescue of XJ11129 by a Modified GA Method
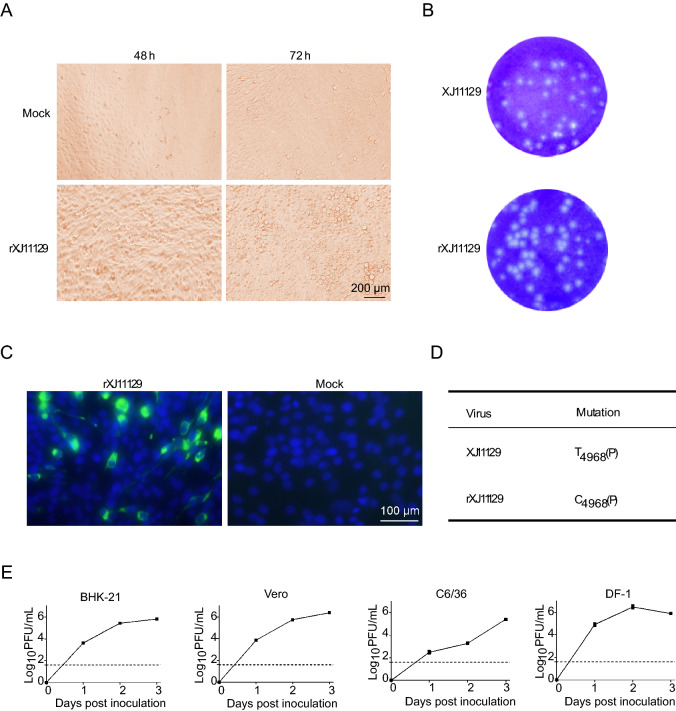
Infectious clones are powerful tools for flavivirus study. Here, a modified GA method was employed to rapidly rescue XJ11129. The viral genome was divided into three fragments, as shown in Fig. 2A. The vector fragment contained the CMV promoter, the hepatitis delta virus ribozyme sequence and the SV40 poly (A) signal and was used for the GA reaction, which ensured the transcription of XJ11129 in eukaryotic cells. All four fragments were obtained by PCR amplification and used for the GA reaction. Although no expected DNA bands were seen, the amounts of the four DNA fragments were significantly reduced, and large quantities of the DNA samples remained in the wells of the gel, confirming the occurrence of the GA reaction (Fig. 2B, 2C). To rescue the virus, the GA products were transfected directly into BHK-21 cells instead of into bacteria. Seventy-two hours post transfection, obvious CPEs were observed in the cells of the GA group, while no CPEs were observed in the cells of the control group (Fig. 3A). The plaque morphology of rXJ11129 was examined in BHK-2 cells. rXJ11129 formed plaques with an average size similar to that of the parental virus (Fig. 3B). The IFA results showed that the rescued virus could effectively infect BHK-21 cells and successfully express virus-specific proteins (Fig. 3C). A sequencing assay was conducted and showed that only one silent mutation emerged at position 4968 in NS3 (T to C) (Fig. 3D). These results indicate that GA is a rescue strategy for rapid acquisition of viruses and that the rXJ11129 clone rescued by the GA method has high genetic similarity to the parental viruses.
Fig. 3.
Rescue and identification of XJ11129 in vitro. A Visible cytopathic effect (CPE) of BHK-21 cells in 2–3 days after infection with recued virus. B Plaque morphology of rescued XJ11129 and the parental XJ11129. Plaques were observed 3 days post-infection after staining with 0.2% crystal violet. C The GA products were inoculated to BHK-21 cells for incubation of 96 h. The infected cells were observed with a fluorescence microscope. D Differences between the sequences of rXJ11129 with its parental virus XJ11129. The numeral subscript after nucleotide is the location of whole viral genome. E Growth kinetics of rXJ11129. The experiments were performed with BHK-21 cells, Vero cells, C6/36 cells and DF-1 cells. Viral titres were determined on BHK-21 cells by plaque assay at the indicated times. The dotted lines represent the limit of sensitivity of the plaque assay. The error bars indicate the range in values of two independent experiments.
Replication of rXJ11129 in Cultured Cells
WNV has a wide range of hosts, including mosquitoes, birds, mammals and so on. To verify whether XJ11129 can infect these hosts, the growth kinetics of rXJ11129 were assessed in four cell lines: mouse cells (BHK-21), avian cells (DF-1), primate cells (Vero) and mosquito cells (C6/36). rXJ11129 exhibited efficient replication ability in all four cell lines. In BHK-21, Vero and C6/36 cells, the rXJ11129 titer peaked at 72 h, at up to 105.81, 106.39, and 105.42 PFU/mL, respectively. However, in DF-1 cells, rXJ11129 replicated to a peak titer of 106.49 PFU/mL at 48 h post infection (Fig. 3E). Taken together, these results clearly show that rXJ11129 can replicate in the above four types of cells and demonstrates the highest replication efficiency in avian cells (DF-1).
The Virulence of rXJ11129 in BALB/c Mice Was Similar to That of NY99
XJ11129 and NY99 belong to clade la of lineage 1. To characterize the virulence of XJ11129, a virulence assay was conducted in a mouse model, and NY99 was used as the control strain.
To determine the neuroinvasiveness of rXJ11129, groups of 6-week-old BALB/c mice were inoculated intraperitoneally with the corresponding dose of rXJ11129 or NY99. As shown in Table 1, 105 PFU of rXJ11129 resulted in 100% mortality in mice, with an average survival time (AST) of 10.6 days. However, inoculation of 105 PFU of NY99 resulted in 80% mortality, with an AST of 9.75 days. Neither morbidity nor mortality was observed in mice following inoculation with 103 PFU of rXJ11129 or NY99. The LD50 of rXJ11129 was slightly higher than that of NY99 (6310 PFU vs 5623 PFU), but the difference between the two viruses was not statistically significant.
Table 1.
Comparison of rXJ11129 with NY99 in nerve-invasiveness.
| Virus | Dose (log10 PFU) | Mortality (No. survival/No. text) | ASTa (Day) | LD50 (PFU) |
|---|---|---|---|---|
| rXJ11129 | 5 | 0 (0/5) | 10.6 | 6310 |
| 4 | 20 (1/5) | 10.75 | ||
| 3 | 100 (5/5) | – | ||
| NY99 | 5 | 20 (1/5) | 9.75 | 5623 |
| 4 | 20 (1/5) | 11 | ||
| 3 | 100 (5/5) | – |
AST average survival time, – not available.
To characterize the neurovirulence of rXJ11129, groups of 6-week-old BALB/c mice were inoculated intracranially with 10 PFU or 1 PFU of each virus via the i.c route. All mice in every group showed symptoms such as a hunched posture, mental distress, and hind limb paralysis and died within 8 days (Fig. 4). No significant difference was found between the two WNV strains. The above results indicate that rXJ11129 exhibits both neuroinvasiveness and neurovirulence in mice, and its virulence in mice is equivalent to that of NY99.
Fig. 4.
Neurovirulence tests of rXJ11129. Mice received either rXJ11129 or NY99 at 1 PFU (A) or 10 PFU (B), and mortality was recorded for 21 days.
Discussion
Reports about WNV in birds and humans and the isolation of viruses from mosquitoes have confirmed the spread of WNV in China (Li et al. 2013; Lu et al. 2014; Cao et al. 2017, 2019). The serious public and human health threat posed by WNV urgently requires us to further understand the biological characteristics of WNV isolates from China and to determine whether these isolates from China exhibit high virulence. In this study, we determined that XJ11129 clusters into linage 1a, which includes numerous highly virulent strains, and has high genetic similarity with the virulent strain NY99. XJ11129 was quickly rescued via a modified GA method and was found to replicate efficiently in mammalian, mosquito and avian cells in vitro and to exhibit high virulence in vivo. Our study not only developed a new tool for WNV rescue but also provided solid experimental evidence that WNV isolates from China exhibit high virulence.
Among the 9 lineages of WNV, lineage 1 is distributed worldwide in distinct regions. Strains belonging to lineage 1a are attributed to major outbreaks in Europe, Africa, and the Americas associated with neurological disease. XJ11129 clustered into linage 1a and shared high nucleotide similarity (97.1%) and amino acid similarity (99.4%) with strain NY99, raising substantial concern about its pathogenicity, especially in humans. Comparison of XJ11129 with NY99 identified 23 amino acid differences distributed across the viral proteins (Fig. 1C). To date, investigations have been conducted on specific mutations that show close relationships with virulence and replication abilities and are distributed in the viral structural prM/M (Hanna et al. 2005) and E (Whiteman et al. 2010; Zhang et al. 2006; Beasley et al. 2005) genes as well as the NS genes NS1 (Whiteman et al. 2011, 2015), NS2A (Liu et al. 2006), NS3 (Shiryaev et al. 2009), NS4A, NS4B (Munoz-Jordan et al. 2005) and NS5 (Zhou et al. 2007).
Infectious cDNA clones of numerous WNV strains have been successfully developed for the study of viral replication and pathogenesis (Shi et al. 2002; Bahuon et al. 2012; Suthar et al. 2012; Pavitrakar et al. 2015). However, the instability of cloning might lead to random substitutions and to bacterial toxicity of viral proteins, potentially making the production of WNV cDNA molecules in bacterial vectors difficult and time consuming (Aubry et al. 2015). Various approaches have been employed to overcome this problem, including the use of very low copy number plasmids (Zhao et al. 2005), bacterial artificial chromosomes (Yun et al. 2003), mutating cryptic bacterial promoter sites in the flavivirus genome (Pu et al. 2011), and a yeast recombination system to assemble full-length clones (Kelly et al. 2010). In addition, numerous bacteria-free approaches have been developed to avoid the need for a bacterial host and, thus, the need to overcome the problems related to the use of bacteria; examples include circular polymerase extension reactions (Edmonds et al. 2013), infectious sub-genomic amplicons (Aubry et al. 2014) and GA (Siridechadilok et al. 2013).
The GA reaction has been widely used in the construction of clones, especially in viral infectious clones (Siridechadilok et al. 2013; Pan et al. 2018; Luo et al. 2019). The GA method decreases the construction time from months to days and results in seamless ligation, avoiding limitations of DNA assembly via restriction enzymes, which greatly simplifies genetic engineering of viral genomes (Gibson et al. 2009). In this study, to avoid using traditional, time-consuming methods for generating infectious clones of XJ11129, we modified the GA method to quickly rescue XJ11129. To ensure amplification efficiency and accuracy, the DNA fragments used for GA are limited to 5000 bp; thus, the genome of XJ11129 was divided into three fragments in this study (Fig. 2). In a study by Siridechadilok et al., the genome of dengue virus (DENV) was divided into two or even 11 fragments, and viruses were successfully rescued (Siridechadilok et al. 2013), showing that the number and length of DNA fragments have no critical effect on virus rescue.
The GA reaction not only is used to rescue viral clones from the parental virus but also facilitates genetic mapping and screening of mutations. For example, both primers for viral genome amplification that contain the target mutation and exogenous DNA fragments that contain the required overlap sequence, can be used in the GA reaction to obtain the target sequence. Finally, mutants and modified viruses were rescued by the transfection of GA products into sensitive cells. Here, XJ11129 was recovered successfully in days using the GA method (Fig. 3A–3C). Sequencing of the recovered virus identified only one silent mutation, at position 4589, suggesting the high accuracy of XJ11129 recovery using the GA method (Fig. 3D). The biological properties of the rescued virus were examined further. WNV has a wide range of hosts, including mosquitoes, birds, and mammals such as humans. Thus, to identify the potential host of the virus, we examined the proliferation characteristics of XJ11129 in cells derived from different hosts. rXJ11129 replicated efficiently in mammalian (BHK-21 and Vero), mosquito (C6/36) and avian (DF-1) cells in vitro, indicating that it can potentially infect these hosts (Fig. 3E). NY99 has high neurovirulence and invasiveness and was selected in this study as a benchmark for high virulence. We evaluated the neurovirulence (Fig. 4) and invasiveness (Table 1) of rXJ11129 and found that rXJ11129 exhibited high neurovirulence and neuroinvasiveness similar to those of NY99.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Key Research and Development Project of China (2016YFD0500304), the National Science and Technology Major Project of China (2018ZX09711003), and the National Natural Science Foundation of China (81621005 and 31770190). C.F.Q. was supported by the National Science Fund for Distinguished Young Scholars (81925025), the Innovative Research Group (81621005) from the NSFC, and the Innovation Fund for Medical Sciences (2019-I2M-5–049) from the Chinese Academy of Medical Sciences.
Author Contributions
CFQ and Huanyu Wang designed the experiments. YG, Hongjiang Wang, SX carried out the experiments with the help of CZ, SF, MC, FL. YG, Hongjiang Wang and HZ analyzed the data. YG and Hongjiang Wang wrote the paper. CFQ, Huanyu Wang, XL and YD checked and finalized the manuscript. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflicts of interest.
Animal and Human Rights Statement
The research protocol was approved by the Ethics Committee of Beijing Institute of Microbiology and Epidemiology of Academy of Military Medical Sciences (AMMS).
Footnotes
Yan Guo, Hongjiang Wang, Songtao Xu contributed equally to this work.
Contributor Information
Huanyu Wang, Email: wanghy@ivdc.chinacdc.cn.
Cheng-Feng Qin, Email: qincf@bmi.ac.cn.
References
- Aubry F, Nougairede A, de Fabritus L, Querat G, Gould EA, de Lamballerie X. Single-stranded positive-sense RNA viruses generated in days using infectious subgenomic amplicons. J Gen Virol. 2014;95:2462–2467. doi: 10.1099/vir.0.068023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry F, Nougairede A, Gould EA, de Lamballerie X. Flavivirus reverse genetic systems, construction techniques and applications: a historical perspective. Antivir Res. 2015;114:67–85. doi: 10.1016/j.antiviral.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahuon C, Despres P, Pardigon N, Panthier JJ, Cordonnier N, Lowenski S, Richardson J, Zientara S, Lecollinet S. IS-98-ST1 West Nile virus derived from an infectious cDNA clone retains neuroinvasiveness and neurovirulence properties of the original virus. PLoS One. 2012;7:e47666. doi: 10.1371/journal.pone.0047666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley DW, Whiteman MC, Zhang S, Huang CY, Schneider BS, Smith DR, Gromowski GD, Higgs S, Kinney RM, Barrett AD. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J Virol. 2005;79:8339–8347. doi: 10.1128/JVI.79.13.8339-8347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Fu S, Lv Z, Tang C, Cui S, Li X, Gao X, Li M, Cao Y, Lei W, He Y, Wang H, Liang G. West Nile virus infection in suspected febrile typhoid cases in Xinjiang. China. Emerg Microbes Infect. 2017;6:e41. doi: 10.1038/emi.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Fu S, Lu Z, Tang C, Gao X, Li X, Lei W, He Y, Li M, Cao Y, Wang H, Liang G. Detection of West Nile Virus infection in viral encephalitis cases, China. Vector Borne Zoonotic Dis. 2019;19:45–50. doi: 10.1089/vbz.2018.2275. [DOI] [PubMed] [Google Scholar]
- Chancey C, Grinev A, Volkova E, Rios M. The global ecology and epidemiology of West Nile virus. Biomed Res Int. 2015;2015:376230. doi: 10.1155/2015/376230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coia G, Parker MD, Speight G, Byrne ME, Westaway EG. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988;69(Pt 1):1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- Donadieu E, Bahuon C, Lowenski S, Zientara S, Coulpier M, Lecollinet S. Differential virulence and pathogenesis of West Nile viruses. Viruses. 2013;5:2856–2880. doi: 10.3390/v5112856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds J, van Grinsven E, Prow N, Bosco-Lauth A, Brault AC, Bowen RA, Hall RA, Khromykh AA. A novel bacterium-free method for generation of flavivirus infectious DNA by circular polymerase extension reaction allows accurate recapitulation of viral heterogeneity. J Virol. 2013;87:2367–2372. doi: 10.1128/JVI.03162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Hanna SL, Pierson TC, Sanchez MD, Ahmed AA, Murtadha MM, Doms RW. N-linked glycosylation of west nile virus envelope proteins influences particle assembly and infectivity. J Virol. 2005;79:13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeha LE, Sila CA, Lederman RJ, Prayson RA, Isada CM, Gordon SM. West Nile virus infection: A new acute paralytic illness. Neurology. 2003;61:55–59. doi: 10.1212/01.wnl.0000073617.08185.0a. [DOI] [PubMed] [Google Scholar]
- Kelly EP, Puri B, Sun W, Falgout B. Identification of mutations in a candidate dengue 4 vaccine strain 341750 PDK20 and construction of a full-length cDNA clone of the PDK20 vaccine candidate. Vaccine. 2010;28:3037. doi: 10.1016/j.vaccine.2009.10.084. [DOI] [PubMed] [Google Scholar]
- Kinney RM, Huang CY, Whiteman MC, Bowen RA, Langevin SA, Miller BR, Brault AC. Avian virulence and thermostable replication of the North American strain of West Nile virus. J Gen Virol. 2006;87:3611–3622. doi: 10.1099/vir.0.82299-0. [DOI] [PubMed] [Google Scholar]
- Komar N. West Nile Virus: epidemiology and ecology in North America. Adv Virus Res. 2003;61:185–234. doi: 10.1016/s0065-3527(03)61005-5. [DOI] [PubMed] [Google Scholar]
- Lanciotti SR. Origin of the West Nile Virus responsible for an outbreak of encephalitis in the Northeastern United States. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- Li XL, Fu SH, Liu WB, Wang HY, Lu Z, Tong SX, Li ZX, Nasci RS, Kosoy O, Cui Y, Liang GD. West nile virus infection in Xinjiang, China. Vector Borne Zoonotic Dis. 2013;13:131–133. doi: 10.1089/vbz.2012.0995. [DOI] [PubMed] [Google Scholar]
- Lim S, Shi P-Y. West Nile virus drug discovery. Viruses. 2013;5:2977–3006. doi: 10.3390/v5122977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Wang XJ, Clark DC, Lobigs M, Hall RA, Khromykh AA. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J Virol. 2006;80:2396–2404. doi: 10.1128/JVI.80.5.2396-2404.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Fu SH, Cao L, Tang CJ, Zhang S, Li ZX, Tusong M, Yao XH, Zhang HL, Wang PY, Wumaier M, Yuan XY, Li MH, Zhu CZ, Fu LP, Liang GD. Human infection with West Nile Virus, Xinjiang, China, 2011. Emerg Infect Dis. 2014;20:1421–1423. doi: 10.3201/eid2008.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Zhang P, Ma X, Wang Q, Lu J, Liu B, Zhao W, Allain JP, Li C, Li T. A rapid strategy for constructing novel simian adenovirus vectors with high viral titer and expressing highly antigenic proteins applicable for vaccine development. Virus Res. 2019;268:1–10. doi: 10.1016/j.virusres.2019.05.008. [DOI] [PubMed] [Google Scholar]
- McDonald E, Martin SW, Landry K, Gould CV, Lehman J, Fischer M, Lindsey NP. West Nile virus and other domestic nationally notifiable arboviral diseases—United States, 2018. Am J Transplant. 2019;19:2949–2954. doi: 10.15585/mmwr.mm6831a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash D, Mostashari F, Fine A, Miller J, O’Leary D, Murray K, Huang A, Rosenberg A, Greenberg A, Sherman M, Wong S, Layton M, West Nile Outbreak Response Working G The outbreak of West Nile virus infection in the New York City area in 1999. N Engl J Med. 2001;344:1807–1814. doi: 10.1056/NEJM200106143442401. [DOI] [PubMed] [Google Scholar]
- Pan H, Yan Y, Zhang J, Zhao S, Feng L, Ou J, Cao N, Li M, Zhao W, Wan C, Ismail AM, Rajaiya J, Chodosh J, Zhang Q. Rapid construction of a replication-competent infectious clone of human adenovirus type 14 by gibson assembly. Viruses. 2018;10:568. doi: 10.3390/v10100568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitrakar DV, Ayachit VM, Mundhra S, Bondre VP. Development and characterization of reverse genetics system for the Indian West Nile virus lineage 1 strain 68856. J Virol Methods. 2015;226:31–39. doi: 10.1016/j.jviromet.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Petersen LR, Carson PJ, Biggerstaff BJ, Custer B, Borchardt SM, Busch MP. Estimated cumulative incidence of West Nile virus infection in US adults, 1999-2010. Epidemiol Infect. 2013;141:591–595. doi: 10.1017/S0950268812001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu SY, Wu RH, Yang CC, Jao TM, Tsai MH, Wang JC, Lin HM, Chao YS, Yueh A. Successful propagation of flavivirus infectious cDNAs by a novel method to reduce the cryptic bacterial promoter activity of virus genomes. J Virol. 2011;85:2927–2941. doi: 10.1128/JVI.01986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, Tilgner M, Lo MK, Kent KA, Bernard KA. Infectious cDNA clone of the epidemic west nile virus from New York City. J Virol. 2002;76:5847–5856. doi: 10.1128/JVI.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev SA, Chernov AV, Aleshin AE, Shiryaeva TN, Strongin AY. NS4A regulates the ATPase activity of the NS3 helicase: a novel cofactor role of the non-structural protein NS4A from West Nile virus. J Gen Virol. 2009;90:2081–2085. doi: 10.1099/vir.0.012864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Gomutsukhavadee M, Sawaengpol T, Sangiambut S, Puttikhunt C, Chin-inmanu K, Suriyaphol P, Malasit P, Screaton G, Mongkolsapaya J. A simplified positive-sense-RNA virus construction approach that enhances analysis throughput. J Virol. 2013;87:12667–12674. doi: 10.1128/JVI.02261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar MS, Brassil MM, Blahnik G, Gale M., Jr Infectious clones of novel lineage 1 and lineage 2 West Nile virus strains WNV-TX02 and WNV-Madagascar. J Virol. 2012;86:7704–7709. doi: 10.1128/JVI.00401-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CW, Tee HK, Lee MH, Sam IC, Chan YF. Enterovirus A71 DNA-launched infectious clone as a robust reverse genetic tool. PLoS One. 2016;11:e0162771. doi: 10.1371/journal.pone.0162771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meulen KM, Pensaert MB, Nauwynck HJ. West Nile virus in the vertebrate world. Arch Virol. 2005;150:637–657. doi: 10.1007/s00705-004-0463-z. [DOI] [PubMed] [Google Scholar]
- Wang HJ, Li XF, Ye Q, Li SH, Deng YQ, Zhao H, Xu YP, Ma J, Qin ED, Qin CF. Recombinant chimeric Japanese encephalitis virus/tick-borne encephalitis virus is attenuated and protective in mice. Vaccine. 2014;32:949–956. doi: 10.1016/j.vaccine.2013.12.050. [DOI] [PubMed] [Google Scholar]
- Wengler G, Castle E, Leidner U, Nowak T, Wengler G. Sequence analysis of the membrane protein V3 of the flavivirus West Nile virus and of its gene. Virology. 1985;147:264–274. doi: 10.1016/0042-6822(85)90129-1. [DOI] [PubMed] [Google Scholar]
- Whiteman MC, Li L, Wicker JA, Kinney RM, Huang C, Beasley DWC, Chung KM, Diamond MS, Solomon T, Barrett ADT. Development and characterization of non-glycosylated E and NS1 mutant viruses as a potential candidate vaccine for West Nile virus. Vaccine. 2010;28:1075–1083. doi: 10.1016/j.vaccine.2009.10.112. [DOI] [PubMed] [Google Scholar]
- Whiteman MC, Wicker JA, Kinney RM, Huang CY-H, Solomon T, Barrett ADT. Multiple amino acid changes at the first glycosylation motif in NS1 protein of West Nile virus are necessary for complete attenuation for mouse neuroinvasiveness. Vaccine. 2011;29:9710. doi: 10.1016/j.vaccine.2011.09.036. [DOI] [PubMed] [Google Scholar]
- Whiteman MC, Popov V, Sherman MB, Wen J, Barrett AD. Attenuated West Nile virus mutant NS1130-132QQA/175A/207A exhibits virus-induced ultrastructural changes and accumulation of protein in the endoplasmic reticulum. J Virol. 2015;89:1474–1478. doi: 10.1128/JVI.02215-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Ferenczi E, Moussa K, Eliott D, Matiello M. Clinical spectrum of west nile virus neuroinvasive disease. Neurohospitalist. 2020;10:43–47. doi: 10.1177/1941874419868636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun SI, Kim SY, Rice CM, Lee YM. Development and application of a reverse genetics system for Japanese encephalitis virus. J Virol. 2003;77:6450–6465. doi: 10.1128/JVI.77.11.6450-6465.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehender G, Ebranati E, Bernini F, Presti AL, Rezza G, Delogu M, Galli M, Ciccozzi M. Phylogeography and epidemiological history of West Nile virus genotype 1a in Europe and the Mediterranean basin. Infect Genet Evol. 2011;11:646–653. doi: 10.1016/j.meegid.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Zhang S, Li L, Woodson SE, Huang CY-H, Kinney RM, Barrett ADT, Beasley DWC. A mutation in the envelope protein fusion loop attenuates mouse neuroinvasiveness of the NY99 strain of West Nile virus. Virology. 2006;353:35–40. doi: 10.1016/j.virol.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Date T, Li Y, Kato T, Miyamoto M, Yasui K, Wakita T. Characterization of the E-138 (Glu/Lys) mutation in Japanese encephalitis virus by using a stable, full-length, infectious cDNA clone. J Gen Virol. 2005;86:2209–2220. doi: 10.1099/vir.0.80638-0. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.