Abstract
Enterovirus A71 (EV-A71) is one of the etiological pathogens leading to hand, foot, and mouth disease (HFMD), which can cause severe neurological complications. The neuropathogenesis of EV-A71 infection is not well understood. The mislocalization and aggregation of TAR DNA-binding protein 43 (TDP-43) is the pathological hallmark of amyotrophic lateral sclerosis (ALS). However, whether TDP-43 was impacted by EV-A71 infection is unknown. This study demonstrated that TDP-43 was cleaved during EV-A71 infection. The cleavage of TDP-43 requires EV-A71 replication rather than the activated caspases due to viral infection. TDP-43 is cleaved by viral protease 3C between the residues 331Q and 332S, while mutated TDP-43 (Q331A) was not cleaved. In addition, mutated 3C which lacks the protease activity failed to induce TDP-43 cleavage. We also found that TDP-43 was translocated from the nucleus to the cytoplasm, and the mislocalization of TDP-43 was induced by viral protease 2A rather than 3C. Taken together, we demonstrated that TDP-43 was cleaved by viral protease and translocated to the cytoplasm during EV-A71 infection, implicating the possible involvement of TDP-43 in the pathogenesis of EV-A71infection.
Electronic supplementary material
The online version of this article (10.1007/s12250-020-00262-x) contains supplementary material, which is available to authorized users.
Keywords: Enterovirus A71 (EV-A71), TAR DNA-binding protein 43 (TDP-43), 3C protease, 2A protease
Introduction
Enterovirus A71 (EV-A71) is a globally important neurotropic virus in the genus of Enterovirus of Picornaviridae family (Lei et al. 2017; Wang C et al. 2017). EV-A71 is one of the causative pathogens that leads to the epidemic of hand, foot, and mouth disease (HFMD) commonly affecting young children, especially in Asian-Pacific region (Chang et al. 2016). From 2008 to 2013, there were about 9 million HFMD cases reported in China (Takahashi et al. 2016). Although typical EV-A71 infection is mild and self-limiting, severe neurological complications, such as brainstem encephalitis, aseptic meningitis, and acute flaccid paralysis can sometimes occur with severe consequence (Caine et al. 2016; Chen et al. 2019; Ju et al. 2020; Lee 2016; Teoh et al. 2016). The neuropathogenesis of EV-A71 infection is not completely understood. It is believed that the inflammatory response initiated by the cytokine storm in the central nervous system plays an important role (Feng et al. 2016; Huang et al. 2017; Zhao et al. 2017).
EV-A71 is a small, non-enveloped, single-stranded, positive-sensed RNA virus with a genome of 7.4 kb (Lei et al. 2017; Wang C et al. 2017). During viral replication, the genome of EV-A71 encodes a primary polyprotein, which is cleaved by the viral proteases 2A and 3C and matures into viral structural and non-structural proteins (Li B et al. 2017; Li J et al. 2017; Wang et al. 2013). The proteases of enteroviruses cleave not only viral polyproteins, but also cellular proteins such as MAVS, MDA5, and Gasdermin D (Feng et al. 2014; Lei et al. 2017; Wang et al. 2013). These virus proteases have been demonstrated to contribute to a large spectrum of disrupted cellular functions during EV-A71 infection, including inhibited interferon response, pyroptosis, suppressed endoplasmic reticulum-associated degradation pathway, and promoted ERK activity (Duan et al. 2017; Lei et al. 2017; Wang C et al. 2017; Wang T et al. 2017).
TAR DNA-binding protein 43 (TDP-43) is a nuclear RNA- and DNA-binding protein with multiple functions involved in transcription, RNA processing, mRNA stability, and translation (Gao et al. 2019; McCluskey et al. 2009; Tudor et al. 2010). The cytoplasmic mislocalization and aggregation of TDP-43 in the motor neurons and the surrounding cells is a key pathological hallmark of amyotrophic lateral sclerosis (ALS), a fatal neurodegenerative disorder characterized by the progressive loss of motor neurons, followed by muscle paralysis (Donde et al. 2020; Riku et al. 2014). The cleavage and cytoplasmic aggregation of TDP-43 is the pathological hallmark of the abnormal neurons of ALS, implicating the role of TDP-43 in the pathogenesis of ALS (Berning and Walker 2019; Shimonaka et al. 2016). Coxsackievirus B3 (CVB3), a species of enterovirus which can also cause neurological complications (Holmes et al. 2016), has been shown to cleave TDP-43 and induce its cytoplasmic accumulation in the stress granules (Fung et al. 2015). However, it is unknown if EV-A71 infection could impact the integrity and distribution of TDP-43. In this study, we found that TDP-43 was cleaved and mislocalized during EV-A71 infection.
Materials and Methods
Cell Culture and Virus
HeLa cells were gifts from Professor Fengmin Zhang (Department of Microbiology, Harbin Medical University, Harbin, China). SH-SY5Y cells were obtained from American Type Culture Collection. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher, Shanghai, China) supplemented with 10% fetal bovine serum (FBS, Bioindustry, Israel), penicillin (100 U/mL), and streptomycin (10 μg/mL). EV-A71 BrCr strain was provided by the Center for Disease Control of Heilongjiang Province of China (Harbin, China). Virus was propagated in HeLa cells. Virus titer was measured by tissue culture infective dose (TCID50).
Construction of Plasmids
pEGFP-TDP-43 and pEGFP-TDP-43-mut, in which 331Q was mutated to 331A, were purchased from Viewsolid (Beijing, China). pEGFP-2A, pEGFP-3C, and pEGFP-3C-mut were constructed based on pEGFP-C1 (Clonetech) as described previously (Guo et al. 2014; Wu et al. 2014).
Transfection
HeLa cells were grown in 12-well plate for 24 h to reach 60% confluency. Transfection solution was prepared with plasmids mixed with Lipofectamine 2000 (Thermo Fisher) in serum-free DMEM. Cells were incubated with transfection mixture for 4 h and then grown in the fresh medium containing 10% FBS and antibiotics. Cells were harvested at 24 h after transfection for further analysis. Each transfection was performed in triplicate.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted by TRIzol reagent (Thermo Fisher) according to the manufacturer’s protocol. 1 μg of total RNA was reverse-transcribed using TransScript All-in-One SuperMix (TransGen, Beijing, China) in a total of 20 μL reverse transcription system. For quantitative real-time PCR (qRT-PCR), TransStart Top Green qPCR SuperMix was used on a LightCycler (Roche). PCR reaction was carried out for 35 cycles according to the cycling conditions: denaturation at 94 °C for 5 s, annealing at 55 °C for 15 s, and extension at 72 °C for 1 min. The relative RNA amount was measured with 2-CT threshold cycle method and normalized to the amount of GAPDH. Reactions were carried out in triplicate. Primers were synthesized by Genewiz (Suzhou, China). Primer sequences are as listed: for EV-A71, forward primer 5′-CCCCTGAATGCGGCTAAT-3′ and reverse primer 5′-CAATTGTCACCATAAGCAGCCA-3′; for GAPDH, forward primer 5′-CTGGGCTACACTGAGCACC-3′ and reverse primer 5′-AAGTGGTCGTTGAGGGCAATG-3′.
Western Blot
Cells were harvested and proteins were extracted by RIPA buffer (Biotopped, Beijing, China) containing 1% phenylmethanesulfonyl fluoride (PMSF) (Biotopped). Proteins were quantified by Bradford assay, separated by 12% polyacrylamide gel (SDS-PAGE), and then transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Kenilworth, NJ). The membrane was blocked with 0.1% Tween-20 in phosphate-buffered saline (PBS) containing 5% bovine serum albumin (BSA) and then incubated with primary antibodies at 4 °C overnight. The membrane was washed three times with 0.1% Tween-20 in PBS and incubated with anti-rabbit IgG for 2 h at room temperature. Immunoblot was visualized with FluorChem R system (ProteinSimple, Santa Clara, CA). The results were analyzed by Image J and quantified by Alpha View 3.4.0 (ProteinSimple). Rabbit anti-enterovirus 3D polyclonal antibody was prepared by the Department of Microbiology of Harbin Medical University. Anti-GFP was purchased from Santa Cruz (SC-9996). Antibody against TDP-43 was purchased from Abcam (ab109535). Antibodies against caspase-3 (19677-1-AP), GAPDH (10494-1-AP), β-actin (66009-1-Ig), β-tubulin (10068-1-AP), and secondary antibodies were purchased from Proteintech (Wuhan, China).
Immunofluorescence Microscopy
HeLa cells cultured in the glass-bottom dish for 24 h. Cells were washed with PBS three times, fixed with 4% paraformaldehyde for 15 min and permeabilized with 1% Triton-X 100 for 20 min at room temperature (RT). After blocking with 1% BSA for 30 min at RT, cells were incubated with anti-TDP-43 antibody diluted by 1:500 at 4 °C overnight and then incubated with fluorescence-conjugated anti-rabbit IgG (H + L) antibodies (Proteintech) at RT for 1 h. Cells were viewed by fluorescence microscope (ECLIPSE Ni, Nikon, Japan) and CV1000 confocal system (Yokogawa, Japan).
Statistical Analysis
All experiments were repeated three times. Data were expressed as mean ± SD and analyzed by Student’s t test. P < 0.05 was defined as statistically significant.
Results
TDP-43 is Cleaved in EV-A71-Infected Cells
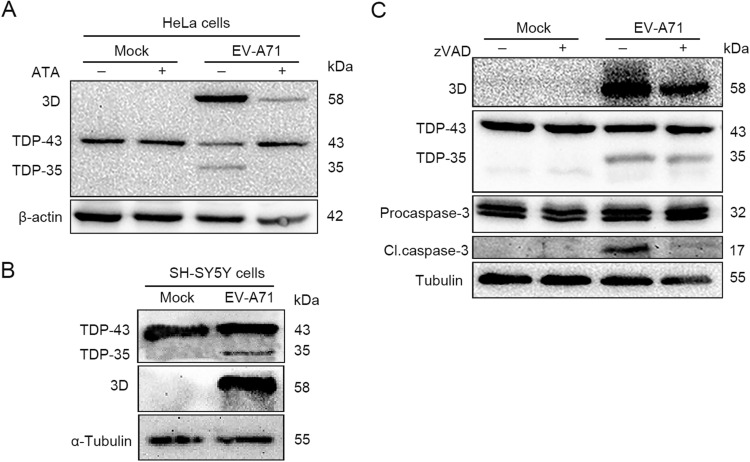
It has been reported that TDP-43 was cleaved and aggregated in the cytoplasmic stress granules during the infection of CVB3 (Fung et al. 2015). Since both CVB3 and EV-A71 belong to genus Enterovirus and share similar features in viral structure and constituents, we postulated that EV-A71 infection may also lead to the cleavage of TDP-43. To this end, HeLa cells and SH-SY5Y were cultured in 12-well plates to 60%–70% confluence and infected with EV-A71 at 1 × 104 TCID50 for 24 h. As shown in Fig. 1, a ~35 kDa fragment of TDP-43 was identified in both HeLa and SH-SY5Y cells infected with EV-A71 (Fig. 1A, 1B), while the addition of aurintricarboxylic acid (Sigma-Aldrich), which has been demonstrated to function as the inhibitor of the 3D RNA-dependent RNA polymerase of EV-A71 (Hung et al. 2010), abolished the cleavage of TDP-43 (Fig. 1A). Since the cleavage and pathological aggregation of TDP-43 in neurodegenerative diseases are largely attributed to the activation of caspases (Rohn 2009; Zhang et al. 2007), the impact of pan-caspase inhibitor on the cleavage of TDP-43 was studied. As shown in Fig. 1C, EV-A71 infection led to the cleavage of TDP-43 in the presence or absence of pan-caspase inhibitor Z-VAD-FMK (20 µmol/L, Beyotime, China), indicating that the TDP-43 cleavage is not dependent on the activated caspase during EV-A71 infection. These results suggested that the cleavage of TDP-43 was induced by EV-A71 replication.
Fig. 1.
TDP-43 is cleaved in the cells infected with EV-A71. A–B HeLa cells and SH-SY5Y were cultured in 12-well plates to 60%–70% confluence and infected with EV-A71 at 1 × 104 TCID50 for 24 h. TDP-43 proteins were analyzed by Western blotting. HeLa cells were cultured in 12-well plates and infected or mock-infected EV-A71 for 24 h in the medium containing 10 µmol/L ATA (aurintricarboxylic acid, 3D inhibitor), the inhibitor of 3D polymerase of enteroviruses (A) or 20 μmol/L pan-caspase inhibitor Z-VAD-FMK (C). Cell lysates were subjected to Western blot analysis.
TDP-43 is Cleaved by the 3C Protease of EV-A71
Enteroviral protease 2A and 3C are responsible for the cleavage of the viral polyprotein translated from the viral genome (Cai et al. 2013; Guo et al. 2014). These proteases also capable of cleaving cellular proteins such as MAVS, eIF4G, and AUF1 during viral infection (Feng et al. 2014; Wong et al. 2013; Wu et al. 2014). To determine whether viral proteases are responsible for the cleavage of TDP-43, HeLa cells were transfected with pEGFP-2A or pEGFP-3C. Control cells were transfected with pEGFP-C1, which was constructed to express EGFP as described previously (Wu et al. 2014). As shown in Fig. 2, the cleaved fragment (~35kDa) of TDP-43 was observed in the cells expressing EGFP-3C or infected with EV-A71, while cleaved TDP-43 was not observed in the cells overexpressing EGFP-2A (Fig. 2A), indicating that TDP-43 was cleaved by viral protease 3C rather than 2A. We also found that the cleaved TDP-43 was accumulating along with the expression of 3C protease (Fig. 2B).
Fig. 2.
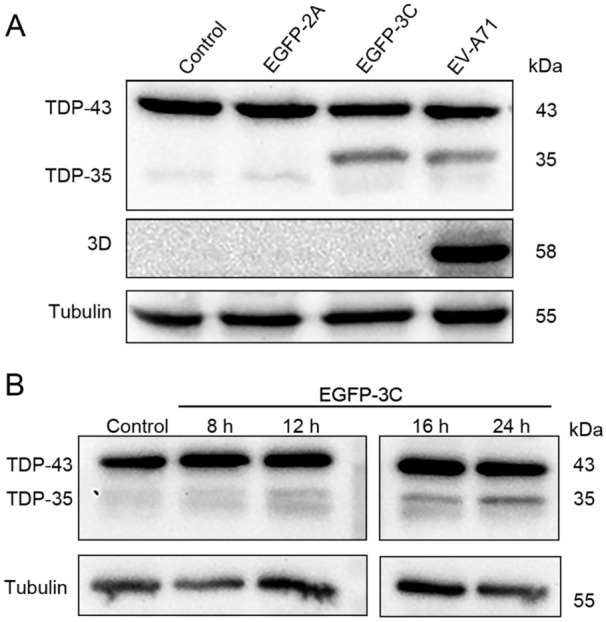
TDP-43 is cleaved by the 3C protease of EV-A71. A HeLa cells were cultured in 12-well plates to 60% confluency and infected with EV-A71 or transfected with pEGFP-2A or pEGFP-3C for 24 h. Control cells were treated with PBS. Cell lysates were subjected to Western blot analysis. B Cells were transfected with pEGFP-3C for various times. Control cells were transfected with pEGFP-C1. Cell lysates were analyzed by Western blotting.
Mutated 3C Protease Does Not Cleaves TDP-43
To confirm that 3C is the viral protease that causes the cleavage of TDP-43, HeLa cells were co-transfected with either pEGFP-3C or pEGFP-3C-mut and pEGFP-TDP-43. pEGFP-3C-mut was expressing a mutant 3C which lacks the protease activity due to the mutation of 147 cysteine to alanine (Fig. 3A). The cleaved EGFP-TDP-43 fusion protein (27 kDa + 43 kDa = 70 kDa), designated as EGFP-TDP-35 (27 kDa + 35 kDa = 62 kDa), was identified in the cells expressing EGFP-3C but not in the cells expressing EGFP-3C-mut (Fig. 3B). These data show that TDP-43 was cleaved by the protease 3C of EV-A71.
Fig. 3.
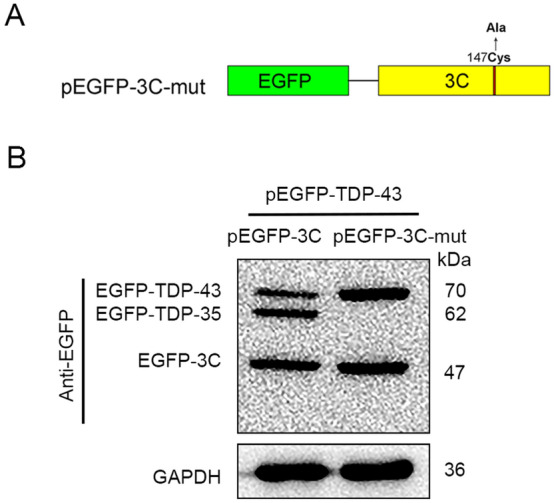
TDP-43 is not cleaved by the mutated 3C protease. A The construct expressing the fusion protein of EGFP with a mutated 3C (designated as pEGFP-3C-mut), which lacks the protease activity, was generated. B Cells were co-transfected with either pEGFP-TDP-43 and pEGFP-3C or pEGFP-TDP-43 and pEGFP-3C-mut for 24 h. Cell lysates were analyzed by Western blotting in which anti-EGFP antibody was used.
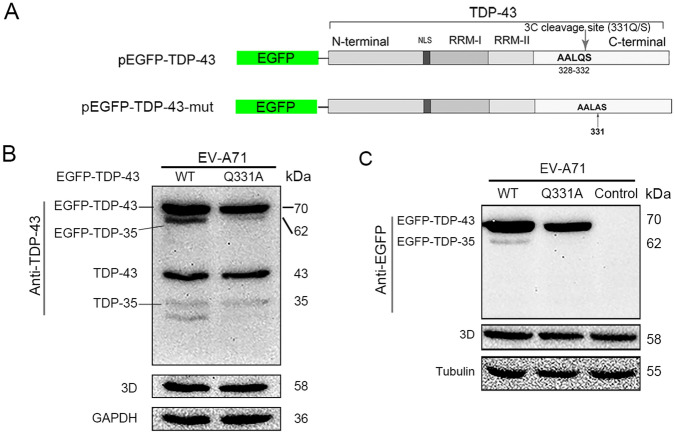
The Cleavage Site of TDP-43 is Close to Its C-terminal
The potential cleavage site of the protease 3C in TDP-43 was analyzed (Fig. 4A). To confirm the cleavage site of TDP-43, the construct expressing EGFP-TDP-43 or a mutant TDP-43 (Q331A) was generated. Cells were transfected with either pEGFP-TDP-43 or pEGFP-TDP-43-mut (Q331A) for 24 h and infected with EV-A71 for 24 h. As shown Fig. 4B, EV-A71 infection induced the cleavage of EGFP-TDP-43, designated as EGFP-TDP-35 (with a molecular weight of 62 kDa), while EGFP-TDP-35 was not observed in the cells expressing EGFP-TDP43-mut, demonstrating that TDP-43 was cleaved between 331Q and 332S. Endogenous TDP-43 and its cleaved form (TDP-35) were also observed in the cells expressing either wild type or mutant TDP-43, further demonstrating that the cleavage of TDP-43 was the result of EV-A71 infection.
Fig. 4.
TDP-43 is cleaved at a site which is close to its C-terminal. The constructs expressing EGFP fusion proteins containing TDP-43 or mutated TDP-43 were generated. The construct containing mutated TDP-43 was designated as pEGFP-TDP-43-mut, in which 331Q is mutated to 331A. The domains of TDP-43 protein and the potential cleavage site of protease 3C of the enteroviruses are indicated (A). Cells were transfected with either pEGFP-TDP-43 (wild type) or pEGFP-TDP-43-mut (Q331A) for 24 h and then infected with EV-A71 for 24 h (B and C). Control cells were transfected with pcDNA3.1 (C). Cell lysates were subjected to Western blotting using anti-TDP-43 antibody (B) or anti-EGFP antibody (C).
We also noted that a fragment of TDP-43 with a size smaller than TDP-35 appeared in the cells transfected with pEGFP-TDP-43 and then infected with EV-A71 (Fig. 4B, left lane). The occurrence of this fragment, which is obviously originated from the endogenous TDP-43, indicates that it is very likely that there is another cleavage site in TDP-43 except that between 331Q and 332S. However, this extra TDP-43 fragment (smaller than TDP-35) was not observed in the cells transfected with pEGFP-TDP-43-mut followed by the infection of EV-A71 (Fig. 4B, right lane), suggesting that virus-induced TDP-43 cleavage at this site may not as efficient as that between 331Q and 332S.
The cleavage of TDP-43 in EV-A71-infected cells was also evaluated by Western blotting with anti-EGFP antibody (Fig. 4C). EFGP-TDP-35 was identified in the cells expressing EGFP-TDP-43, but not in the cells expressing EGFP-TDP-43-mut or control cells, which were transfected with pcDNA3.1 (Fig. 4C). Collectively, these results demonstrated that TDP-43 was cleaved between 331Q and 332S, which is close to its C-terminal.
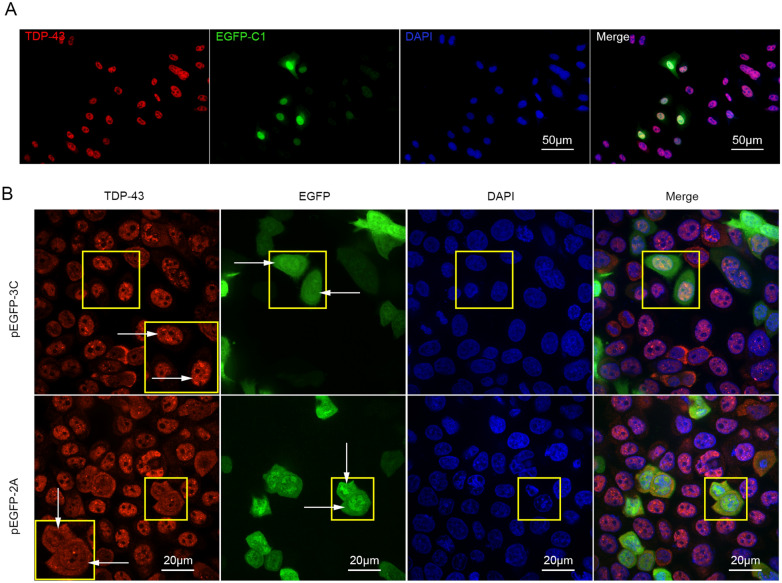
The Cytoplasmic Translocation of TDP-43 is Not the Result of Its Cleavage
Study has shown that TDP-43 was translocated into the cytoplasmin in CVB3-infected cells (Fung et al. 2015). Our previous study has demonstrated that EV-A71 infection induced the accumulation of proteins in the form of cytoplasmic stress granules. Thus, we asked the question whether the intracellular localization of TDP-43 is altered by EV-A71 infection. As shown in Fig. 5, TDP-43 was localized almost entirely in the nucleus of control cells (Fig. 5A) or in the cells expressing protease 3C of EV-A71 (Fig. 5B, upper panel), while cytoplasmic TDP-43 was identified in the cells expressing 2A (Fig. 5B, lower panel). These findings demonstrate that the cytoplasmic translocation of TDP-43 was induced by protease 2A rather than protease 3C, suggesting that the cleavage of TDP-43 does not contribute to its mislocalization in EV-A71-infected cells.
Fig. 5.
The cytoplasmic translocation of TDP-43 is caused by protease 2A of EV-A71. HeLa cells were transiently transfected with pEGFP-C1 (A), pEGFP-2A (B), and pEGFP-3C (B), respectively, for 24 h. Cells were harvested for immunofluorescence staining using anti-TDP-43 antibody (Alexa-fluor-594). Cell nuclei were stained with DAPI. Fluorescence images were taken by fluorescence microscope (A) and CV1000 confocal system (B). Yellow squares in the upper panel of B indicate the cells expressing EGFP-3C. Yellow squares in the lower panel of B indicate the cells expressing EGFP-2A. The larger square in the upper right or lower right panel of B is the magnified image enclosed by the smaller square in the same panel. The distribution of TDP-43 was indicated by arrows in the upper right and lower right panels of B. Arrows in the upper-middle of B indicate the nuclei of the cells expressing EGFP-3C, while arrows in the lower-middle of B indicate the cytoplasm of the cells expressing EGFP-2A.
Discussion
To reveal virus-host interaction is essential for understanding the pathogenesis of viral diseases and developing potential treatment. The pathogenesis of neurological injury caused by EV-A71 infection is incompletely understood (Huang et al. 1999; Teoh et al. 2016). As an RNA and DNA-binding protein, TDP-43 is widely involved in a variety of cellular processes. The pathological aggregation of TDP-43 is the key molecular event of ALS and frontotemporal lobar degeneration (FTLD) (Hart and Gitler 2012; Riku et al. 2016). The cleavage and cytoplasmic aggregation of TDP-43 have been observed in the cells infected with CVB3, another neurotropic enterovirus (Fung et al. 2015). In this study, we demonstrated that TDP-43 was cleaved by the protease 3C of EV-A71 and translocated into the cytoplasm, implicating the potential role of TDP-43 in pathogenesis of EV-A71 infection.
Similar to the report in which TDP-43 was found to be cleaved and aggregated in the cytoplasmic stress granules during CVB infection (Fung et al. 2015), this study shows that TDP-43 was cleaved by protease 3C and translocated into the cytoplasm induced by protease 2A of EV-A71. However, we did not study whether TDP-43 or the cleaved TDP-43 was participating in the formation of stress granules in EV-A71-infected cells.
The proteases 2A and 3C of enteroviruses are essential for viral replication by cleaving the viral polyprotein into viral structural and non-structural proteins. Virus protease 2A and 3C also cleave cellular proteins such as MDA5, RIG-I, MAVS, and TRIF to suppress innate immunity (Feng et al. 2014; Lei et al. 2010; Lei et al. 2011; Wang et al. 2013). Previous studies show that the cleavage site for protease 3C of enteroviruses is often between Q and G or Q and S in the target sequence of AXXQG or AXXQS (Sun et al. 2016). In this study, we demonstrated that TDP-43 was cleaved by 3C protease between the residue 331Q and 332S in the sequence of AALQS. When this sequence was mutated (331Q was mutated to 331A), 3C protease failed to cleave TDP-43.
As a highly conserved nuclear DNA/RNA binding protein, TDP-43 is involved in all aspects of RNA metabolism including the control of transcription, splicing, mRNA transport, and translation (Fiesel et al. 2012; Freibaum et al. 2010). It is believed that TDP-43 may function as a part of ribonucleoprotein complexes (Colombrita et al. 2012; Ule 2008). Evidence suggests that nuclear RNA-binding proteins are utilized to promote efficient viral replication. Heterogeneous ribonucleoprotein C (hnRNP C) is able to stimulate poliovirus RNA synthesis (Ertel et al. 2010), and hnRNP A1 promotes the translation of EV-A71 through the interaction with type I internal ribosome entry site (IRES) (Levengood et al. 2013; Tolbert et al. 2017). TDP-43 has been described to interact with hnRNP A1, A2/B1, A3, and C2 (Buratti et al. 2005; Romano et al. 2014).
Although we show that TDP-43 was cleaved by the protease 3C of EV-A71, how the cleaved TDP-43 would impact viral replication and cellular functions remains to be studied. Our data show that neither knockdown of TDP-43 with siRNA nor TDP-43 overexpression altered the replication of EV-A71 (Supplementary Figure S1 and Figure S2). Although we did not investigate the cellular impact of TDP-43 cleavage during EV-A71 infection, it is logical to predict that processes associated with RNA metabolism could be affected due to the critical role played by TDP-43 in this aspect.
EV-A71 infection primarily impacts the neural system of young children. However, up to now, there is no evidence about the cleavage and the cytoplasmic translocation of TDP-43 in the patients infected with EV-A71. Theiler’s murine encephalomyelitis virus (TMEV), which belongs to the Cardiovirus genus of the Picornaviridae family, is the rodent pathogen of the central nervous system (Gerhauser et al. 2019). One report has shown that the infection of TMEV induced the cleavage of TDP-43 with the production of the cleaved fragment, TDP-35. Although the cleavage site in TDP-43 was not investigated (Masaki et al. 2019), we predict that TDP-43 is likely cleaved at the same site as in the case of EV-A71 infection, since TMEV also encodes a viral protease 3C with cysteine protease activity (GenBank: AAA47930.1). Moreover, the cytoplasmic mislocalization of TDP-43 was also seen in the cells infected with TMEV (Masaki et al. 2019). Collectively, the previous report and our results suggest that the cleavage and cytoplasmic mislocalization of TDP-43 might be a common feature for the infection of picornaviruses. In vivo study is needed to further evaluate the significance of the altered TDP-43 in the pathogenesis of EV-A71.
This study shows that the cytoplasmic localization of TDP-43 was induced by protease 2A rather than 3C of EV-A71. It has been demonstrated that protease 2A of enteroviruses cleaves nucleoporins in the nuclear pore complex (NPC), leading to the selective removal of the N-terminal phenylalanine-glycine (FG)-rich domains of the nucleoporins (Park et al. 2015; Watters and Palmenberg 2011). Although the overall structure of NPC remains relatively unaffected in the cells infected with enteroviruses, the permeability of nuclear pore is increased (Watters et al. 2017; Wobst et al. 2017). Thus, it is likely that the cytoplasmic localization of TDP-43 is the consequence of the increased permeability of the nuclear pores induced by 2A protease of EV-A71. However, it is unknown concerning the integrity of TDP-43 (cleaved or non-cleaved) which one is localized in the cytoplasm.
In summary, the present study demonstrated that TDP-43 was cleaved by the protease 3C in EV-A71-infected cells, while viral protease 2A induced the cytoplasmic localization of TDP-43. Our data imply that the cleavage and mislocalization of TDP-43 might contribute to the pathogenesis of EV-A71 infection.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by the National Natural Foundation of China (81672007 and 81971920 to Wenran Zhao, 81871652 to Zhaohua Zhong, and 81772188 to Yan Wang) and Health and Family Planning Commission of Heilongjiang Province (2017-158 to Xiaoman Wo). We are grateful to the help of Dr. Ying Wu (School of Life Sciences, Northeast Forestry University, Harbin, China) in viewing the images of fluorescence microscope.
Author Contributions
WZ and ZZ conceived the experiments, analyzed the data. WZ wrote the manuscript. XW, YY, and YX performed the majority of the laboratory work. Yao Wang and YC prepared and performed the immunofluorescence visualization. SZ and Yan Wang prepared of plasmids. LL and XZ prepared Enterovirus A71 for this study.
Compliance with Ethical Standards
Conflicts of interest
There is no conflicting interest in this work.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Xiaoman Wo and Yuan Yuan contributed equally to this work.
Contributor Information
Zhaohua Zhong, Email: zhongzh@hrbmu.edu.cn.
Wenran Zhao, Email: zhaowr@hrbmu.edu.cn.
References
- Berning BA, Walker AK. The pathobiology of TDP-43 C-terminal fragments in ALS and FTLD. Front Neurosci. 2019;13:335. doi: 10.3389/fnins.2019.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- Cai Q, Yameen M, Liu W, Gao Z, Li Y, Peng X, Cai Y, Wu C, Zheng Q, Li J, Lin T. Conformational plasticity of the 2A proteinase from enterovirus 71. J Virol. 2013;87:7348–7356. doi: 10.1128/JVI.03541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine EA, Moncla LH, Ronderos MD, Friedrich TC, Osorio JE. A single mutation in the VP1 of enterovirus 71 is responsible for increased virulence and neurotropism in adult interferon-deficient mice. J Virol. 2016;90:8592–8604. doi: 10.1128/JVI.01370-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PC, Chen SC, Chen KT. The current status of the disease caused by enterovirus 71 infections: epidemiology, pathogenesis, molecular epidemiology, and vaccine development. Int J Environ Res Public Health. 2016;13:890. doi: 10.3390/ijerph13090890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Hu L, Xu F, Liu CJ, Li J. A case report of a teenager with severe hand, foot, and mouth disease with brainstem encephalitis caused by enterovirus 71. BMC Pediatr. 2019;19:59. doi: 10.1186/s12887-019-1428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, Silani V, Ratti A. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger rnas and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem. 2012;287:15635–15647. doi: 10.1074/jbc.M111.333450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donde A, Sun M, Jeong YH, Wen X, Ling J, Lin S, Braunstein K, Nie S, Wang S, Chen L, Wong PC. Upregulation of ATG7 attenuates motor neuron dysfunction associated with depletion of TARDBP/TDP-43. Autophagy. 2020;16:672–682. doi: 10.1080/15548627.2019.1635379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Zhu M, Xiong Q, Wang Y, Xu C, Sun J, Wang C, Zhang H, Xu P, Peng Y. Regulation of enterovirus 2A protease-associated viral IRES activities by the cell’s ERK signaling cascade: implicating ERK as an efficiently antiviral target. Antiviral Res. 2017;143:13–21. doi: 10.1016/j.antiviral.2017.03.018. [DOI] [PubMed] [Google Scholar]
- Ertel KJ, Brunner JE, Semler BL. Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates. J Virol. 2010;84:4229–4242. doi: 10.1128/JVI.02198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Langereis MA, Lork M, Nguyen M, Hato SV, Lanke K, Emdad L, Bhoopathi P, Fisher PB, Lloyd RE, van Kuppeveld FJ. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J Virol. 2014;88:3369–3378. doi: 10.1128/JVI.02712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M, Guo S, Fan S, Zeng X, Zhang Y, Liao Y, Wang J, Zhao T, Wang L, Che Y, Wang J, Ma N, Liu L, Yue L, Li Q. The preferential infection of astrocytes by enterovirus 71 plays a key role in the viral neurogenic pathogenesis. Front Cell Infect Microbiol. 2016;6:192. doi: 10.3389/fcimb.2016.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel FC, Weber SS, Supper J, Zell A, Kahle PJ. TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 2012;40:2668–2682. doi: 10.1093/nar/gkr1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung G, Shi J, Deng H, Hou J, Wang C, Hong A, Zhang J, Jia W, Luo H. Cytoplasmic translocation, aggregation, and cleavage of TDP-43 by enteroviral proteases modulate viral pathogenesis. Cell Death Differ. 2015;22:2087–2097. doi: 10.1038/cdd.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang L, Yan T, Perry G, Wang X. TDP-43 proteinopathy and mitochondrial abnormalities in neurodegeneration. Mol Cell Neurosci. 2019;100:103396. doi: 10.1016/j.mcn.2019.103396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhauser I, Hansmann F, Ciurkiewicz M, Loscher W, Beineke A. Facets of theiler’s murine encephalomyelitis virus-induced diseases: an update. Int J Mol Sci. 2019;20:448. doi: 10.3390/ijms20020448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zhong X, Lin L, Wu S, Wang T, Chen Y, Zhai X, Wang Y, Wu H, Tong L, Han Y, Pan B, Peng Y, Si X, Zhang F, Zhao W, Zhong Z. A 3C(pro)-dependent bioluminescence imaging assay for in vivo evaluation of anti-enterovirus 71 agents. Antiviral Res. 2014;101:82–92. doi: 10.1016/j.antiviral.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Hart MP, Gitler AD. ALS-associated ataxin 2 polyq expansions enhance stress-induced caspase 3 activation and increase TDP-43 pathological modifications. J Neurosci. 2012;32:9133–9142. doi: 10.1523/JNEUROSCI.0996-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes CW, Koo SS, Osman H, Wilson S, Xerry J, Gallimore CI, Allen DJ, Tang JW. Predominance of enterovirus B and echovirus 30 as cause of viral meningitis in a UK population. J Clin Virol. 2016;81:90–93. doi: 10.1016/j.jcv.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Huang CC, Liu CC, Chang YC, Chen CY, Wang ST, Yeh TF. Neurologic complications in children with enterovirus 71 infection. N Engl J Med. 1999;341:936–942. doi: 10.1056/NEJM199909233411302. [DOI] [PubMed] [Google Scholar]
- Huang Q, Wang Y, Si C, Zhao D, Wang Y, Duan Y. Interleukin-35 modulates the imbalance between regulatory T cells and T helper 17 cells in enterovirus 71-induced hand, foot, and mouth disease. J Interferon Cytokine Res. 2017;37:522–530. doi: 10.1089/jir.2017.0080. [DOI] [PubMed] [Google Scholar]
- Hung HC, Chen TC, Fang MY, Yen KJ, Shih SR, Hsu JT, Tseng CP. Inhibition of enterovirus 71 replication and the viral 3d polymerase by aurintricarboxylic acid. J Antimicrob Chemother. 2010;65:676–683. doi: 10.1093/jac/dkp502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Y, Tan Z, Huang H, Chen M, Tan Y, Zhang C, Wang J, Wang H, Chen M. Clinical and epidemiological characteristics of coxsackievirus a6- and enterovirus 71-associated clinical stage 2 and 3 severe hand, foot, and mouth disease in Guangxi, Southern China, 2017. J Infect. 2020;80:121–142. doi: 10.1016/j.jinf.2019.09.021. [DOI] [PubMed] [Google Scholar]
- Lee KY. Enterovirus 71 infection and neurological complications. Korean J Pediatr. 2016;59:395–401. doi: 10.3345/kjp.2016.59.10.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X, Liu X, Ma Y, Sun Z, Yang Y, Jin Q, He B, Wang J. The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation and type I interferon responses. J Virol. 2010;84:8051–8061. doi: 10.1128/JVI.02491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X, Sun Z, Liu X, Jin Q, He B, Wang J. Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by toll-like receptor 3. J Virol. 2011;85:8811–8818. doi: 10.1128/JVI.00447-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X, Zhang Z, Xiao X, Qi J, He B, Wang J. Enterovirus 71 inhibits pyroptosis through cleavage of gasdermin D. J Virol. 2017;91:e01069. doi: 10.1128/JVI.01069-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levengood JD, Tolbert M, Li ML, Tolbert BS. High-affinity interaction of hnRNP A1 with conserved RNA structural elements is required for translation and replication of enterovirus 71. RNA Biol. 2013;10:1136–1145. doi: 10.4161/rna.25107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Yue Y, Zhang Y, Yuan Z, Li P, Song N, Lin W, Liu Y, Gu L, Meng H. A novel enterovirus 71 (ev71) virulence determinant: the 69th residue of 3C protease modulates pathogenicity. Front Cell Infect Microbiol. 2017;7:26. doi: 10.3389/fcimb.2017.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gao F, Hao SB, Cheng D, Zhang WQ, Lin B, Zhao L, Yu XJ, Wang ZY, Wen HL. Contribution of 3CD region to the virulence of enterovirus 71. Biomed Environ Sci. 2017;30:767–771. doi: 10.3967/bes2017.103. [DOI] [PubMed] [Google Scholar]
- Masaki K, Sonobe Y, Ghadge G, Pytel P, Roos RP. TDP-43 proteinopathy in theiler’s murine encephalomyelitis virus infection. PLoS Pathog. 2019;15:e1007574. doi: 10.1371/journal.ppat.1007574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey LF, Elman LB, Martinez-Lage M, Van Deerlin V, Yuan W, Clay D, Siderowf A, Trojanowski JQ. Amyotrophic lateral sclerosis-plus syndrome with TAR DNA-binding protein-43 pathology. Arch Neurol. 2009;66:121–124. doi: 10.1001/archneur.66.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park N, Schweers NJ, Gustin KE. Selective removal of FG repeat domains from the nuclear pore complex by enterovirus 2A(pro) J Virol. 2015;89:11069–11079. doi: 10.1128/JVI.00956-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riku Y, Watanabe H, Yoshida M, Tatsumi S, Mimuro M, Iwasaki Y, Katsuno M, Iguchi Y, Masuda M, Senda J, Ishigaki S, Udagawa T, Sobue G. Lower motor neuron involvement in TAR DNA-binding protein of 43 kDa-related frontotemporal lobar degeneration and amyotrophic lateral sclerosis. JAMA Neurol. 2014;71:172–179. doi: 10.1001/jamaneurol.2013.5489. [DOI] [PubMed] [Google Scholar]
- Riku Y, Watanabe H, Yoshida M, Mimuro M, Iwasaki Y, Masuda M, Ishigaki S, Katsuno M, Sobue G. Marked involvement of the striatal efferent system in TAR DNA-binding protein 43 kDa-related frontotemporal lobar degeneration and amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2016;75:801–811. doi: 10.1093/jnen/nlw053. [DOI] [PubMed] [Google Scholar]
- Rohn TT. Cytoplasmic inclusions of TDP-43 in neurodegenerative diseases: a potential role for caspases. Histol Histopathol. 2009;24:1081–1086. doi: 10.14670/hh-24.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M, Buratti E, Romano G, Klima R, Del Bel Belluz L, Stuani C, Baralle F, Feiguin F. Evolutionarily conserved heterogeneous nuclear ribonucleoprotein (hnRNP) A/B proteins functionally interact with human and drosophila TAR DNA-binding protein 43 (TDP-43) J Biol Chem. 2014;289:7121–7130. doi: 10.1074/jbc.M114.548859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimonaka S, Nonaka T, Suzuki G, Hisanaga S, Hasegawa M. Templated aggregation of TAR DNA-binding protein of 43 kDa (TDP-43) by seeding with TDP-43 peptide fibrils. J Biol Chem. 2016;291:8896–8907. doi: 10.1074/jbc.M115.713552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Chen S, Cheng A, Wang M. Roles of the picornaviral 3C proteinase in the viral life cycle and host cells. Viruses. 2016;8:82. doi: 10.3390/v8030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Liao Q, Van Boeckel TP, Xing W, Sun J, Hsiao VY, Metcalf CJ, Chang Z, Liu F, Zhang J, Wu JT, Cowling BJ, Leung GM, Farrar JJ, van Doorn HR, Grenfell BT, Yu H. Hand, foot, and mouth disease in china: modeling epidemic dynamics of enterovirus serotypes and implications for vaccination. PLoS Med. 2016;13:e1001958. doi: 10.1371/journal.pmed.1001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh HL, Mohammad SS, Britton PN, Kandula T, Lorentzos MS, Booy R, Jones CA, Rawlinson W, Ramachandran V, Rodriguez ML, Andrews PI, Dale RC, Farrar MA, Sampaio H. Clinical characteristics and functional motor outcomes of enterovirus 71 neurological disease in children. JAMA Neurol. 2016;73:300–307. doi: 10.1001/jamaneurol.2015.4388. [DOI] [PubMed] [Google Scholar]
- Tolbert M, Morgan CE, Pollum M, Crespo-Hernandez CE, Li ML, Brewer G, Tolbert BS. HnRNP A1 alters the structure of a conserved enterovirus IRES domain to stimulate viral translation. J Mol Biol. 2017;429:2841–2858. doi: 10.1016/j.jmb.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor EL, Galtrey CM, Perkinton MS, Lau KF, De Vos KJ, Mitchell JC, Ackerley S, Hortobagyi T, Vamos E, Leigh PN, Klasen C, McLoughlin DM, Shaw CE, Miller CC. Amyotrophic lateral sclerosis mutant vesicle-associated membrane protein-associated protein-B transgenic mice develop TAR-DNA-binding protein-43 pathology. Neuroscience. 2010;167:774–785. doi: 10.1016/j.neuroscience.2010.02.035. [DOI] [PubMed] [Google Scholar]
- Ule J. Ribonucleoprotein complexes in neurologic diseases. Curr Opin Neurobiol. 2008;18:516–523. doi: 10.1016/j.conb.2008.09.018. [DOI] [PubMed] [Google Scholar]
- Wang B, Xi X, Lei X, Zhang X, Cui S, Wang J, Jin Q, Zhao Z. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog. 2013;9:e1003231. doi: 10.1371/journal.ppat.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sun M, Yuan X, Ji L, Jin Y, Cardona CJ, Xing Z. Enterovirus 71 suppresses interferon responses by blocking janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling through inducing karyopherin-alpha1 degradation. J Biol Chem. 2017;292:10262–10274. doi: 10.1074/jbc.M116.745729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wang B, Huang H, Zhang C, Zhu Y, Pei B, Cheng C, Sun L, Wang J, Jin Q, Zhao Z. Enterovirus 71 protease 2Apro and 3Cpro differentially inhibit the cellular endoplasmic reticulum-associated degradation (ERAD) pathway via distinct mechanisms, and enterovirus 71 hijacks ERAD component p97 to promote its replication. PLoS Pathog. 2017;13:e1006674. doi: 10.1371/journal.ppat.1006674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters K, Palmenberg AC. Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J Virol. 2011;85:10874–10883. doi: 10.1128/JVI.00718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters K, Inankur B, Gardiner JC, Warrick J, Sherer NM, Yin J, Palmenberg AC. Differential disruption of nucleocytoplasmic trafficking pathways by rhinovirus 2A proteases. J Virol. 2017;91:e02472. doi: 10.1128/JVI.02472-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobst HJ, Delsing L, Brandon NJ, Moss SJ. Truncation of the TAR DNA-binding protein 43 is not a prerequisite for cytoplasmic relocalization, and is suppressed by caspase inhibition and by introduction of the a90v sequence variant. PLoS ONE. 2017;12:e0177181. doi: 10.1371/journal.pone.0177181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Si X, Angeles A, Zhang J, Shi J, Fung G, Jagdeo J, Wang T, Zhong Z, Jan E, Luo H. Cytoplasmic redistribution and cleavage of AUF1 during coxsackievirus infection enhance the stability of its viral genome. FASEB J. 2013;27:2777–2787. doi: 10.1096/fj.12-226498. [DOI] [PubMed] [Google Scholar]
- Wu S, Wang Y, Lin L, Si X, Wang T, Zhong X, Tong L, Luan Y, Chen Y, Li X, Zhang F, Zhao W, Zhong Z. Protease 2A induces stress granule formation during coxsackievirus b3 and enterovirus 71 infections. Virol J. 2014;11:192. doi: 10.1186/s12985-014-0192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Xu YF, Dickey CA, Buratti E, Baralle F, Bailey R, Pickering-Brown S, Dickson D, Petrucelli L. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci. 2007;27:10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhang Z, Zhang Y, Feng M, Fan S, Wang L, Liu L, Wang X, Wang Q, Zhang X, Wang J, Liao Y, He Z, Lu S, Yang H, Li Q. Dynamic interaction of enterovirus 71 and dendritic cells in infected neonatal rhesus macaques. Front Cell Infect Microbiol. 2017;7:171. doi: 10.3389/fcimb.2017.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.