Abstract
Preeclampsia (PE) is a multi-system disorder that is specific to human pregnancy. Inadequate oxygenation of uterus and placenta is considered as one of the leading causes for the disease. MicroRNA-210(miR-210) is one of the prime molecules that has emerged in response to hypoxia. The objective of this study was to determine miR-210 expression patterns in plasma from severe PE and mild PE patients, and how that affects the expression of miR-210 target genes. The expression levels of miR-210 were validated using reverse transcription-quantitative PCR in plasma of severe PE (15) and mild PE (15) patients in comparison to controls subjects (15) with normal pregnancy. Then, the association between miR-210 and its downstream genes was validated by using human miR-210 targets RT2 profiler PCR Array. Both the categories (mild and severe) showed significantly high miR-210 expression levels. Also out of the 84 hypoxia miR-210 associated genes screened using mRNA, 18 genes were found to be differentially expressed in severe PE whereas 16 genes in mild PE cases with varying magnitude. All the genes in both the PE groups were found downregulated in comparison to controls. These downregulated genes expressed in both the cases were shown to be participating in immunosuppression, apoptosis, cell growth, signaling, angiogenesis, DNA repair. This study provides novel data on the genes that work downstream of miR-210 and how dysregulated expression of miR-210 can affect their expression and in turn functioning which can be associated with PE risk and severity. This study is the very first to determine the effect of miR-210 expression levels on associated genes in plasma samples.
Keywords: Gene expression, Hypoxia, microRNA-210, Plasma, Preeclampsia
Abbreviations: ACOG, American College of Obstetricians and Gynecologists; cDNA, Complementary DNA; HIF, hypoxia inducible factor; KCMF1, potassium channel modulatory factor 1; mRNA, messenger RNA; miRNA, Micro RNA; PCR, polymerase chain reaction; PE, preeclampsia; RT-PCR, quantitative real time PCR
1. Introduction
Preeclampsia (PE) is a leading cause of maternal and fetal morbidity worldwide [1]. The disease is characterized by proteinuria and hypertension [2], approximately affecting 2–8% of pregnant women [3,4]. Several risk factors contribute to the PE pathogenesis, including pregnancy age, times, hypertension, anemia, overweight/obesity, urinary tract infection, and hereditary factors [5,6].
PE pathology is classified into mild and severe. According to ACOG, mild PE is defined by increased maternal systolic blood pressure exceeding 140 mm Hg, and/or diastolic blood pressure exceeding 90 mm Hg on two occasions separated by 6-h apart, with a significant proteinuria (≥300 mg of protein in a 24-h urine specimen or ≥1+ by dipstick) following 20 weeks of gestation [7]. On the other hand, severe PE is characterized by an increase in the systolic blood pressure exceeding 160 mm Hg, and/or diastolic blood pressure exceeding 110 mm Hg on at least 2 occasions 6-h apart, with either evidence of mild proteinuria, or mild hypertension plus severe proteinuria (≥2 g/24-h or ≥2+ by dipstick) following 20 weeks of gestation [7]. Coupled with clinical pressures surrounding the risk of under-diagnosis, this has resulted in a dilution of diagnostic specificity for PE [8]. Thus, a need to clarify the underlying pathogenic mechanisms of PE, to unambiguously distinguish the condition and open the doors to early prognostic and therapeutic measures.
The pathogenesis of PE is not clearly understood, although it has been generally believed that an abnormal development of the placenta at early gestation may be the critical cause of this disease [9]. Specifically, impaired trophoblast invasion and aberrant spiral arterial remodeling are thought to decrease uteroplacental perfusion and excessive placental hypoxia/ischemia, causing the release of soluble factors or cell debris which further damage the maternal endothelium [10]. Recently, accumulating evidence has indicated that the abnormal expression of miRNAs in the placenta is associated with preeclampsia [9,11,12]. MicroRNAs are 22- to 24-nucleotide noncoding RNAs that regulate gene expression by seed sequence pairing with the 3′-untranslated region (UTR) of target mRNAs, leading to repressed translation or induced mRNA cleavage of the target genes [13]. MiRNAs have been identified as essential mediators of numerous cellular processes [14], potentially including the response to hypoxia. Moreover, they are reported to play critical roles in the regulation of uteroplacental activities which participate in the pathogenesis of PE.
Studies have revealed that miR-29b promotes PE via regulating the apoptosis and invasion of trophoblasts [15]. MiR-126 is downregulated in the placentas of PE patients, and it correlates with the decreased expression of vascular endothelial growth factor (VEGF) [16]. MiR-210, a member of hypoxia-induced miRs, also known as hypoxia-miRs, is ubiquitously expressed in a variety of cells including mammary epithelial and trophoblast cells [17]. MiR-210 is specifically induced by hypoxia-inducible factor-1α during hypoxia [18], and regulates many hypoxia response pathways, including cell survival, angiogenesis, mitochondrial metabolism, and DNA repair [19]. MiR-210 can target and suppress many genes, including the cell cycle regulator E2F transcription factor 3, the receptor tyrosine kinase ligand ephrinA3, the DNA repair protein RAD52, and the iron-sulfur cluster assembly proteins 1/2 [14,20]. Recent studies revealed that upregulated expression of miR-210 is seen in patients with preeclampsia [21] and demonstrated that hypoxia-inducible miR-210 might participate in the occurrence of preeclampsia [22]. The repressive effect of miR-210 on primary trophoblast cell invasion has also been reported [22]. Besides, the study of Luo et al. [9] revealed that miR-210 could be a potential serum biomarker for preeclampsia. Their study was the first to report that aberrant miR-210 expression may contribute to the occurrence of preeclampsia by interfering with KCMF1-mediated signaling in the human placental trophoblast cells. A recent study has shown an association between mRNA/miRNA network involving miR-210, miR-20, iron sulfur-cluster assembly, hypoxia-inducible factor 1α, cytochrome c oxidase-10, phosphatase, tensin homolog and DNA repair protein RAD52 expression levels and potential intrauterine insults like fetal growth restriction [10]. Despite these findings, the details of the role played by miR-210 in preeclampsia development remain elusive.
An aberrant miR-210 expression is present not only in solid tumors and harmed organs but is secreted into the circulation, allowing detection in plasma [23]. It should be noted that all the studies on the effect of miR-210 on gene expression to date have focused on only severe PE, and parallel studies involving mild PE have not been performed. Our earlier studies show a marked variation in the extent of differential expression of miR-210 [21]. We speculate this difference can in turn show variation in its effect on gene expression, according to PE severity and on its pathogenesis. The present study addresses the miR-210 expression patterns in plasma from severe PE patients compared to mild PE patients, and how the difference affects the differential expression of miR-210 target genes in both cases. It aims at evaluating potential hypoxia-induced pathways that may be involved in PE pathophysiology.
2. Materials and methods
2.1. Ethics approval and consent to participate
Informed consent for sample collection were obtained from all participants. The project was approved by the Ethics Committee of Salmaniya Medical Complex and Arabian Gulf University, Kingdom of Bahrain (Ethical approval number/Project Number: 32-PI-01/15). The study was performed following the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments. All the design, analysis, interpretation of data, drafting, and revisions followed the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies, available through the Enhancing the Quality and Transparency of Health Research (EQUATOR) network.
2.2. Sample collection
The study was carried out on 30 Arab women with a confirmed diagnosis of PE, who were recruited into the study between November 2017 and January 2018. Among them, 15 was presented with mild PE and another 15 presented with severe PE. Blood pressure, gestational age, and urine protein were assessed to screen PE, which was classified as per ACOG guidelines [7]. Mild PE patients had systolic blood pressure ≥140 mm Hg, diastolic blood pressure ≥90 mm Hg on 2 occasions separated by 6 h, and proteinuria (≥300 mg of protein in a 24-h urine specimen or ≥1+ by dipstick) [7]. On the other hand, severe PE was confirmed if one or more of these conditions were met: persistent systolic blood pressure ≥160 mm Hg and/or diastolic blood pressure ≥110 mm Hg on least at 2 occasions 6 h apart; proteinuria (≥2 g/24 h or ≥2+ by dipstick); and serum creatinine ≥106 mmol/l (except patients whose serum creatinine was increased before the test), low platelet count (<105/ml); increased lactate dehydrogenase; elevated liver enzyme function (ALT, AST), and persistent upper abdominal pain [7]. A total of 15 healthy Arab women were included as controls. Inclusion criteria for the control group included no previous history of hypertension, cardiovascular disease, hepatitis, kidney disease, diabetes, and any evidence of intrapartum infection or other complications of pregnancy such as fetal anomalies or chromosomal abnormalities. All women included in the study were in their third trimester of pregnancy. Venous blood samples of study subjects were collected in 4-ml EDTA-K2 tubes, which were kept at room temperature for 30 min, and then centrifuged at 1000 g for 10 min to collect plasma, which was stored at −40 °C until use.
2.3. Total RNA isolation for extraction of miRNA and mRNA
Total RNA was isolated from plasma using miRNeasy serum/plasma kit, as per the manufacturer protocol (Qiagen, Valencia, CA). Synthetic C. elegans miRNA cel-miR-39 (Qiagen, Valencia, CA), was added to denatured samples following inhibition of endogenous RNase activity, to normalize variation in RNA isolation from different samples, as described [21]. miRNA was eluted with 110 μl elution solution. Similarly, mRNA extraction was done using QIAamp RNA blood Minikit from fresh blood (Qiagen, Valencia, California, USA), according to the manufacturer's protocol. Briefly Given the requirements for highly pure and optimal RNA amounts, all RNA samples will be screened for purity by measuring optical density (OD) at 260 nm (OD260) and 280 nm (OD280). miRNA or mRNA concentration (μg/ml) will be calculated based on the OD260 reading.
2.4. Real-time reverse transcription-polymerase chain reaction for miR-210 expression assay
MiR-210 expression in control, as well as sPE and mPE samples, was performed using 2 μg of total RNA was used for miRNA reverse transcription using the TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, lnc., Foster City, CA, USA). Then, the expression levels of miRNA were determined on an ABI PRISM 7900HT PCR system (Applied Biosystems, Foster City, CA, USA) with the following system: for the quantification, U6 was used to normalize miRNA. Primers of miR-210 were designed as follows: miR-210-forward,5-GTGCAGGGTCCGAGGT-3, and miR-210-reverse: 5-CTGTGCGTGTGACAGCGGCTGA-3; U6-forward,5-CTCGCTTCGGCAGCACA-3, and U6-reverse,5-ACGCTTCACGAATTTGCGT-3. Then, the fold changes of miR-210 were calculated using the 2–ΔΔCt method [24]. Each sample was analyzed in duplicate, and the mean value was set as the final expression value. The final results were reported as relative expression by setting the expression values of miR-210 in controls as 1 and the expression value of other samples was calculated relative to this control [25].
2.5. Reverse transcription real-time PCR for RT2 profiler PCR arrays
To carry out the assay of gene targets of miR-210, the conversion of mature mRNA to cDNA was done by RT2 First Strand Kit (Qiagen, Valencia, California, USA), according to the manufacturer's protocol. Briefly, 5 μg of RNA was taken along with 3 μl of buffer and the volume was made up to 10 μl using RNase-free water. Then 10 μl of reverse transcription mix was added to it followed by incubation at 42 °C, for 15 min and then immediately reaction was stopped by incubating it at 95 °C for 5 min. The reaction was done in a PCR thermocycler. Once the reaction was complete then 82 μl of RNase-free water was added and subjected to real-time PCR for PCR array studies. Gene expression detection was be carried out on human miR-210 targets RT2 profiler PCR Array using RT2 SYBR Green PCR Kit (Qiagen, Valencia, California, USA) according to the manufacturer's protocol. RT2 profiler PCR Array enabled assessment of multiple cDNA samples using real-time PCR. Each Array plate, apart from 84 genes includes 5 housekeeping genes, a genomic DNA control, reverse-transcription controls, and contain a positive PCR control (Human miR-210 Targets gene list, Qiagen, Valencia, California, USA). Briefly, the reaction mixture for a 96-well plate was; 1350 μl of 2 X SYBR Green Master Mix, 1248 μl of RNase-free water and 102 μl of template cDNA. A total of 25 μl of reaction mixture was added to each well of 96-well plate. The reaction was run on Applied Biosystems StepOne Plus PCR system as follows: 1 cycle was run at 95 °C for 10 min and then 40 cycles were run with each cycle consisting of 15 s at 95 °C, 30–40 s at 55 °C and 30 s at 72 °C. Open-array real-time qPCR analysis software was used to analyze and review the amplification plots and threshold cycle values obtained. The ΔCT for each pathway-focused gene was calculated using the CT values for the gene of interest (GOI) and the housekeeping gene used for normalization (HKG) using the formula: ΔCT = CTGOI – CTAVG HKG. For every gene, ΔΔCT was calculated using the formula: ΔΔCT = ΔCT (group 2) - ΔCT (group 1), where group 1 is the control and group 2 is the sPE or mPE samples. Finally, the fold change for each gene from group 1 and group 2 as 2(-ΔΔCT). Data analysis was performed with DataAssist™ Software (Applied Biosystems) through global mean normalization of all genes.
2.6. Statistical analysis
In the current study, SPSS 20.0 software (IBM, SPSS, Chicago, IL, USA) was used to perform statistical analysis. The characteristics of study subjects were compared using Student t-test. The significance of gene amplifications was determined by Mann-Whitney U test for 2 group comparison, and Kruskal-Wallis test for multiple comparisons; P < 0.01 was considered statistically significant.
3. Results
3.1. Clinical characteristics of study subjects
The main clinical characteristics for the study groups are summarized in Table 1. All subjects in each category were Bahraini Arabs and all pregnancies were primiparas. There was no significant difference between maternal age and pre-pregnancy body mass index or smoking among the study groups and controls. However, the mPE and sPE group differed significantly in gestational age at delivery, birth weight, blood pressure protein content in urine, mode of delivery and number of a female fetus. Studies have shown that the sex of the fetus impacts the maternal immune and in turn, determines the risk of developing preeclampsia [26].
Table 1.
Clinical characteristics of normal and preeclamptic patients.
| Characteristics | Controls (n = 15) | Mild Preeclamptic patients (n = 15) | Severe Preeclamptic patients (n = 15) |
|---|---|---|---|
| Maternal Age (years) | 30 (25–35) | 32 (29–35) | 33 (29–37) |
| Prepregnancy body mass weight (Kg/m2) | 24 (22–26) | 26 (21–31) | 25 (22–29) |
| Gestational age at delivery (weeks) | 39 (37–41) | 32 (29–35) | 30 (27–33) |
| Birth weight (Kg) | 3.3 (3.1–3.5) | 2.7 (2.1–3.3) | 2.4 (2.1–2.7) |
| Systolic Blood Pressure (mmHg) | 105 (90–120) | 143 (140–146) | 172 (165–179) |
| Diastolic Blood Pressure (mmHg) | 71 (65–77) | 96 (90–102) | 112 (95–129) |
| Proteinuria (g/24 h) | Normal | 2.2 (1.8–2.6) | 3.4 (3.0–3.8) |
| Mode of delivery | 10 vaginal (67%) | 7 vaginal (46%) | 4 vaginal (27%) |
| (Cesarean/Vaginal) | 5 cesarean (33%) | 8 cesarean (54%) | 11 cesarean (73%) |
| Female fetus (n) | 5 (33%) | 7 (46%) | 10 (66%) |
| Smoker | 0 (0%) | 0 (0%) | 0 (0%) |
Values expressed as median (range) whereas for mode of delivery, fetus gender and smoking the values are as percentages.
n = number.
3.2. Relative expression of microRNA-210 using real time-PCR
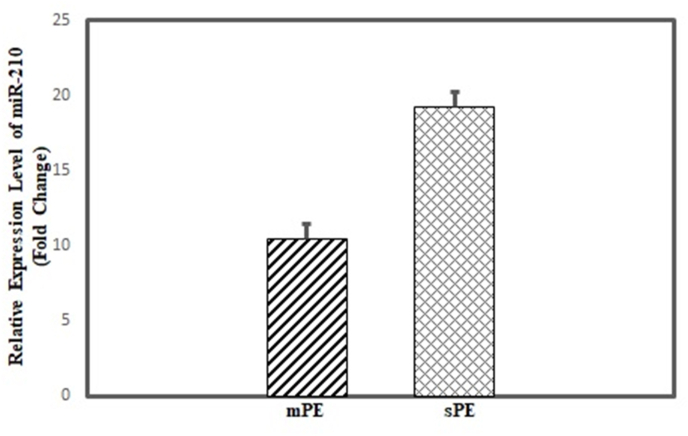
Total miRNA isolated from plasma of controls, mild PE and severe PE was subjected to real-time PCR using specific miR-210 primers. Based on P˂0.01 and fold change of above 2 cut off, both the sample category (mPE and sPE) showed significant relative expression in comparison to control samples (Fig. 1). The mPE showed a fold change of 10.43 (P = 0.005) whereas sPE samples showed an even higher expression with a fold change of 19.20 (P = 0.003). This indicates that miRNA-210 is highly upregulated in preeclamptic patients as compared to control subjects.
Fig. 1.
Relative expression of microRNA-210 using real time-PCR.
Relative expression of microRNA-210 measured in mild preeclampsia (mPE)
patient (15) and severe preeclampsia (sPE) patient (15) plasma samples. The x axis shows the sample category whereas the y axis indicates the expression levels of microRNA-210 in each category with respect to control samples (15). The expression levels plotted are the mean of all the samples under each category. The error bars show standard error of the mean (Mean ± SEM).
3.3. Gene targets of microRNA-210 profiling according to PE severity versus the controls
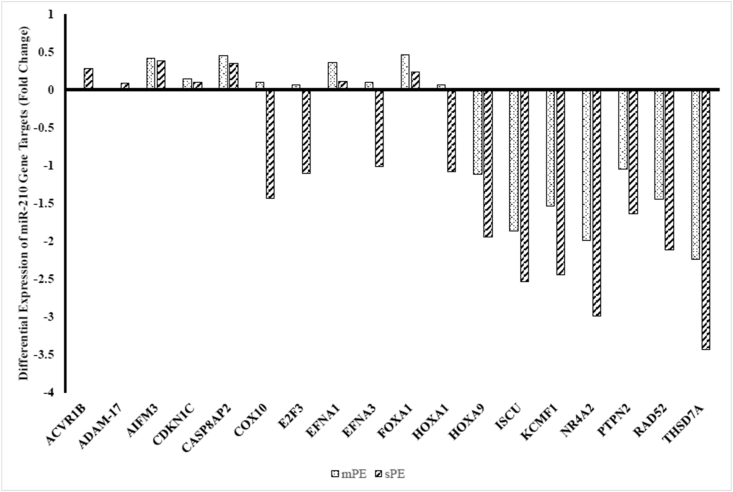
To carry out assay of gene targets of miR-210, a pathway focused RT2 profiler PCR Array was used which allowed assessment of multiple cDNA samples using mRNA in real-time PCR. Out of the 84 hypoxia miR-210 associated genes screened, 39 were correctly amplified in plasma of sPE cases whereas 36 genes in mPE cases versus control subjects (Ct ˂31). A total of 45 samples were screened out of which 15 were of control subjects and 15 of mPE and 15 sPE cases. A good correlation of Ct values in controls and cases was seen, thereby reinforcing the study consistency and reliability. Raw data obtained were analyzed by online RT2 PCR array data analysis software (Qiagen, Valencia, CA). The two case groups were compared with the control women. All the genes amplified in both the PE groups were found as downregulated in comparison to controls. Based on a conservative selection criterion (fold change ˂ 0.5 and p-value ˂ 0.01), 18 genes were found to be differentially expressed in sPE whereas 16 genes in mPE cases with varying magnitude (Fig. 2,Table 2). Overall, the genes were more downregulated in severe cases than mild ones and their symbol and function is described in Table 3.
Fig. 2.
Differentially expressed microRNA-210 target genes in severe and mild preeclampsia patients versus controls.
All the genes amplified in both the preeclampsia groups i.e., mild and severe, were found downregulated in comparison to controls. A total of 18 genes were found to be differential expressed in severe PE whereas 16 genes in mild PE cases with varying magnitude. The x axis shows all the genes that were differentially expressed in both the cases whereas as y axis indicate their fold change with respect to control samples. The expression of genes ACVR1B and ADAM-17 was not observed for mild PE.
Table 2.
Differentially expressed microRNA-210 target genes in severe and mild preeclampsia plasma samples versus controls.
| Gene Symbol | Mild PE Fold change |
P ˂ 0.01* | Severe PE Fold change |
P ˂ 0.01* |
|---|---|---|---|---|
| ACVR1B | NA | NA | 0.283 | 0.0094 |
| ADAM-17 | NA | NA | 0.089 | 0.0084 |
| AIFM3 | 0.421 | 0.0054 | 0.378 | 0.0042 |
| CDKN1C | 0.144 | 0.0034 | 0.100 | 0.0076 |
| CASP8AP2 | 0.445 | 0.0069 | 0.345 | 0.0091 |
| COX10 | 0.097 | 0.0053 | −1.44 | 0.0071 |
| E2F3 | 0.065 | 0.0042 | −1.10 | 0.0095 |
| EFNA1 | 0.356 | 0.0036 | 0.111 | 0.0081 |
| EFNA3 | 0.094 | 0.0065 | −1.02 | 0.0091 |
| FOXA1 | 0.466 | 0.0063 | 0.236 | 0.0072 |
| HOXA1 | 0.064 | 0.0025 | −1.08 | 0.0079 |
| HOXA9 | −1.12 | 0.0075 | −1.94 | 0.0027 |
| ISCU | −1.86 | 0.0093 | −2.54 | 0.0052 |
| KCMF1 | −1.54 | 0.0034 | −2.44 | 0.0054 |
| NR4A2 | −1.99 | 0.0068 | −2.99 | 0.0019 |
| PTPN2 | −1.05 | 0.0039 | −1.64 | 0.0062 |
| RAD52 | −1.45 | 0.0025 | −2.12 | 0.0047 |
| THSD7A | −2.24 | 0.0017 | −3.43 | 0.0053 |
*P value is for fold change observed for each gene amplified. A conservative selection criterion (fold change ˂ 0.5 and p value ˂ 0.01) was considered.
NA: Not Applicable.
Table 3.
Details of the genes differentially expressed in preeclamptic patients (mild and severe) as compared to controls.
| Gene Bank | Gene Symbol | Description |
|---|---|---|
| NM_004302 | ACVR1B | Activin A receptor, type IB |
| NM_003183 | ADAM-17 | metalloproteinase domain gene |
| NM_144704 | AIFM3 | Apoptosis-inducing factor, mitochondrion-associated, 3 |
| NM_001362 | CDKN1C | Cyclin Dependent Kinase Inhibitor 1C |
| NM_012115 | CASP8AP2 | Caspase 8 associated protein 2 |
| NM_001303 | COX10 | COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase |
| NM_001949 | E2F3 | E2F transcription factor 3 |
| NM_182685 | EFNA1 | Ephrin-A1 |
| NM_004952 | EFNA3 | Ephrin-A3 |
| NM_004496 | FOXA1 | Forkhead box A1 |
| NM_005522 | HOXA1 | Homeobox A1 |
| NM_152739 | HOXA9 | Homeobox A9 |
| NM_014301 | ISCU | Iron-sulfur cluster scaffold homolog |
| NM_020122 | KCMF1 | Potassium channel modulatory factor 1 |
| NM_005126 | NR4A2 | Nuclear receptor subfamily 4, group A, member 2 |
| NM_002827 | PTPN2 | Protein tyrosine phosphatase, non-receptor type 2 |
| NM_134424 | RAD52 | RAD52 homolog |
| NM_015204 | THSD7A | Thrombospondin, type I, domain containing 7A |
4. Discussion
Preeclampsia has been coined “the disease of theories” since its precise origin remains elusive. Although, fetal sonography is considered an important clinical tool for monitoring of the fetus and the mother [27]. Differential expression of miRNAs in preeclamptic placentae and plasma has been reported by several groups [9,21] and the molecular mechanisms by which these miRNAs participate in the regulation of placental cell function has elucidated some aspects of the pathogenesis of preeclampsia related to angiogenesis, apoptosis, or cell cycle of placental cells [28]. Since a single miRNA is very often involved in several molecular pathways in pregnancy, we used miR-210 as a model for the study of miRNA overexpression in PE. Among hypoxia-regulated miRNA, miR-210 has been reported to be involved in several diseases. Like McCormick et al. [29] demonstrated that miR-210 is a target of HIF-1 and HIF-2 and is closely correlated with the prognosis of patients with renal cancer. MiR-210 is among the top members of a list of miRNAs with predominant overexpression in preeclamptic placentae [9] and maternal plasma at early to mid-gestation [21]. Elucidation of the underlying mechanisms by which miR-210 exerts its effects is likely to provide a better understanding of the pathophysiology of PE and uncover new targets for therapeutic intervention. In the current study, we explored the expression patterns of miR-210 in plasma from sPE patients and mPE patients, and how the difference in relative expression can affect the differential expression of miR-210 target genes in both cases.
Our pilot study was concentrated on the expression of miR-210 gene targets in plasma samples since increased miRNA expression suggests the downregulation of potential target mRNAs which may contribute to PE pathophysiology. Using a specific primer and real-time PCR, we confirmed that miR-210 expression was upregulated in plasma samples of sPE and mPE as compared to control samples. Interestingly, previous studies have shown that miR-210 is predominantly elevated in placental and plasma samples of patients with PE [12,30]. Many pathway studies have contributed that miR-210 is specifically induced by hypoxia-inducible factor-1α during hypoxia and regulates many hypoxia response pathways [18]. Furthermore, we investigated an array of 84 genes that are associated with miR-210 and in both the cases (sPE and mPE), genes were found to be downregulated with a degree of variation. Based on a strict criterion, 18 genes (ACVR1B, ADAM-17, AIFM3, CDKN1C, CASP8AP2, COX10, E2F3, EFNA1, EFNA3, FGFRL1, FOXA1, HOXA1, HOXA9, ISCU, KCMF1, NR4A2, PTPN1, RAD52, THSD7A) were differentially expressed in sPE and 16 genes (AIFM3, CDKN1C, CASP8AP2, COX10, E2F3, EFNA1, EFNA3, FOXA1, HOXA1, HOXA9, ISCU, KCMF1, NR4A2, PTPN2, RAD52, THSD7A) in mPE samples in comparison to controls. Gene type expressed in both the cases was the same except ACVR1B and ADAM-17 were not seen expressed in the mPE samples.
ACVR1B is an activin A receptor, type 1B gene, downregulated in sPE samples of our study which codes for transmembrane serine/threonine kinase activin type-1 receptor forming an activin receptor complex with activin receptor type-2 (ACVR2A or ACVR2B) [31,32]. The receptor complex interacts with activin and regulates many physiological processes including neuronal differentiation and neuronal survival, extracellular matrix production, immunosuppression [32]. Since the ACVR1B portion of the receptor complex act as downstream mediators of activin signal, therefore, downregulation of its gene would result in overall blockage of the physiological activities which include many processes responsible for physiology of normal pregnancy. ACVR1B gene has been known to be associated with PE [33].
ADAM-17 is a metalloproteinase domain gene that controls various placental cellular activities. Like our findings, its downregulation has also been reported in placental samples of preeclamptic patients and other pregnancy-associated complications in a previous study by Mayor-Lynn and group [25]. The AIFM3 gene codes for a protein known as apoptosis-inducing factor mitochondrion-associated 3 which induces apoptosis [34]. Although AIFM3 is widely expressed in various tissues, the function of AIFM3 in the occurrence and development progress of PE has not been reported till date. This is the first study which reports its differential expression in association with PE. It is widely known that AIFM3 is a direct target of miR-210 which have been related to the proliferation of human hepatoma cells [35]. Another apoptosis-associated gene screened in our study is CASP8AP2 which is described as caspase 8 associated protein 2 [36]. It acts as a downstream mediator for CASP8-induced activation of Necrosis Factor-kappa-β and histone gene transcription and progression through the S phase [36]. Differential expression of CASP8AP2 has been extensively explored in association with PE and our study provides supporting evidence to the findings of previous studies [37].
CDKN1C gene known as cyclin dependent kinase inhibitor 1C, helps in regulating growth and cell cycle. Association of CDKN1C with PE has been earlier reported by Enquobahrie et al. [38]. However, our findings were in contrast to their findings as they showed upregulation of the gene. Another gene expression investigated by Enquobahrie et al. in PE was downregulation of the NR4A2 gene, which is a nuclear receptor subfamily 4, group A, member 2, is involved in cell growth and proliferation, connective tissue development and function. Observations of our study for NR4A2 expression in both mPE and sPE is similar to that of Enquobahrie et al. study [38].
COX10 a cytochrome c oxidase assembly protein, RAD52 a DNA repair protein-coding gene and ISCU an iron-sulfur cluster scaffold homolog works downstream of miR-210 is part of the placental hypoxia-responsive network. Tight regulation of this network is required to prevent the development of maternal-fetal complications [10]. RAD52 is a key player in DNA double-strand break repair and homologous recombination [39]. COX10 gene is involved in the mitochondrial heme biosynthetic pathway and catalyzes the conversion of protoheme (heme B) to heme O [40]. The ISCU gene encodes a component of the iron-sulfur (Fe–S) cluster scaffold which act as cofactors for enzymes, including those that regulate metabolism, and oxidative stress response [41]. All these genes mediate the effects of miR-210 participating in oxidative stress and mitochondrial metabolism, angiogenesis [42], cell survival [43], DNA repair [10], and cell migration and invasion [30]. All these genes have not been experimentally demonstrated till date to be associated with PE and ours is the first study to report so.
Previous studies have revealed that miR-210 could repress trophoblast cell invasion and migration via modulating KCMF1 [14], EFNA3, HOXA9 [30], and THSD7A [9]. In the current study, downregulation of all these genes along with additional genes belonging to the same families ie E2F3-E2F transcription factor 3, EFNA1-Ephrin-A1 (belonging to ephrin family along with EFNA3) and HOXA1-Homeobox A1(belonging to homeobox family along with HOXA9) was observed in both mPE and sPE samples. KCMF1 gene is known to code a 42-kDa zinc finger protein with a high serine and threonine content. Luo and group were the first to report significantly lower levels of KCMF1 in preeclamptic placenta tissues than in gestational week–matched normal placentas, which was inversely correlated with the level of miR-210 [14]. Their data indicate that aberrant miR-210 expression may contribute to the occurrence of PE by interfering with KCMF1-mediated signaling in the human placenta [14]. A similar study by the same group on placental tissues and HTR8/SVneo cells showed that hypoxia-inducible miR-210 contributes to PE via targeting thrombospondin type I domain-containing 7A (THSD7A) [9]. THSD7A is expressed in placenta vasculature and umbilical vein endothelial cells; promotes focal adhesion assembly and inducing angiogenesis [44]. Also, a study by Zhang et al. showed repression in HOXA9 and EFNA3 in response to upregulated miR-210 in mild and severe preeclampsia placental samples [30]. Another gene related to trophoblast cells is PTPN2, found differentially downregulated in both cases of our study. It is a protein tyrosine phosphatase, non-receptor type 2 and an important member of the protein-tyrosine phosphatase (PTP) family [45]. PTPN2 is involved in the regulation of T-cells and its related biological process [45]. The study of Li et al. showed the hypoxia induces the elevation of miR-210 which then promotes the invasion of trophoblast cells as well as the failure of arterial remodeling, by downregulating PTPN2, leading to the occurrence and development of PE via the PDG-FR-Akt signaling pathway [45]. Similarly, FOXA1 is fork head box A1 gene that encodes a member of the forkhead class of DNA-binding proteins that interact with chromatin [46]. Our findings are supported by the study of Zhu et al., 2020 who observed expression of FOXA1 in the placentas of early-onset preeclampsia was significantly lower than of normal pregnant women. Further, in vitro, functional studies showed that silencing FOXA1 increased apoptosis and inhibited the migration and invasion of trophoblast cells [47].
Taken together, the present study demonstrated that upregulated miR-210 plays a critical role in the development of PE by targeting and downregulating several different genes that otherwise play a vital role in maintaining the normal physiology of pregnancy. Hence both miR-210 and the downstream genes could serve as potential biomarkers and therapeutic targets of PE. Some limitations restrict the power of this study. Firstly, the small sample size which possibly can limit the detection of an actual number of miR-210 associated genes. Thus a bigger sample size is needed to replicate and our findings and validate the involvement of specific genes identified in our stuy. Also, as a dynamic event, a single measure of RNA species that is dependent on half-life, degradation rate and stability may not describe adequately the full picture of gene expression in PE. Despite few shortcomings, our study has several strengths. It is the first to determine the effect of miR-210 expression levels on the expression of the associated gene in plasma samples. Whatever studies done previously have been on placental samples. Additionally, this study is the only one till date that has been done on PE samples categorized according to the severity (mPE and sPE). We were also able to report a few new genes that have not been earlier experimentally proven to be associated with PE like AIFM3, COX10, RAD52, ISCU, E2F3, EFNA1 HOXA1.
5. Conclusion
In conclusion, we report here that high expression levels of miR-210 are found in the plasma of PE patients varying in its relative expression depending on the severity of the disease. We further identify a number of genes targeted by miR-210 which were differentially downregulated depending on the PE type. Our study can complement elucidating the biological function of miR-210, the in-house manager of the hypoxia pathway. Understanding the expression profile of genes affected by miR-210 is a step forward towards understanding molecular events that are associated with PE. This may provide the basis for designing new prevention and treatment strategies to improve reproductive outcomes.
Funding
This work was supported by a research grant (Project Number: 32-PI-01/15) awarded to the authors by the College of Medicine and Medical Sciences Research and Ethics Committee, Arabian Gulf University, Kingdom of Bahrain.
Patient consent for publication
Informed consent for publication were obtained from all participants of this study.
Author's contributions
Deeba S. Jairajpuri: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing-Original Daft, Visualization, Supervision, Project administration, Funding acquisition. Zainab H. Malalla: Data Curation, Methodology. Sameh Sarray: Formal Analysis, Writing-Review and Editing. Naeema Mahmood: Resources, Investigation.
CRediT authorship contribution statement
Deeba S. Jairajpuri: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing – original draft, Visualization, Supervision, Project administration, Funding acquisition. Zainab H. Malalla: Data curation, Methodology. Sameh Sarray: Formal analysis, Writing – review & editing. Naeema Mahmood: Resources, Investigation.
Declaration of competing interest
The authors have no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2021.03.001.
Contributor Information
Deeba S. Jairajpuri, Email: deebasj@agu.edu.bh.
Zainab H. Malalla, Email: zainmal@live.com.
Sameh Sarray, Email: samehmss@agu.edu.bh.
Naeema Mahmood, Email: naeemamahmood@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Eastwood K., Patterson C., Hunter A., Mccance D.R., Young I., Holmes V. Evaluation of the predictive value of placental vascularization indices derived from 3-dimensional power Doppler whole placental volume scanning for prediction of pre-eclampsia: a systematic review and meta-analysis. Placenta. 2017;51:89–97. doi: 10.1016/j.placenta.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed R., Dunford J., Mehran R., Robson S., Kunadian V. Pre-eclampsia and future cardiovascular risk among women: a review. J. Am. Coll. Cardiol. 2009;63:1815–1822. doi: 10.1016/j.jacc.2014.02.529. [DOI] [PubMed] [Google Scholar]
- 3.Duley L. The global impact of pre-eclampsia and eclampsia. Semin. Perinatol. 2009;33:130–137. doi: 10.1053/j.semperi.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Abalos E., Cuesta C., Grosso A., Chou D., Say L. Global and regional estimates of preeclampsia and eclampsia:a systematic review. Eur. J. Obstet. Gynecol. Reprod. Biol. 2013;170:1–7. doi: 10.1016/j.ejogrb.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Liu S., Joseph K., Liston R., Bartholomew S., Walker M., León J., Kirby R., Sauve R., Kramer M. Incidence, risk factors, and associated complications of eclampsia. Obstet. Gynecol. 2011;118:987–994. doi: 10.1097/AOG.0b013e31823311c1. [DOI] [PubMed] [Google Scholar]
- 6.Olaya-Garay S., Velasquez-Trujillo P., Vigil-De G. Blood pressure in adolescent patients with pre-eclampsia and eclampsia. Int. J. Gynaecol. Obstet. 2017;138:335–339. doi: 10.1002/ijgo.12237. [DOI] [PubMed] [Google Scholar]
- 7.ACOG practice bulletin Diagnosis and management of preeclampsia and eclampsia. Obstet. Gynecol. 2002;99:159–167. doi: 10.1016/s0029-7844(01)01747-1. [DOI] [PubMed] [Google Scholar]
- 8.Sheikh A., Small H., Currie G., Delles C. Systematic review of micro-RNA expression in pre-eclampsia identifies a number of common pathways associated with the disease. PloS One. 2016;11(8) doi: 10.1371/journal.pone.0160808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo R., Wang Y., Xu P., Cao G., Zhao Y., Shao X., Li Y.X., Chang C., Peng C., Wang Y. Hypoxia-inducible miR-210 contributes to preeclampsia via targeting thrombospondin type I domain containing 7A. Sci. Rep. 2016;6:19588. doi: 10.1038/srep19588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vrijens K., Tsamou M., Madhloum N., Gyselaers W., Nawrot T. Placental hypoxia-regulating network in relation to birth weight and ponderal index: the ENVIRONAGE Birth Cohort Study. J. Transl. Med. 2018;16:2. doi: 10.1186/s12967-017-1375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betoni J., Der r K., Pahl M., Rogers L., Muller C., Packard R., Carey D., Kuivaniemi H., Tromp G. MicroRNA analysis in placentas from patients with preeclampsia: comparison of new and published results. Hypertens. Pregnancy. 2013;32:321–339. doi: 10.3109/10641955.2013.807819. [DOI] [PubMed] [Google Scholar]
- 12.Ura B., Feriotto G., Monasta L., Bilel S., Zweyer M., Celeghini C. Potential role of circulating microRNAs as early markers of preeclampsia, Taiwan. J. Obstet. Gynecol. 2014;53:232–234. doi: 10.1016/j.tjog.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Bartel D. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo R., Shao X., Xu P., Liu Y., Wang Y., Zhao Y., Liu M., Ji L., Li Y., Chang C., Qiao J., Peng C., Wang Y. MicroRNA-210 contributes to preeclampsia by downregulating potassium channel modulatory factor 1, Hypertension. 2014;64:839–845. doi: 10.1161/HYPERTENSIONAHA.114.03530. [DOI] [PubMed] [Google Scholar]
- 15.Li P., Guo W., Du L., Zhao J., Wang Y., Liu L., Hu Y., Hou Y. MicroRNA-29b contributes to pre-eclampsia through its effects on apoptosis, invasion and angiogenesis of trophoblast cells. Clin. Sci. 2013;124:27–40. doi: 10.1042/CS20120121. [DOI] [PubMed] [Google Scholar]
- 16.Hong F., Li Y., Xu Y. Decreased placental miR-126 expression and vascular endothelial growth factor levels in patients with pre-eclampsia. J. Int. Med. Res. 2014;42:1243–1251. doi: 10.1177/0300060514540627. [DOI] [PubMed] [Google Scholar]
- 17.Koushki M., Amiri D., Omidi-Ardali H., Rezaei T. Assessment of correlation between miR-210 expression and pre-eclampsia risk: a meta-analysis. Rep. Biochem. Mol. Biol. 2018;7:94–101. [PMC free article] [PubMed] [Google Scholar]
- 18.Kulshreshtha R., Ferracin M., Wojcik S., Garzon R., Alder H., Agosto-Perez F., Davuluri R., Liu C., Croce C., Negrini M., Calin G., Ivan M. A microRNA signature of hypoxia. Mol. Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X., Le Q.T., Giaccia A.J. MiR-210–micromanager of the hypoxia pathway. Trends Mol. Med. 2010;16:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crosby M.E., Kulshreshtha R., Ivan M., Glazer P.M. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Canc. Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jairajpuri D.S., Malalla Z.H., Mahmood N., Almawi W.Y. Circulating microRNA expression as predictor of preeclampsia and its severity. Gene. 2017;627:543–548. doi: 10.1016/j.gene.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Anton L., Olarerin-George A.O., Schwartz N., Srinivas S., Bastek J., Hogenesch J.B., Elovitz M.A. MiR-210 inhibits trophoblast invasion and is a serum biomarker for preeclampsia. Am. J. Pathol. 2013;183:1437–1445. doi: 10.1016/j.ajpath.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrie C., Gal S., Dunlop H., Pushkaran B., Liggins A., Pulford K., Banham A., Pezzella F., Boultwood J., Wainscoat J., Hatton C., Harris A. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 24.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Mayor-Lynn K., Toloubeydokhti T., Cruz A.C., Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod. Sci. 2011;18:46–56. doi: 10.1177/1933719110374115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor B.D., Ness R.B., Klebanoff M.A., Tang G., Roberts J., Hougaard D., Louring-Skogstrand K., Haggerty C. The impact of female fetal sex on preeclampsia and the maternal immune milieu. Pregnancy Hypertens. 2018;12:53–57. doi: 10.1016/j.preghy.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visnovsky J., Kudela E., Nachajova M., Danko J. The examination of superior mesenteric artery circulation in fetus during pregnancy. J. Matern. Fetal Neonatal Med. 2015;10:1–5. doi: 10.3109/14767058.2015.1038991. [DOI] [PubMed] [Google Scholar]
- 28.Fu G., Brkic J., Hayder H., Peng C. MicroRNAs in human placental development and pregnancy complications. Int. J. Mol. Sci. 2013;14:5519–5544. doi: 10.3390/ijms14035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCormick R.I., Blick C., Ragoussis J., Schoedel J., Mole D.R., Young A.C., Selby P.J., Banks R.E., Harris A.L. MiR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br. J. Canc. 2013;108:1133–1142. doi: 10.1038/bjc.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Fei M., Xue G., Zhou Q., Jia Y., Li L., Xin H., Sun S. Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia, new insights into molecular mechanisms for the disease. J. Cell Mol. Med. 2012;16:249–259. doi: 10.1111/j.1582-4934.2011.01291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He W.W., Gustafson M.L., Hirobe S., Donahoe P.K. Developmental expression of four novel serine/threonine kinase receptors homologous to the activin/transforming growth factor-beta type II receptor family. Dev. Dynam. 1993;196:133–142. doi: 10.1002/aja.1001960207. [DOI] [PubMed] [Google Scholar]
- 32.Yndestad A., Ueland T., Øie E., Florholmen G., Halvorsen B., Attramadal H., Simonsen S., Frøland S., Gullestad L., Christensen G., Damås J., Aukrust P. Elevated levels of activin A in heart failure: potential role in myocardial remodeling. Circulation, Circulation. 2004;109:1379–1385. doi: 10.1161/01.CIR.0000120704.97934.41. [DOI] [PubMed] [Google Scholar]
- 33.Zhu X.M., Han T., Sargent I.L., Yin G.W., Yao Y.Q. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am. J. Obstet. Gynecol. 2009;200:661–667. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 34.Zheng A., Zhang L., Song X., Wang Y., Minjie W., Feng J. Clinical implications of a novel prognostic factor AIFM3 in breast cancer patients. BMC Canc. 2019;19:451. doi: 10.1186/s12885-019-5659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang K., Myers K.A. The role of hypoxia-induced miR-210 in cancer progression. Int. J. Mol. Sci. 2015;16:6353–6372. doi: 10.3390/ijms16036353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barcaroli D., Bongiorno-Borbone L., Terrinoni A., Hofmann T.G., Rossi M., Knight R.A., Matera A.G., Melino G., De Laurenzi V. FLASH is required for histone transcription and S-phase progression. Proc. Natl. Acad. Sci. U.S.A. 2006;103:14808–14812. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hubel C.A. Oxidative stress in the pathogenesis of preeclampsia. Proc. Soc. Exp. Biol. Med. 1999;222:22–35. doi: 10.1177/153537029922200305. [DOI] [PubMed] [Google Scholar]
- 38.Enquobahrie D.A., Meller M., Rice K., Psaty B.M., Siscovick D.S., Williams M.A. Differential placental gene expression in preeclampsia. Am. J. Obstet. Gynecol. 2008;199:566 e1–e11. doi: 10.1016/j.ajog.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortensen U.H., Lisby M., Rothstein R. Rad52. Curr. Biol. 2009;19:R676–R677. doi: 10.1016/j.cub.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Antonicka H., Leary S.C., Guercin G.H., Agar J.N., Horvath R., Kennaway N.G., Harding C.O., Jaksch M., Shoubridge E.A. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 2003;12:2693–2702. doi: 10.1093/hmg/ddg284. [DOI] [PubMed] [Google Scholar]
- 41.Kim J.H., Tonelli M., Markley J.L. Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase. Proc. Natl. Acad. Sci. U.S.A. 2012;109:454–459. doi: 10.1073/pnas.1114372109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan S.Y., Zhang Y.Y., Hemann C., Mahoney C.E., Zweier J.L., Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster Assembly proteins ISCU1/2. Cell Metabol. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z., Sun H., Dai H., Walsh R., Imakura M., Schelter J., Burchard J., Dai X., Chang A., Diaz R., Joseph R., Marszalek J., Bartz S., Carleton M., Cleary M., Linsley P., Grandori C. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 44.Kuo M.W., Wang C.H., Wu H.C., Chang S.J., Chuang Y.J. Soluble THSD7A is an N-glycoprotein that promotes endothelial cell migration and tube formation in angiogenesis. PloS One. 2011;6 doi: 10.1371/journal.pone.0029000. doi, 10.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J., Wu G., Cao Y., Hou Z. Roles of miR-210 in the pathogenesis of pre-eclampsia. Arch. Med. Sci. 2019;15:183–190. doi: 10.5114/aoms.2018.73129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albergaria A., Paredes J., Sousa B., Milanezi F., Carneiro V., Bastos J., Costa S., Vieira D., Lopes N., Lam E., Lunet N., Schmitt F. Expression of FOXA1 and GATA-3 in breast cancer: the prognostic significance in hormone receptor-negative tumours. Breast Cancer Res. 2009;11:R40. doi: 10.1186/bcr2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu J., Wei Y., Wang Z., Jie Q., Sun F., Li Q., Long P., Huang Y., Yu Y., Ma Y. Down-regulated FOXA1 in early-onset pre-eclampsia induces apoptosis, and inhibits migration and invasion of trophoblast cells. J. Gene Med. 2020 doi: 10.1002/jgm.3273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.