Abstract
Prenyltransferase (PTase) enzymes play crucial roles in natural product biosynthesis by transferring isoprene unit(s) to target substrates, thereby generating prenylated compounds. The prenylation step leads to a diverse group of natural products with improved membrane affinity and enhanced bioactivity, as compared to the non-prenylated forms. The last two decades have witnessed increasing studies on the identification, characterization, enzyme engineering, and synthetic biology of microbial PTase family enzymes. We herein summarize several examples of microbial soluble aromatic PTases for chemoenzymatic syntheses of unnatural novel prenylated compounds.
Keywords: Microbial prenyltransferase, Biosynthesis, Enzyme engineering, Prenylation, Synthetic biology
Abbreviations: DHN, dihydroxynaphthalene; DMAPP, dimethylallyl diphosphate; DMATS, dimethylallyltryptophan synthase; DMSPP, dimethylallyl S-thiolodiphosphate; FPP, farnesyl diphosphate; GFPP, geranyl farnesyl diphosphate; GPP, geranyl diphosphate; GSPP, geranyl S- thiolodiphosphate; IPP, isopentenyl pyrophosphate; PTase, prenyltransferase; PPP, phytyl pyrophosphate; RiPP, ribosomally synthesized and posttranslationally modified peptide; THN, 1,3,6,8-tetrahydroxynaphthalene
1. Introduction
Prenylation modifications play pivotal roles in both primary and secondary metabolism in living creatures. Natural products modified by prenyl moiety(ies) have improved lipophilicity, as compared to the corresponding non-prenylated molecules, leading to higher affinity to the cell membranes and augmented interactions with cellular targets [1]. Owing to the significantly enhanced biological activities by prenylation, this group of natural products has attracted intensive attention from the scientific community. For example, dimericbiscognienyne A (1), an unusual prenylated meroterpenoid dimer of biscognienyne A (2) from the endolichenic fungus Biscogniauxia sp., shows short-term memory enhancement activity in Alzheimer's disease model flies (Fig. 1) [2]. Hypatone A (3) and hyphenrone K (4), from the plant Hypericum patulum, are reportedly the most potent Cav3.1 agonist and antagonist, respectively [3]. Furthermore, hapalindole A (5), from the cyanobacterium Hapalosiphon sp., displays potent immunomodulatory activity [4]. Due to the remarkable biological activities of prenylated natural products, the identification and characterization of the prenyltransferase (PTase) enzymes responsible for the prenylation step(s) and the engineered biosyntheses of these types of molecules have been hot topics for decades.
Fig. 1.
Examples of natural products modified by prenyl moieties.
PTases utilize various prenyl donors, including dimethylallyl (C5), geranyl (C10), farnesyl (C15), geranylgeranyl (C20), and geranylfarnesyl (C25) pyrophosphates [1]. The prenylation types include normal (C-1 of prenyl donor connects to the prenyl acceptor) and reverse (C-3 of prenyl donor connects to the prenyl acceptor) prenylations, C-prenylation, O- prenylation [1], N-prenylation [5,6], and S-prenylation [7]. The microbial PTases involved in natural product biosyntheses are classified into three groups: (a) the UbiA-type membrane-binding proteins, predicted to contain seven transmembrane segments, (b) the dimethylallyltryptophan synthase (DMATS)-type, and (c) the ABBA-type. The latter two types of PTases both have the αββα PT folds, which led to the term ABBA. Although the three enzyme groups catalyze Friedel-Crafts alkylations of aromatic substrates with diverse prenyl donors, they exhibit distinct characteristics.
The Mg2+-dependent UbiA PTase superfamily, a type of PTase have been reported from bacterial, fungal, plant, and animal origin, is a group of membrane-bound PTases involved in the biosyntheses of a variety of small molecules, such as dolichols, ubiquinones, hemes, plant chlorophylls, vitamins E and K, and structural lipids, as well as fungal meroterpenoids (Fig. 2C) [[8], [9], [10]]. The ABBA-type and DMATS-type PTases from bacteria and fungi are typically soluble proteins possessing the αββα PT folds, but they share no sequence or structural similarity with the membrane-bound UbiA-type PTases (Fig. 2A and B) [8]. In contrast, the ABBA-type and DMATS-type PTases are involved in secondary metabolite biosynthesis, and lack the aspartate-rich motifs [(N/D)DXXD] for metal-binding. The ABBA-type PTases are mainly found in bacteria, especially Actinomycetes and Cyanobacteria, as exemplified by cyanobactins, which are ribosomally synthesized and post-translationally modified peptides (RiPPs) comprising six to eight amino acids, produced by many cyanobacteria [11]. The DMATS-type of PTase can be considered as a specific group of ABBA-type PTases. The DMATS-type of PTase specifically accept tryptophan, tyrosine or indole ring-containing aromatic compounds as native substrates. Notably, it is reported that some terpene cyclases also showed prenyltransfer activities, which expanded the knowledge and scope of PTase [12].
Fig. 2.
The crystal structures of (A) ABBA-type PTase NphB (1ZDY), (B) DMATS-type PTase AtaPT (5KCL), (C) UbiA-type PTase ApUbiA (4OD4), and (D) the catalytic pathway and tested substrates for NphB (the prenylated sites are highlighted).
This review focuses on the recently reported microbial soluble aromatic PTases for engineered biosynthesis, and continues the literature coverage from previous review articles [1,[13], [14], [15], [16]].
2. The ABBA-type aromatic PTases
2.1. Hydroxynaphthalene PTase NphB
The antioxidant meroterpenoid naphterpin (6) is produced by the mevalonate and polyketide hybrid biosynthetic pathway in Streptomyces sp. CL190 [17]. Thus, the malonate-derived intermediate 1,3,6,8-tetrahydroxynaphthalene (THN, 7) undergoes several enzyme reaction steps, including geranylation, oxidative dearomatization, and chlorination, to give 8, which further undergoes an α-hydroxyketone rearrangement to produce 9 (Fig. 2C) [18,19]. Interestingly, in vitro assays demonstrated that NphB, which catalyzes the geranylation at C-4 of 7, has unusually relaxed substrate specificity for aromatic compounds to give C- or O- prenylated products [20]. The acceptable substrates for NphB include 1,6- dihydroxynaphthalene (1,6-DHN, 10), 2,7-DHN (11), and 4-hydroxyphenylpyruvate (12), as well as a variety of plant polyphenols, such as naringenin (13), daidzein (14), genistein (15), resveratrol (16), and olivetol (17) (Fig. 2C). Most of the enzyme reaction products contain the geranyl group in the ortho-C position of the phenolic hydroxy group. Interestingly, when 2,7-DHN (11) was used as the substrate, the di-prenylated product 1,6-digeranyl-2,7-DHN was produced along with the monoprenylated product, 1-geranyl-2,7-DHN.
NphB is a 33 kDa soluble monomeric protein with 307 residues. The overall structure of NphB was solved by single crystal X-ray diffraction at 1.4 Å, and represents the first X-ray structure of the ABBA-type PTase family [17]. The single domain structure of NphB harbors a novel cylindrical β-sheet PT barrel fold, consisting of ten anti-parallel β-strands assembled around a central solvent-filled core. At the more polar “upper” side of the barrel, the Mg2+ is coordinated in an octahedral manner by four equatorially occupied water molecules and two axially positioned oxygens from the carboxylate group of Asp62 and the α-phosphate of GPP. The magnesium ion-coordinating mode of NphB is quite different from that of the UbiA-type PTases [5]. Specifically, the UbiA-type PTases possess a conserved Mg2+ ion-binding motif with the sequence (N/D)DXXD, which is also conserved in the IPP synthases. In contrast, the ABBA-type PTases are usually metal ion-independent, and the negative charges of dimethylallyl pyrophosphate (DMAPP) are negated by the positively charged residues, while the oxygen atoms form hydrogen bonds with tyrosine residues. In this regard, NphB is an exception and requires a Mg2+ ion for its catalytic activity. In NphB, the Mg2+ ion is bound in an octahedral coordination comprising four equatorial water molecules, the carboxylate of the Asp62 residue, and bridged and non-bridged oxygens of the α-phosphate of DMSPP [17].
2.2. Indolactam PTases TleC and MpnD
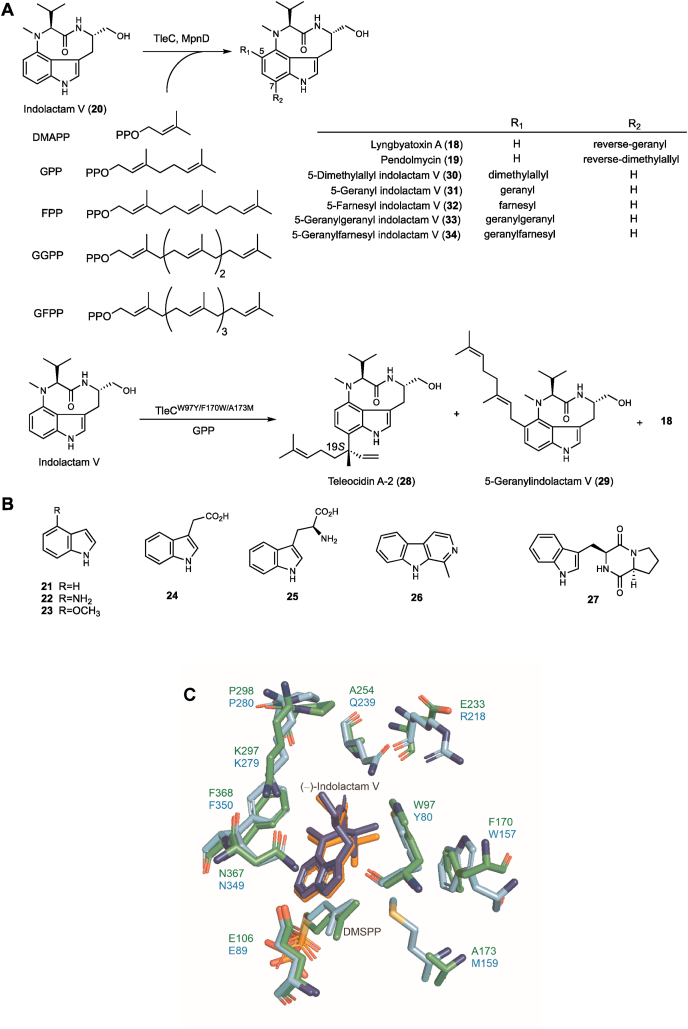
TleC from Streptomyces blastmyceticus and MpnD from Marinactinospora thermotolerans are responsible for the “reverse” prenylations of indolactam V (20) at the C-7 indole position with GPP and DMAPP, respectively, in the biosyntheses of lyngbyatoxin A (teleocidin A-1, 18) and pendolmycin (19) (Fig. 3A) [[21], [22], [23]]. In sharp contrast to NphB, both TleC and MpnD (sharing 38% amino acid sequence identity) display rigid substrate preferences towards the prenyl acceptors. Thus, among the tested substrates, such as indolactam V (20), indole (21), 4-aminoindole (22), 4-methoxyindole (23), indole acetic acid (24), l-tryptophan (25), harmane (26), and brevianamide F (27), only indolactam V was accepted (Fig. 3B). Instead, these two enzymes accept prenyl donors with different chain lengths, varying from the smallest DMAPP (C5) to the largest GFPP (C25), to produce a series of indolactam V derivatives in which the C-7 or C-5 indole position is prenylated (30–34). The enzyme kinetics experiments clearly demonstrated that TleC and MpnD prefer to transfer the C10 GPP and C5 DMAPP as the prenyl donors, respectively. The strict preference for the acceptor substrates and the relaxed specificity for the prenyl donors observed for TleC and MpnD demonstrated the striking difference from the conventional ABBA-type PTases, which usually exhibit reversed substrate specificity toward the prenyl acceptors and donors [23].
Fig. 3.
(A) The substrate specificities of the MpnD, TleC, and TleC mutants. (B) The tested substrates for TleC and MpnD. (C) Superimpositions of the active site residues of TleC (green) and MpnD (cyan) with (−)-indolactam V and DMSPP.
The structural bases for the unusual substrate scope of TleC and MpnD were revealed by their X-ray crystallographic structures: the apo- and ternary complex structures with (−)-indolactam V and the DMAPP analogue, dimethylallyl S-thiophosphate (DMSPP) (Fig. 3C) [23]. Thus, the presence of tight hydrogen bond networks for the substrate binding in the TleC and MpnD ternary complex structures clarified their strict prenyl acceptor substrate selectivity and exclusion of other indole derivatives. A comparison of the active site structures of TleC and MpnD further suggested the high similarity of the DMSPP binding mode with the α- and β-phosphates anchored by positively charged residues, and the different active site accommodations of the prenyl side chains. In particular, three amino acid residues, Trp97, Phe170, and Ala173 in TleC and Tyr80, Trp157, and Met159 in MpnD, work together to control the chain length selectivity of the prenyl donor and the regioselectivity of the prenylation reactions (Fig. 3C). Notably, Trp97 of TleC rotates to generate a large pocket to hold the prenyl side chain, while this rotation was not observed for the corresponding Tyr80 in MpnD, due to the steric hindrance between Tyr80 and Met159.
With this structural information in hand, a panel of mutants was constructed and characterized, including TleCA173M, TleCW97Y/A173M, and TleCW97Y/F170W/A173M, and MpnDM159A. The rationale behind these mutations is that, as mentioned above, the key residues Trp97, Phe170, and Ala173 in TleC, and the corresponding residues Tyr80, Trp157, and Met159 in MpnD play important roles in controlling the prenyl chain length by modulating the size and shape of the substrate binding pocket. The results demonstrated that the TleCA173M mutant preferred the smaller DMAPP over GPP, while the MpnDM159A mutant exhibited improved prenylation activity for the C10 GPP to yield lyngbyatoxin A and decreased prenylation activity for the C5 DMAPP. Intriguingly, the TleCW97Y/A173M double mutant and TleCW97Y/F170W/A173M triple mutant catalyzed the production of teleocidin A-2 (C-19-epimer of lyngbyatoxin A, 28) and 5-geranylindolactam V (29), in addition to lyngbyatoxin A (Fig. 3A).
3. The DMATS-type aromatic PTases
3.1. Diketopiperazine indole PTase FtmPT1
The fungal indole PTs utilize DMAPP to catalyze the prenylation of the indole moiety in tryptophan-containing molecules and have various substrate specificities and regiospecificities, thereby generating numerous structurally diverse and biologically active prenyl indole alkaloids. FtmPT1 from Aspergillus fumigatus is an indole PTase that naturally catalyzes the C-2 prenylation of the indole residue in the diketopiperazine brevianamide F (27) with DMAPP, to produce tryprostatin B (35) (Fig. 4) [24]. The structural analysis of FtmPT1 revealed that the Tyr205 residue forms a hydrogen bond with the keto group of brevianamide F [25]. The saturation-mutagenesis of this residue demonstrated that 15 mutants produced the regularly C3- prenylated brevianamide F, but not the C2-prenylated product [26]. Furthermore, three d-amino acid-containing substrate analogues, cyclo-d-Trp-d-Pro, cyclo-l-Trp-d-Pro, and cyclo-d-Trp-l-Pro, were used to test the substrate scopes of two selected mutants, Y205 N and Y205L. Interestingly, the C3-reverse prenylated products were obtained as the major products, in sharp contrast to the C2- and C3-normally prenylated derivatives with cyclo-l-Trp-l-Pro.
Fig. 4.
The catalytic reactions of (A) FtmPT1 and its Y205X mutant with cyclo-L-Trp-L-Pro, and (B) FtmPT1 and its two possible mechanisms with (E)-4-(1H-indol-3-yl)but-3-en-2-one.
Notably, FtmPT1 not only exhibits broad substrate specificity toward various tryptophan-containing cyclodipeptides, but also accepts other aromatic compounds such as tyrosine, hydroxynaphthalenes, and flavonoids [24]. In a project seeking to generate unnatural, novel products by using fungal indole PTases, FtmPT1 also accepted the indole derivative (E)-4-(1H-indol-3-yl)but-3-en-2-one (36) as a substrate to give the side-chain prenylated product 37 (Fig. 4B) [27]. Observations of the tested substrates led to the conclusion that, for the side chain part outside the aromatic ring, the presence of an α,β-unsaturated ketone moiety is needed, and for the aromatic ring, the indole ring is preferred over the benzene ring. Two possible mechanisms, both driven by the deprotonation of the indole nitrogen by Glu102, were proposed for the FtmPT1-catalyzed prenylation of 36 (Fig. 4B) [27]. Path A is the “normal-type” prenylation, where C-1 of the dimethylallyl carbocation is attacked by the C-9 position of 36. Path B adopts a “reverse-type” prenylation, where C-3 of the dimethylallyl carbocation is attacked by the C-3 position of the indole ring, followed by product formation by subsequent rearrangement and aromatization steps. However, the exact prenylation mechanism remains to be determined.
3.2. Tryptophan PTase FgaPT2
The PTase FgaPT2, identified from the fungus Aspergillus fumigatus, is responsible for the C-4 prenylation of tryptophan in the first step of ergot alkaloid biosyntheses (Fig. 5A). The complex structure of FgaPT2 with l-tryptophan and DMSPP was determined at 2.2 Å resolution [28]. The Lys174 residue of FgaPT2 was proposed to act as the base that abstracts a proton from the prenyl-attached arenium intermediate (38) in the final rearomatization step [29]. The FgaPT2K174A mutant produced C-4 prenylated and C-3 reverse-prenylated l-tryptophan derivatives (39), with the latter as the major product. This study supported the mechanism in which C-3 is prenylated first, and then converted to the C-4 prenylated product by a Cope rearrangement and rearomatization (Fig. 5A). Interestingly, the K174F mutant of FgaPT2 showed C-3 prenylation activity with the unnatural substrate l-tyrosine, and its catalytic efficiency toward l-tyrosine is 5-times higher than that of the wild type [30]. It also catalyzed major C3-prenylation and minor N4-prenylation toward 4-amino-l-phenylalanine. However, this mutant showed extremely low activity toward the natural substrate, l-tryptophan.
Fig. 5.
(A) A Cope rearrangement mechanism for the reaction catalyzed by FgaPT2 WT and the K174A mutant. (B) The enzyme reactions of FgaPT2 and its K174F/R244X mutants.
Interestingly, the Arg244 residue of FgaPT2 was proposed to interact with the tryptophan side chain, leading it to become a hot spot for engineering [31]. Among the Arg244 mutants obtained by saturation mutagenesis, thirteen displayed increased prenylation activities toward tryptophan-containing cyclic dipeptides (Fig. 5B), and the most significant R244L mutant showed 77-fold higher activity as compared to that of the wild-type enzyme. The substrate preferences of these mutants toward tryptophan-containing cyclic dipeptides were also altered. For example, the wild-type, R244A, R244T, and R244Q enzymes prefer cyclo-l-Trp-d-Pro and cyclo-l-Trp-l-Leu as substrates, while the R244G mutant prefers cyclo-l-Trp-l-Leu and cyclo-l-Trp-l-Trp. Overall, FgaPT2K174F and FgaPT2R244X successfully altered the regiospecificity of prenylation toward tryptophan-containing cyclic dipeptides, from tryptophan C4-normal prenylation to C3-reverse prenylation.
3.3. Promiscuous aromatic PTase AtaPT
AtaPT is a 424 amino acid residue DMATS-type aromatic PTase from the fungus Aspergillus terreus [32]. This enzyme reportedly exhibits wide substrate promiscuity regarding both aromatic acceptors and prenyl donors (Fig. 6). AtaPT can utilize prenyl donors with chain lengths from C5 to C20, including phytyl pyrophosphate (PPP), to yield monoprenylated to triprenylated products with C–C and/or C–O bonds. The substrate promiscuity of AtaPT was investigated by using three prenyl donors, DMAPP, GPP, and FPP, and 106 potential aromatic acceptors, including not merely the tryptophan-containing dipeptides but also many drug-like aromatic compounds, such as quinoline alkaloids, benzophenones, and flavonoid glycosides. In total, 72 aromatic compounds were prenylated by all three prenyl substrates. Interestingly, some of the monoprenylated products in turn served as substrates for further modification by AtaPT to produce di- and/or triprenylated products. This broad substrate promiscuity of AtaPT is one of the most diverse substrate scopes among the characterized PTases.
Fig. 6.
(A) The substrate promiscuity of AtaPT and the G326 M mutant. (B) Some selected aromatic acceptors for AtaPT (the prenylated sites are highlighted). (C) The apo structure of AtaPT. (D) The complex structures of AtaPT-GSPP and (+)-butyrolactone II.
Structurally speaking, AtaPT shares the common DMATS superfamily features with five consecutive αββα units harboring a central active site. In AtaPT, the prenyl donor binding site and the tyrosine shield, comprising four tyrosine residues that stabilize the carbocation intermediate, are highly conserved as compared with other members. However, the less conserved aromatic acceptor binding pocket, surrounded by relatively small chain hydrophobic residues, is more spacious and hydrophobic than those of other DMATS superfamily members. These findings also clarify the basis for the broad substrate promiscuity of AtaPT.
Crystallographic analyses and molecular simulation studies of AtaPT complexed with substrate(s), including DMSPP, GSPP, (+)- and (−)-butyrolactone II (40), and genistein (15), indicated the conformational differences between the apo and complex structures (Fig. 6C and D). The conformational fluctuation of the tyrosine shield region led to the identification of two key residues, Try190 and Try268, which play a crucial role in controlling the space where the prenyl acceptor substrate binds. Further mutagenesis studies revealed that Gly326 contributes to the prenyl donor selectivity. The G326M mutation dramatically reduced the activity toward GPP and FPP, and enhanced the activity toward DMAPP. The counterpart residues for controlling the prenyl donor size for FgaPT2, AnaPT, FtmPT1, and CdpNPT are Met328, Val340, Met364, and Met349, respectively.
This gatekeeping residue rule led to the identification of the corresponding residues in other PTases, including I337 in BrePT, Thr351 in CdpC2PT, and Phe335 in CdpC3PT (Fig. 7A) [33]. Upon switching these bulky residues to a small glycine, the mutant PTases FtmPT1M364G, BrePTI337G, CdpNPTM349G, CdpC2PTT351G, CdpC3PTF335G, FgaPT2M328G, and FgaPT2M328G/R244Q displayed significantly improved catalytic efficiencies toward installing a geranyl moiety on tryptophan-containing cyclic dipeptides (Fig. 7B). Using these engineered enzymes, 42 geranylated derivatives were obtained from 15 cyclic dipeptides. Notably, the regioselectivity of the geranylation positions of all seven indole carbon positions of cyclo-l-Trp-l-Trp can be controlled by using these engineered enzymes as catalysts. This study provided a leading example, by engineering the gatekeeping residues and thereby activating or improving the geranylation of tryptophan-containing cyclic dipeptides to generate a library of prenylated cyclic dipeptides.
Fig. 7.
(A) The native enzymatic reactions of BrePT, CdpC3PT, and FgaPT2. (B) The geranylated positions catalyzed by the engineered dimethylallyl transferases with gatekeeping residue mutations.
4. Hapalindoles PTases AmbP1 and AmbP3
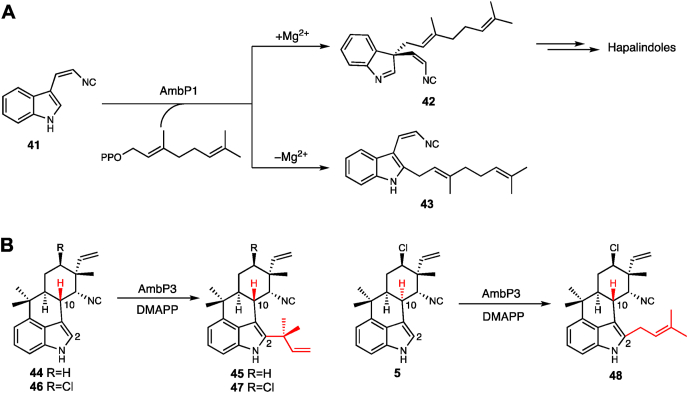
Hapalindole-type alkaloids, such as hapalindole A (5) (Fig. 1), are a large group of indole monoterpenoids exclusively produced by Stigonematalean cyanobacteria [34]. Their structural diversity and complexity, as well as broad-spectrum antimicrobial and antitumor activities, make them intriguing molecules for bio- and organic synthetic studies [35,36]. The biosynthetic gene cluster amb for ambiguine production encodes the ABBA-type aromatic PTase AmbP1 and the DMATS-type aromatic PTase AmbP3 (Fig. 8) [[37], [38], [39], [40], [41], [42]]. The two proteins have been well characterized in terms of their substrate specificities, metal ion dependence, and crystallographic structures.
Fig. 8.
The prenyltransfer reactions catalyzed by (A) AmbP1 and (B) AmbP3.
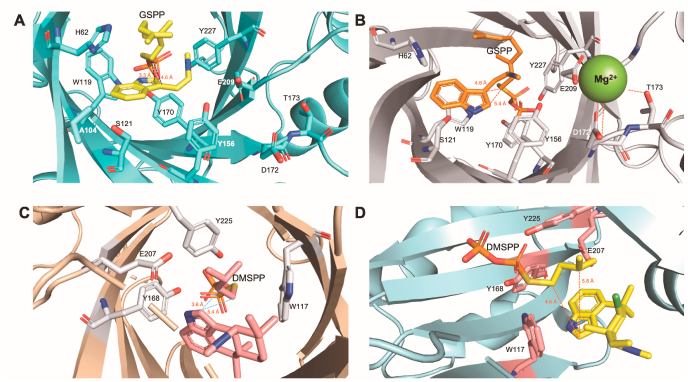
The aromatic PTase AmbP1 transferred the C10 geranyl group to C-3 of 41, in the presence of Mg2+ under basic conditions (pH 8.0–9.0) (Fig. 8A) [38]. However, the competitive formation of the C-2 geranylated side product 43 was detected in the absence of Mg2+ under basic or acidic pH conditions (pH 6.0). The different Mg2+ ion-dependent and -independent regioselectivities were further investigated by the X-ray structures of AmbP1. Interestingly, the apo structure of the ABBA-type AmbP1 contains a Mg2+ ion without coordination to the central β-barrel, and instead it is located at a marginal position. The complex structures of AmbP1 with 41 and GSPP, obtained from soaking experiments at pH 6.5 (Mg2+- free, 2.6 Å, Fig. 9A) and pH 8.0 (Mg2+-bound, 2.0 Å, Fig. 9B), respectively, revealed the rationale for the Mg2+-dependent regioselectivity. In the Mg2+-free structure, the C-1 position of GSPP was 3.3 Å from the C-2 of 41, and 4.6 Å from the C-3 of 41, indicating the preference of C-2 prenylation over C-3. In the Mg2+-bound structure, the Mg2+ ion was close to the isonitrile group of 41, and chelated by hydrogen bonds with Asp172, Thr173, Gly208, and Glu209. The orientation of 41 was altered, and the distances from C-1 of GSPP to C-2 and C-3 of 41 were 5.4 and 4.6 Å, respectively, indicating a preferred C-3 prenylation reaction. Further mutation studies of Glu209 (E209A and E209L) confirmed its role in the Mg2+ ion binding and catalytic cavity formation.
Fig. 9.
The active sites of (A) AmbP1 with 41 and GSPP, (B) AmbP1 with 41, GSPP, and Mg2+, (C) AmbP3 with 44 and DMSPP, and (D) AmbP3 with 5 and DMSPP.
The other PTase, AmbP3, catalyzes either normal or reverse prenylation depending on the hapalindole substrate (Fig. 8B) [39]. Enzymatic studies revealed that the DMATS-type AmbP3 accepts hapalindoles U (44) and G (46) with the 10R configuration as substrates, and the reverse prenylations using DMAPP took place at their C-2 positions to yield ambiguines H (45) and A (47), respectively. Interestingly, when the 10S-hapalindole A (5) was used as the prenyl acceptor, its C-2 position was prenylated normally to give 48. The complex structures of AmbP3 with DMSPP/44 (Fig. 9C) and DMSPP/5 (Fig. 9D), each obtained at 2.0 Å and hereafter respectively designated as HU and HA, clarified their different catalytic mechanisms. Both of the hapalindoles are supported by hydrophobic amino acids. In the HU structure, the indole ring of 44, Trp117, and Tyr168 form a cation shield, which is important to stabilize the cation intermediate after the phosphate removal from DMAPP, while in the HA structure, the role of Tyr168 is adopted by Tyr225. Furthermore, the distances from the C-2s of 44 and 5 to the C-1 and C-3 of DMSPP clearly showed the preference of reverse or normal prenylation. In the HU structure, the aforementioned distances were 3.6 Å and 5.4 Å, respectively, suggesting preferred reverse prenylation, while in the HA structure, the respective distances were 5.8 Å and 4.6 Å, suggesting preferred normal prenylation. As a result, the plasticity to accept DMAPP and two stereoisomeric hapalindoles was revealed by the complex crystal structures, representing the first X-ray model of a PTase that catalyzes both normal and reverse prenylations.
5. Cyanobactin PTases
5.1. Peptide tryptophan PTase KgpF
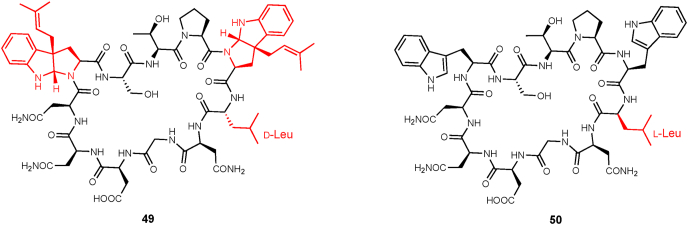
Kawaguchipeptins A (49) and B (50) are two cyclic undecapeptides with similar amino acid sequences [backbone sequence: cyclic-(WLNGDNNWSTP)] identified from the cyanobacterium Microcystis aeruginosa NIES-88 (Fig. 10) [43,44]. The two compounds are members of the cyanobactins, a class of ribosomally-synthesized and post-translationally modified peptides (RiPPs). Kawaguchipeptin A has one d-Leu and two C-3-dimethylallylated tryptophan moieties, while kawaguchipeptin B only has l-amino acid residues without prenyl modifications. KgpF is the PTase responsible for the di-prenylation of the tryptophan ring in 50 [45]. In vitro assays revealed that KgpF exhibits prenylation activity with numerous prenyl acceptors and donors. As for prenyl acceptors, KgpF catalyzes the prenylation of both cyclic and linear peptides as substrates and affords monoprenylated products from linear peptides and DMAPP, while diprenylated products were only detected when the cyclic peptides were used as prenyl acceptors. As for the prenyl donor, KgpF prefers DMAPP over IPP.
Fig. 10.
The structures of kawaguchipeptins A (49) and B (50).
However, with the limited substrates tested, the full catalytic role concerning the stereo- and regio-selectivity of KgpF toward other substrates, especially modified tryptophan, remained unclear. To clarify the substrate scope, many tryptophan derivatives were tested as substrates for KgpF [46]. Interestingly, KgpF displayed activity toward Fmoc-tryptophan (51) (Fig. 11). This finding suggested that the N-terminal tryptophan residues of peptides and tryptophan derivatives with free amino groups are not acceptable substrates of KgpF. The homology model of KgpF revealed that the Tyr69 and Trp137 residues interacted with the N-terminal Fmoc group of Fmoc-tryptophan by π-π interactions. This work provided a basis for engineering KgpF to produce more prenylated tryptophan derivatives and peptide libraries harboring prenylated tryptophan fragments.
Fig. 11.
The proposed catalytic mechanism of KgpF with Fmoc-Trp (51).
5.2. Peptide tyrosine O-PTase PagF
PagF is a tyrosine O-prenyltransferase identified in the biosynthetic pathway of prenylagaramide [cyclic-(INPYLYP), 52] (Fig. 12A) in Oscillatoria agardhii [47]. This enzyme was fully characterized in terms of biochemical activity, cocrystal structure studies with both linear and macrocyclic peptide substrates, substrate scope investigations, and mutagenesis studies [48]. PagF transfers the dimethylallyl group from DMAPP to the hydroxy group of an l-tyrosine residue, and lacks activity with GPP.
Fig. 12.
(A) The structure of cyclic-(INPYLYP) (52). (B) The cocrystal structures of PagF (F222 shown), DMSPP, and Mg2+.
The overall structure of PagF shares the typical features of the ABBA-type enzyme family. It is double-layered, with 10 α-helices covering 10 antiparallel β-strands to form a barrel-like architecture [48]. PagF has a solvent-exposed hydrophobic pocket (Fig. 12B), and prefers larger substrates to shield the hydrophobic active site from the solvent. When the peptide substrate binds within the active site, the Tyr hydroxy group is located at an appropriate distance from the prenyl donor, thus facilitating the allylic carbocation electrophilic attacks on the Tyr oxygen to form the O-prenylated product. Notably, PagF shows a high catalytic preference toward the first tyrosine residue, rather than the second one. This strict regioselectivity seems to be exclusive to the cyanobacterial KgpF homologues, and is distinct from those of the canonical small molecule PTases.
The authors also reported the crystal structure of PirF, which prefers GPP as the prenyl donor. A comparison between PirF and PagF indicated that nearly all of the residues in the active site are highly conserved, except for Phe222. In the cocrystal structure of PagF/DMSPP, the Phe222 residue is located at the top of the barrel, exerting an essential hydrophobic effect to stabilize the allylic carbocation against solvent quenching (Fig. 12A) [49]. Based on these findings, Phe222 mutants were investigated. Surprisingly, the PagFF222A and PagFF222G mutants both showed robust geranyltransferase activities, using GPP as the prenyl donor. In contrast, the PagFWT showed no activity with GPP. The single mutation of PagF thus completely switched the prenyl donor selectivity from DMAPP to GPP. The cocrystal structure of PagFF222A/GPP/Mg2+ clearly revealed that the GPP occupies the space generated by the Phe/Ala replacement, as also supported by the kinetic data of the two variants. This work provided examples of biocatalysts that are strictly tolerant for the allylic donor, but have donor specificity that may be altered entirely.
6. Terpene cyclase with aromatic PTase activity
The aromatic PTases from both bacteria and fungi have been well characterized by extensive efforts. Quite recently, new insights into terpene cyclases (TPCs) that also function as PTases have been reported by Liu and co-workers (Fig. 13) [12]. TPCs, as exemplified by AaTPS and FgGS, exhibited PTase activities toward indole and aromatic substrates by using DMAPP or GPP as the prenyl donor. Both AaTPS and FgGS are class I TPCs identified from the filamentous fungi Alternaria alternata and Fusarium graminearum, respectively. Sequence alignments of these two enzymes with other TPCs suggested that they retain the typical TPC features with the highly conserved aspartate-rich motif D(D/S)XX(D/E) and the Mg2+-binding motif NSE/DTE, which are evolutionarily unrelated to those of the indole PTases that adopt the ABBA fold. Interestingly, the roles of AaTPS and FgGS as a PTC or indole PT can be altered by changing the pH conditions from neutral (pH 7.1) to alkaline (pH 8.6). This substrate specificity investigation demonstrated that AaTPS only accepts DMAPP as the sole prenyl donor, but for the prenyl acceptors, the naphthols are also tolerated in addition to indoles. In contrast, FgGS accepted both DMAPP and GPP as the prenyl donors.
Fig. 13.
The catalytic profiles of AsTPS and FgGS as (A) terpene cyclases (the original function) and (B) prenyltransferases (newly found function).
Crystallographic studies indicated that these bifunctional enzymes utilize the same active site to catalyze the TPC and aromatic PT enzyme reactions. The PPi anion was chelated by the Mg2+ ion bound to the enzyme, and the prenyl moiety was accommodated by the hydrophobic active site. The indole must bind adjacent to the prenyl moiety to facilitate the C–C bond formation. The three-dimensional contour of the hydrophobic active site plays important roles in controlling the orientation of the indole ring, thereby guiding the regiospecificity of the PT reaction.
Physiologically speaking, excess concentrations of IPP/DMAPP are cytotoxic, as revealed by several studies [12]. Therefore, it is likely that in response to the accumulation of IPP/DMAPP, the cryptic aromatic PTase function is activated to reduce the intracellular concentration of isoprenoid precursors, thus alleviating the cytotoxicity. Based on these findings, the cryptic PTase activity may be a universal feature of the TPCs. Since these two enzymes are from different families but exert the same PT activity, they may serve as potential biocatalysts providing more opportunities for engineering PTases.
7. Conclusion
In this review, we summarized recently reported several microbial soluble αββα folds-containing aromatic PTases for the aim of engineered biosynthesis. More insights were gained including their structures, mutant studies, substrate scopes, and their roles as biocatalysts, in particular the DMATS-type of PTases which have been more intensively studied from fungal and also bacterial origin. This type of enzymes play important roles in primary and secondary metabolism.
The rapid developments of bioinformatics and sequencing technology, along with the progress in enzyme engineering methodology, have accelerated our comprehension of the enzymes involved in the key steps of natural product biosynthesis. Continuing efforts will lead to the characterization of novel PT enzymes and the elucidation of their unique reaction mechanisms. The cases reviewed here represent the variations of the substrate scope and the enzyme engineering of soluble PTases from microbial origins, and hopefully will promote the engineered biosyntheses of unnatural novel prenylated compounds with more diverse and complex structures for drug discovery.
CRediT authorship contribution statement
He-Ping Chen: Writing – original draft, Writing – review & editing. Ikuro Abe: Supervision, Conceptualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
We declare we have no conflict of interest.
Acknowledgements
The authors would like to express sincere appreciation to an excellent group of coworkers whose contributions are cited in the text. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (JSPS KAKENHI Grant No. JP16H06443 and JP20H00490), and Japan Science and Technology Agency (JST SICORP Grant No. JPMJSC1701). H.P. C. is a recipient of the JSPS Postdoctoral Fellowship for Foreign Researchers (ID No. P19413).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Mori T. Enzymatic studies on aromatic prenyltransferases. J Nat Med. 2020;74(3):501–512. doi: 10.1007/s11418-020-01393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao H., Chen G.D., Zou J., He R.R., Qin S.Y., Hu D. Dimericbiscognienyne A: a meroterpenoid dimer from Biscogniauxia sp. with new skeleton and its activity. Org Lett. 2017;19(1):38–41. doi: 10.1021/acs.orglett.6b03264. [DOI] [PubMed] [Google Scholar]
- 3.Ye Y.S., Li W.Y., Du S.Z., Yang J., Nian Y., Xu G. Congenetic hybrids derived from dearomatized isoprenylated acylphloroglucinol with opposite effects on Cav3.1 low voltage-gated Ca2+ channel. J Med Chem. 2020;63(4):1709–1716. doi: 10.1021/acs.jmedchem.9b02056. [DOI] [PubMed] [Google Scholar]
- 4.Chilczuk T., Steinborn C., Breinlinger S., Zimmermann-Klemd A.M., Huber R., Enke H. Hapalindoles from the cyanobacterium Hapalosiphon sp. inhibit T cell proliferation. Planta Med. 2020;86(2):96–103. doi: 10.1055/a-1045-5178. [DOI] [PubMed] [Google Scholar]
- 5.Brunson J.K., KcKinnie S.M.K., Chekan J.R., McCrow J.P., Miles Z.D., Bertrand E.M., Bielinski V.A., Luhavaya H., Oborník M., Smith G.J., Hutchins D.A., Allen A.E., Moore B.S. Biosynthesis of the neurotoxin domoic acid in a bloom-forming diatom. Science. 2018;361(6409):1356–1358. doi: 10.1126/science.aau0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abe I., Tanaka H., Abe T., Noguchi H. Enzymatic formation of unnatural cytokinin analogs by adenylate isopentenyltransferase from mulberry. Biochem Biophys Res Commun. 2007;355(3):795–800. doi: 10.1016/j.bbrc.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 7.Charron G., Tsou L.K., Maguire W., Yount J.S., Hang H.C. Alkynyl-farnesol reporters for detection of protein S-prenylation in cells. Mol Biosyst. 2011;7(1):67–73. doi: 10.1039/c0mb00183J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W. Bringing bioactive compounds into membranes: the UbiA superfamily of intramembrane aromatic prenyltransferases. Trends Biochem Sci. 2016;41(4):356–370. doi: 10.1016/j.tibs.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng W., Li W. Structural insights into ubiquinone biosynthesis in membranes. Science. 2014;343(6173):878–881. doi: 10.1126/science.1246774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda Y., Abe I. Biosynthesis of fungal meroterpenoids. Nat Prod Rep. 2016;33(1):26–53. doi: 10.1039/c5np0090d. [DOI] [PubMed] [Google Scholar]
- 11.Gu W., Dong S.H., Sarkar S., Nair S.K., Schmidt E.W. The biochemistry and structural biology of cyanobactin pathways: enabling combinatorial biosynthesis. In: Moore Bradley S., editor. vol. 604. Elsevier; 2018. pp. 113–163. (Methods Enzymol). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He H., Bian G., Herbst-Gervasoni C.J., Mori T., Shinsky S.A., Hou A., Mu X., Huang M., Cheng S., Deng Z., Christianson D.W., Abe I., Liu T. Discovery of the cryptic function of terpene cyclases as aromatic prenyltransferase. Nat Commun. 2020;11(1):3958. doi: 10.1038/s41467-020-17642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkelblech J., Fan A., Li S.M. Prenyltransferases as key enzymes in primary and secondary metabolism. Appl Microbiol Biotechnol. 2015;99(18):7379–7397. doi: 10.1007/s00253-015-6811-y. [DOI] [PubMed] [Google Scholar]
- 14.Fan A.L., Winkelblech J., Li S.M. Impacts and perspectives of prenyltransferases of the DMATS superfamily for use in biotechnology. Appl Microbiol Biotechnol. 2015;99(18):7399–7415. doi: 10.1007/s00253-015-6813-9. [DOI] [PubMed] [Google Scholar]
- 15.Steffan N., Grundmann A., Yin W.B., Kremer A., Li S.M. Indole prenyltransferases from Fungi: a new enzyme goup with high potential for the production of prenylated indole derivatives. Curr Med Chem. 2009;16(2):218–231. doi: 10.2174/092986709787002772. [DOI] [PubMed] [Google Scholar]
- 16.Tanner M.E. Mechanistic studies on the indole prenyltransferases. Nat Prod Rep. 2015;32(1):88–101. doi: 10.1039/C4NP00099D. [DOI] [PubMed] [Google Scholar]
- 17.Kuzuyama T., Noel J.P., Richard S.B. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature. 2005;435(7044):983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miles Z.D., Diethelm S., Pepper H.P., Huang D.M., George J.H., Moore B.S. A unifying paradigm for naphthoquinone-based meroterpenoid (bio)synthesis. Nat Chem. 2017;9(12):1235–1242. doi: 10.1038/nchem.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray L.A.M., McKinnie S.M.K., Pepper H.P., Erni R., Miles Z.D., Cruickshank M.C., López-Pérez B., Moore B.S., George J.H. Total synthesis establishes the biosynthetic pathway to the naphterpin and marinone natural products. Angew Chem Int Ed. 2018;57(34):11009–11014. doi: 10.1002/anie.201804351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumano T., Richard S.B., Noel J.P., Nishiyama M., Kuzuyama T. Chemoenzymatic syntheses of prenylated aromatic small molecules using streptomyces prenyltransferases with relaxed substrate specificities. Bioorg Med Chem. 2008;16(17):8117–8126. doi: 10.1016/j.bmc.2008.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Awakawa T., Zhang L., Wakimoto T., Hoshino S., Mori T., Ito T., Ishikawa J., Tanner M.E., Abe I. A methyltransferase initiates terpene cyclization in teleocidin B biosynthesis. J Am Chem Soc. 2014;136(28):9910–9913. doi: 10.1021/ja505224r. [DOI] [PubMed] [Google Scholar]
- 22.Ma J., Zuo D., Song Y., Wang B., Huang H., Yao Y. Characterization of a single gene cluster responsible for methylpendolmycin and pendolmycin biosynthesis in the deep sea bacterium Marinactinospora thermotolerans. ChemBioChem. 2012;13(4):547–552. doi: 10.1002/cbic.201100700. [DOI] [PubMed] [Google Scholar]
- 23.Mori T., Zhang L., Awakawa T., Hoshino S., Okada M., Morita H., Abe I. Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases. Nat Commun. 2016;7:10849. doi: 10.1038/ncomms10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grundmann A., Li S.M. Overproduction, purification and characterization of FtmP1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology. 2005;151(7):2199–2207. doi: 10.1099/mic.0.27962-0. [DOI] [PubMed] [Google Scholar]
- 25.Jost M., Zocher G., Tarcz S., Matuschek M., Xie X., Li S.M., Stehle T. Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity. J Am Chem Soc. 2010;132(50):17849–17858. doi: 10.1021/ja106817c. [DOI] [PubMed] [Google Scholar]
- 26.Zhou K., Zhao W., Liu X.Q., Li S.M. Saturation mutagenesis on Tyr205 of the cyclic dipeptide C2-prenyltransferase FtmPT1 results in mutants with strongly increased C3-prenylating activity. Appl Microbiol Biotechnol. 2016;100(23):9943–9953. doi: 10.1007/s00253-016-7663-9. [DOI] [PubMed] [Google Scholar]
- 27.Chen J., Morita H., Wakimoto T., Mori T., Noguchi H., Abe I. Prenylation of a nonaromatic carbon of indolylbutenone by a fungal indole prenyltransferase. Org Lett. 2012;14(12):3080–3083. doi: 10.1021/ol301129x. [DOI] [PubMed] [Google Scholar]
- 28.Metzger U., Schall C., Zocher G., Unsöld I., Stec E., Li S.M., Heide L., Stehle T. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc Natl Acad Sci USA. 2009;106(34):14309–14314. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luk L.Y.P., Qian Q., Tanner M.E. A Cope rearrangement in the reaction catalyzed by dimethylallyltryptophan synthase? J Am Chem Soc. 2011;133(32):12342–12345. doi: 10.1021/ja2034969. [DOI] [PubMed] [Google Scholar]
- 30.Zheng L., Mai P., Fan A., Li S.M. Switching a regular tryptophan C4-prenyltransferase to a reverse tryptophan-containing cyclic dipeptide C3-prenyltransferase by sequential site-directed mutagenesis. Org Biomol Chem. 2018;16(36):6688–6694. doi: 10.1039/c8ob01735b. [DOI] [PubMed] [Google Scholar]
- 31.Fan A., Li S.M. Saturation mutagenesis on Arg244 of the tryptophan C4-prenyltransferase FgaPT2 leads to enhanced catalytic ability and different preferences for tryptophan-containing cyclic dipeptides. Appl Microbiol Biotechnol. 2016;100(12):5389–5399. doi: 10.1007/s00253-016-7365-3. [DOI] [PubMed] [Google Scholar]
- 32.Chen R., Gao B., Liu X., Ruan F., Zhang Y., Lou J., Feng K., Wunsch C., Li S.M., Dai J., Sun F. Molecular insights into the enzyme promiscuity of an aromatic prenyltransferase. Nat Chem Biol. 2017;13(2) doi: 10.1038/nchembio.2263. 226-4. [DOI] [PubMed] [Google Scholar]
- 33.Liao G., Mai P., Fan J., Zocher G., Stehle T., Li S.M. Complete decoration of the indolyl residue in cyclo-L-Trp-L-Trp with geranyl moieties by using engineered dimethylallyl transferase. Org Lett. 2018;20(22):7201–7205. doi: 10.1021/acs.orglett.8b03124. [DOI] [PubMed] [Google Scholar]
- 34.Awakawa T., Liu X. Hapalindole-type alkaloid biosynthesis. In: Liu Hung-wen, Begley Tadhg., editors. vol. 2. Elsevier; 2020. pp. 486–499. (Comprehensive Natural Products III: Chemistry and Biology). 2018. [DOI] [Google Scholar]
- 35.Li S., Lowell A.N., Newmister S.A., Yu F., Williams R.M., Sherman D.H. Decoding cyclase-dependent assembly of hapalindole and fischerindole alkaloids. Nat Chem Biol. 2017;13:467–469. doi: 10.1038/nchembio.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newmister S.A., Li S., Garcia-borràs M., Sanders J.N., Yang S., Lowell A.N., Yu F., Smith J.L., Williams R.M., Houk K.N., Sherman D.H. Structural basis of the Cope rearrangement and cyclization in hapalindole bogenesis. Nat Chem Biol. 2018;14:345–351. doi: 10.1038/s41589-018-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hillwig M.L., Zhu Q., Liu X. Biosynthesis of ambiguine indole alkaloids in cyanobacterium Fischerella ambigua. ACS Chem Biol. 2014;9(2):372–377. doi: 10.1021/cb400681n. [DOI] [PubMed] [Google Scholar]
- 38.Awakawa T., Mori T., Nakashima Y., Zhai R., Wong C.P., Hillwig M.L., Liu X., Abe I. Molecular insight into the Mg2+ -dependent allosteric control of indole prenylation by aromatic prenyltransferase AmbP1. Angew Chem Int Ed. 2018;57(23):6810–6813. doi: 10.1002/anie.201800855. [DOI] [PubMed] [Google Scholar]
- 39.Wong C.P., Awakawa T., Nakashima Y., Mori T., Zhu Q., Liu X., Abe I. Two distinct substrate binding modes for the normal and reverse prenylation of hapalindoles by the prenyltransferase AmbP3. Angew Chem Int Ed. 2018;57(2):560–563. doi: 10.1002/anie.201710682. [DOI] [PubMed] [Google Scholar]
- 40.Hillwig M.L., Liu X. A new family of iron-dependent halogenases acts on freestanding substrates. Nat Chem Biol. 2014;10:6–10. doi: 10.1038/nchembio.1625. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell A.J., Zhu Q., Maggiolo A.O., Ananth N.R., Hillwig M.L., Liu X., Boal A.K. Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5. Nat Chem Biol. 2016;12:636–640. doi: 10.1038/nchembio.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hillwig M.L., Zhu Q., Ittiamornkul K., Liu X. Discovery of a promiscuous non-heme iron halogenase in ambiguine alkaloid biogenesis: implication for an evolvable enzyme family for late-stage halogenation of aliphatic carbons in small molecules. Angew Chem Int Ed. 2016;55(19):5780–5784. doi: 10.1002/anie.201601447. [DOI] [PubMed] [Google Scholar]
- 43.Ishida K., Matsuda H., Murakami M., Yamaguchi K. Kawaguchipeptin B, an antibacterial cyclic undecapeptide from the cyanobacterium Microcystis aeruginosa. J Nat Prod. 1997;60(7):724–726. doi: 10.1021/np970146k. [DOI] [PubMed] [Google Scholar]
- 44.Ishida K., Matsuda H., Murakami M., Yamaguchi K. Kawaguchipeptin A, a novel cyclic undecapeptide from cyanobacterium Microcystis aeruginosa (NIES-88) Tetrahedron. 1996;52(27):9025–9030. doi: 10.1016/0040-4020(96)00452-8. [DOI] [PubMed] [Google Scholar]
- 45.Parajuli A., Kwak D.H., Dalponte L., Leikoski N., Galica T., Umeobika U., Trembleau L., Bent A., Sivonen K., Wahlsten M., Wang H., Rizzi E., De Bellis G., Naismith J., Jaspars M., Liu X., Houssen W., Fewer D.P. A unique tryptophan C-prenyltransferase from the kawaguchipeptin biosynthetic pathway. Angew Chem Int Ed. 2016;55(11):3596–3599. doi: 10.1002/anie.201509920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okada M., Sugita T., Akita K., Nakashima Y., Tian T., Li C., Mori T., Abe I. Stereospecific prenylation of tryptophan by a cyanobacterial post-translational modification enzyme. Org Biomol Chem. 2016;14(40):9639–9644. doi: 10.1039/C6OB01759B. [DOI] [PubMed] [Google Scholar]
- 47.Donia M.S., Schmidt E.W. Linking chemistry and genetics in the growing cyanobactin natural products family. Chem Biol. 2011;18(4):508–519. doi: 10.1016/j.chembiol.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hao Y., Pierce E., Roe D., Morita M., McIntosh J.A., Agarwal V., Cheatham T.E., III, Schmidt E.W., Nair S.K. Molecular basis for the broad substrate selectivity of a peptide prenyltransferase. Proc Natl Acad Sci USA. 2016;113(49):14037–14042. doi: 10.1073/pnas.1609869113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Estrada P., Morita M., HaoY, Schmidt E.W., Nair S.K. A single amino acid switch alters the isoprene donor specificity in ribosomally synthesized and post-translationally modified peptide prenyltransferases. J Am Chem Soc. 2018;140(26):8124–8127. doi: 10.1021/jacs.8b05187. [DOI] [PMC free article] [PubMed] [Google Scholar]