Abstract
Roots are essential organs for capturing water and nutrients from the soil. In particular, root system architecture (RSA) determines the extent of the region of the soil where water and nutrients can be gathered. As global climate change accelerates, it will be important to improve belowground plant parts, as well as aboveground ones, because roots are front-line organs in the response to abiotic stresses such as drought, flooding, and salinity stress. However, using conventional breeding based on phenotypic selection, it is difficult to select breeding lines possessing promising RSAs to adapted to abiotic stress because roots remain hidden underground. Therefore, new breeding strategies that do not require phenotypic selection are necessary. Recent advances in molecular biology and biotechnology can be applied to the design-oriented breeding of RSA without phenotypic selection. Here I summarize recent progress in RSA ideotypes as “design” and RSA-related gene resources as “materials” that will be needed in leveraging these technologies for the RSA breeding. I also highlight the future challenges to design-oriented breeding of RSA and explore solutions to these challenges.
Keywords: DRO1, edaphic stress, qSOR1, QTL, root plasticity, root robustness
Introduction
By 2050, the world’s population is expected to grow to 9.7 to 10 billion people (Gupta et al. 2020). This will require an increase in food production, whereas the area of global arable land available for agriculture will not increase proportionally. In addition, the water demand for agriculture is expected to increase, even though the water supply is expected to be insufficient to meet this new demand (Gupta et al. 2020). Furthermore, drought and flooding events in agricultural land may also increase due to global climate change (Bailey-Serres et al. 2019). In order to achieve increased crop production under a cruel environment for crops, development of crops that is resistant to environmental stresses will play an important role.
Since the “Green Revolution” has developed high-yield varieties of rice and wheat, crop breeding for maximum yield in an environment where water and fertilizer are available has primarily focused on improving aboveground plant architecture (Khush 2001). However, it is also important to improve belowground plant architecture to facilitate future increases in crop production in unstable environments where water and nutrients are insufficient (Herder et al. 2010). Many abiotic stresses such as drought, submergence, and nutrient deficiency or excess affect plants through their roots. However, little progress has been made in improving root traits compared to improving aboveground traits.
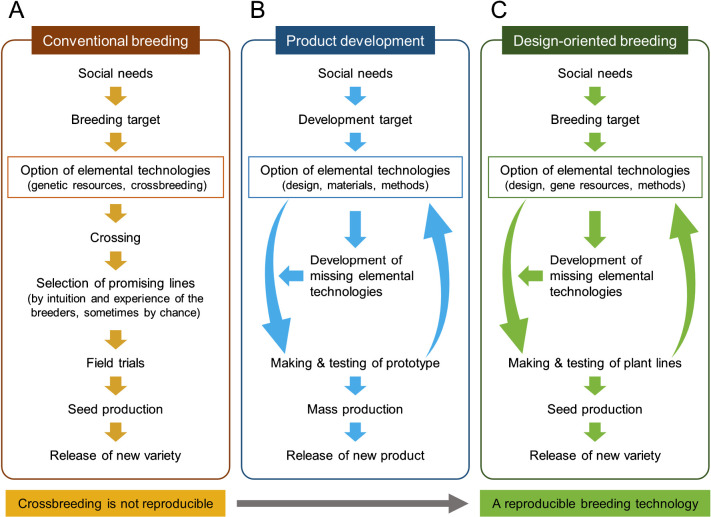
Crossbreeding has been a main driver of crop development to this date. Breeders’ intuition and experience has greatly contributed to the selection of promising breeding lines after crossing (Fig. 1A). Unfortunately, in many cases, breeders cannot directly visualize roots because they remain hidden underground. To overcome this limitation, many researchers are developing technologies that enable high-throughput phenotyping of root traits in the field (Atkinson et al. 2019). At present, there are few low-cost, high-throughput root phenotyping methods available for breeding of root characters in the field (Teramoto et al. 2019, Yoshino et al. 2019). Shovelomics, a method consisting of digging up the root base of plants grown in the field and measuring root characters, is one of the simplest and easiest root phenotyping methods (Trachsel et al. 2011), but provides only limited information on the basal parts of the root, not a picture of the whole root system. Thus, it is still difficult to improve root traits by phenotypic selection during the breeding process.
Fig. 1.
Concept of design-oriented breeding based on the different features between crossbreeding and industrial product development.
How can root traits be improved in an efficient manner? Recent advances in molecular biology and biotechnology have provided many breeding techniques for practical crop breeding, such as DNA marker-assisted selection, genomic selection, and genome editing (Hickey et al. 2019). These techniques are expected to be used for the breeding of root traits that are difficult to conduct phenotypic selection in the field. To do so, a new breeding strategy that does not require root phenotypic selection will be needed. An ideal breeding without phenotypic selection is to design an ideal root system adapted to a target edaphic environment via modification of the genes associated with target root traits.
The developmental process used in industrial manufacturing may be helpful in considering how to conceptualize such a breeding strategy. In developing an industrial product, a prototype product is designed to fulfill a social need, and optimal materials and production methods are selected before the prototype is developed (Fig. 1B). When a required technology is missing, it must be developed before production of the prototype can begin. When the performance of the prototype is acceptable, it is mass produced and then released. Compared to product development, a major problem with conventional breeding is that a common ideotype—i.e., the “design”—is not agreed upon by stakeholders beforehand. Crop design is often done independently by each breeder, and promising breeding lines are likely to be selected by the judgment of individual breeders (Fig. 1A). This process lacks reproducibility. Another problem is that the effects of genetic resources—i.e., the “materials”—can be inconsistent. In crop breeding, a gene that shows an effect on a target trait in one variety will not necessarily have the same effect in another. In contrast, during product development, the same elemental technologies can be used to produce a given product anywhere, with high reproducibility.
Like product development, protocols of design-oriented breeding can be divided into three elemental components: “design”, “materials”, and “methods” (Fig. 1C). Each of these three elements can be understood to correspond to different features of a plant breeding system: “design” corresponds an ideotype, “materials” to gene resources, and “methods” to various breeding techniques, including DNA marker-assisted selection, genomic selection, and genome editing, among others. Of these components, breeding techniques are highly reproducible, although some techniques (e.g., tissue culture) may require modification depending on the species and/or subspecies used. On the other hand, ideotypes and gene resources are not highly reusable for different species. If they could be as consistently applied across species as breeding techniques can be, this would facilitate the development of new crops efficiently by strategies that did not focus on phenotypic selection (Fig. 1C).
In this article, I consider the design-oriented breeding of roots, and summarize the current status and challenges for each elemental technology related to plant root breeding. I focus on “design” and “materials” because “methods” are commonly considered by researchers interested in other, non-root traits. Root traits include root system architecture (RSA), root structure, physiological function, and root-microbe interactions, and it is difficult to cover each of these in a single review. Therefore, I focus only on RSA, which is the main component determining the area from which roots are able to capture water and nutrients from the soil (Kitomi et al. 2018).
Design: what is the plant ideotype with respect to root system architecture?
During crop breeding, each breeder imagines society’s needs in the present or for the future and selects promising lines that have the required traits. The ideotype is a very useful conceptual framework for breeders interested in phenotypic selection. Since the “Green Revolution”, the breeding of wheat and rice for aboveground architectural traits has progressed with the semi-dwarf plant as an ideotype (Khush 2001). Like aboveground architecture, RSA ideotypes have also been proposed by many researchers (Lynch 2019, Meister et al. 2014, Schmidt and Gaudin 2017). However, RSA ideotype breeding is not widely practiced because roots remain hidden underground. In this section, I summarize the representative RSA ideotypes proposed by previous studies and discuss the challenges facing targeted breeding of RSA ideotypes. I also consider two types of soil-related stress: biotic stresses, such as pests and diseases, as well as abiotic stresses, such as drought and salinity. I focus on the main abiotic stresses relevant for agriculture, especially water stresses, such as drought and flooding, and mineral stress, such as nitrogen, phosphorus, and salinity. I introduce the RSA ideotypes by comparing these abiotic stresses in two agroecosystems: regular fields (including both dry fields and upland fields), and paddies.
Drought stress
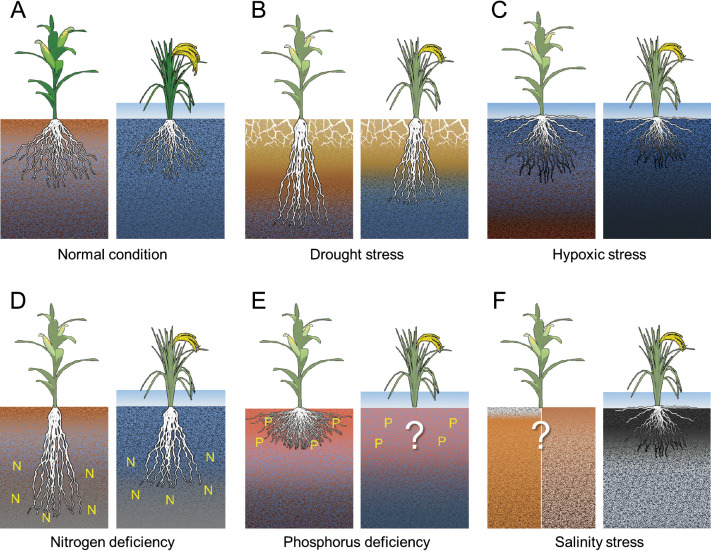
When rainfall is scarce, continued water evaporation from the soil surface results in drought conditions. Relative to the RSA suitable during normal conditions (Fig. 2A), a deeper RSA may be more advantageous under drought conditions brought about by drying from the soil surface (Fig. 2B). Under the surface, those maize varieties with fewer crown and lateral roots, as well as steeper and thicker roots, are better adapted for water uptake from the subsoil (Lynch 2013, Lynch and Wojciechowski 2015). In rice, a deeper rooting phenotype is related to a functional allele at DEEPER ROOTING 1 (DRO1), a quantitative trait locus (QTL) for root growth angle, which limits the impact of drought stress. A comparative study found that a functional DRO1 allele results in increased grain yield, whereas the shallow rooting phenotype resulting from a non-functional allele at DRO1 was susceptible to drought stress (Uga et al. 2013). Paddy fields consist of an impermeable hardpan under a plow layer, depending on soil condition. It has been thought that the ability of roots to penetrate the hardpan and for lateral root formation to efficiently capture water from the plow layer are important plant adaptations for paddy crops (Kano-Nakata et al. 2013, Suralta et al. 2018). However, in many paddies in drought-prone areas around South Asia, this hardpan is not formed (Singh et al. 2017). For those paddies without a hardpan layer, the RSA ideotype idea for avoiding drought stress may be the same as that for regular fields (Fig. 2B).
Fig. 2.
Promising ideotypes of root system architectures adapted to abiotic stresses. In each panel A–F, left and right figures represent regular and paddy fields, respectively. Each panel shows schematic models of the ideal root system architecture under a different abiotic stress condition. Plants shown are maize and rice—i.e., representative monocot crops—grown in regular and paddy fields, respectively. N, nitrogen; P, phosphorous. Question marks in the figure mean that no RSA ideotype have been established based on field studies.
Hypoxic stress caused by flooding
In extreme submergences, such as catastrophic floods, the roots can act as an anchor to prevent a plant from being washed away. When flooding occurs from extensive precipitation such that water covers the soil surface, field crops will experience stress disorders, such as root rot caused by hypoxic stress. In this case, rooting on the soil surface and/or adventitious rooting around the topsoil may be an effective RSA to avoid hypoxic condition (Fig. 2C; Pedersen et al. 2021). While common maize and its wild relatives (teosintes) are susceptible to flooding, some teosinte accessions found in the wetlands of Nicaragua have been found to be adapted to flooding conditions (Mano and Omori 2007). These accessions can develop soil-surface roots (SOR) to access oxygen from the air, even when the plant is flooded. When flooding conditions continue until harvest in paddies without drainage, soil reduction can be caused by hypoxic conditions, resulting in reduced root development (Fageria et al. 2008, Takai and Kamura 1966). In rice, the SOR phenotype has been also found in Indonesian lowland rice belonging to the Bulu ecotype, although common rice ordinarily develops underground roots (Ueno and Sato 1989). SOR formation may have arisen because selection pressures have driven the Bulu ecotype to adapt to severe hypoxic environments (Lafitte et al. 2001). Recently, a QTL associated with SOR formation, quantitative trait locus for SOIL SURFACE ROOTING 1 (qSOR1), has been isolated from one of these Bulu varieties (Kitomi et al. 2020). In a paddy field subject to reducing stress, rice plants with SOR, caused by a loss-of-function allele at qSOR, had higher grain yields than rice without SOR. Thus, regardless of the crop species, the SOR phenotype may be an effective RSA in avoiding hypoxic stress (Fig. 2C).
Nutrient deficiency
There are two types of minerals found in field conditions: water-soluble and water-insoluble minerals. Water-soluble minerals tend to move into the subsoil according to gravity, following water, whereas water-insoluble minerals tend to remain in the topsoil. Of the most important minerals for plant growth, nitrogen (N) is mobile and phosphorus (P) is immobile. Since both minerals are very important for crop production, they are typically added to the fields in large quantities in developed regions. On the other hand, in developing regions, the soil may be deficient in both minerals due to lack of adequate fertilizer application. In both cases, RSA optimization is important because it allows more efficient absorption of each mineral.
In the field, N easily shifts to the subsoil with water. For this reason, RSAs consisting of steeper, longer, fewer, and thicker roots may be most effective for field crop N accumulation in (Fig. 2D). For example, a “steep, cheap, and deep” RSA ideotype for maize has been proposed; this would permit effective uptake of N as well as water from the subsoil (Lynch 2013). In paddy fields with a hardpan under the plow layer, N loss caused by leaching to the subsoil is negligible (Yamamuro 1986). However, deep roots have been reported to affect to yield. In rice, a deep root-type line, showing higher N uptake after the heading stage, had a higher yield compared to a shallow root-type line (Arai-Sanoh et al. 2014). Just as in regular fields, a deep RSA may also be advantageous for N absorption in paddy fields (Fig. 2D).
An RSA ideotype including shallower axial roots, enhanced adventitious rooting, a greater number of axial roots, and a greater dispersion of lateral roots may more effectively capture P from the topsoil under low P soil conditions (Fig. 2E; Lynch 2011). Common beans (Phaseolus vulgaris) showing a shallow RSA phenotype were found to show higher fertility than deep RSA ones in P-deficient soils (Lynch and Brown 2001). The same trend was also observed in maize (Lynch 2019, Zhu et al. 2005). In rice, P deficiency is a major limiting factor in upland and rain-fed lowland fields in several developing regions (Ismail et al. 2007). P treatments applied to paddy fields behave similar those applied to regular fields (Oo et al. 2020). There, rice roots have been found to develop in soil zones characterized by high P concentration (He et al. 2003). These data suggest that shallower RSAs may be advantageous in paddy fields with P deficiency condition just as they are in regular fields (Fig. 2E). However, which RSA ideotype is suitable for P-deficient paddy fields remains unclear and requires further study.
Salinity stress
Salinity stress is one of major abiotic stresses and is expected to have an adverse impact on an estimated 50% of all arable lands worldwide by 2050 (Butcher et al. 2016). To develop salt-tolerant crops, many studies have identified mechanisms of salt tolerance and to isolate related genes (Bailey-Serres et al. 2019, van Zelm et al. 2020). However, no RSA ideotype for salt resistance has been reported to date. If such an RSA existed, it would be expected to enable stable crop production in saline-prone areas, which are estimated to increase in abundance in the future (Rogers and Benfey 2015). Salts present in fertilizer and/or irrigation water can accumulate in the soil surface or subsoil, and the degree to which this happens depends on differences in cultivation conditions including the natural environment and irrigation (Ismail et al. 2007, Munns et al. 2006). Little is known about the ideal RSA for such conditions (Fig. 2F), although inducible development of adventitious roots with cortical aerenchyma may be useful in irrigated saline soils where water input is available (Schmidt and Gaudin 2017). In coastal areas, saltwater intrusion has resulted from sea level rises caused by global warming, resulting in saline damage to crop production (Gopalakrishnan et al. 2019). In coastal paddy fields, long-term saline condition cause not only salinity stress but also reducing stress due to soil structural changes by salts (Qadir and Schubert 2002, Srivastava et al. 2014). Recently, the SOR phenotype has been found to allow rice plants to avoid reducing stress caused by saline conditions (Kitomi et al. 2020). This suggests that the SOR phenotype may be useful in saline paddy fields (Fig. 2F) as well as in flooded paddy fields (Fig. 2C), although the SOR does not strengthen ability of salinity tolerance (Kitomi et al. 2020). The degree to which the SOR phenotype is effective in reducing salt stress in field crops should be clarified in future studies.
Robustness vs plasticity in RSA
Plants have both robustness- and plasticity-oriented responses to environmental changes. With respect to roots, robustness is understood as the ability to continue innate development irrespective of environmental changes, whereas plasticity reflects the ability of the root to adapt to these changes. In the following section, I discuss the advantages and disadvantages of root robustness and plasticity for RSA in response to a hypothetical environmental change to the water environment.
When soil moisture varies strongly over short time scales in the field, a robust RSA would be insensitive to short-lived changes, thereby reducing the cost of mounting a response to environmental variation. On the other hand, a plastic RSA would be sensitive to environmental changes and would involve an adaptive strategy. The extra costs associated with adapting to constantly changing environments may result in decreased crop yield (Schneider and Lynch 2020). With respect to long-term environmental changes, the robust RSA strategy will have an advantage if it is well adapted to the environment. However, if the RSA type is not well adapted to the target environment, plant growth will be compromised by abiotic stress. For example, deep-rooted cultivars could avoid drought while shallow-rooted cultivars would have limited growth (Uga et al. 2013). However, when the deep root-type cultivar is grown under normal moisture conditions, the development of roots that extend to the subsoil without fertilization will be costly and will not enhance seed production. For example, deep rooted upland rice generally has a lower yield than shallow rooted lowland rice (Atlin et al. 2006, Fukai and Cooper 1995). In contrast, the plastic RSA strategy allows plants to response to stress conditions (e.g., drought) by adaptively changing their roots only when drought is experienced. For example, in rice, an introgression line showing plasticity in lateral root length and density under drought conditions showed increased shoot biomass relative to the wild type variety, which lacked RSA plasticity (Kano-Nakata et al. 2013). Thus RSA plasticity gives plants the ability to adapt to the environment, but only at a cost (Schneider and Lynch 2020), and it is not well understood whether RSA plasticity contributes to increase crop yield under stress conditions.
When considering RSA ideotype breeding, it is important to first ask which is easier to use: a robust RSA or plastic RSA? At present, robust RSAs may be preferable. Ideal plant architecture (IPA) for aboveground plant parts is usually determined by robust traits such as culm and panicle size and number (Dong et al. 2017, Jiao et al. 2010). These IPAs are easy to select for ideotype breeding in practice. A robust RSA is probably also desirable for well-defined climates, with distinct wet and dry seasons. For example, deep root RSA phenotypes would be useful during prolonged drought, while shallow root RSA phenotypes would be useful in lowland fields prone to flooding. In contrast, there are few comprehensive field studies that have identified how plasticity may contribute to crop productivity and which morphological changes are advantageous (Schneider and Lynch 2020). Therefore, the method by which RSA plasticity can be utilized as an ideotype is not well understood. However, if the mechanisms responsible for RSA plasticity are elucidated in the field, it could find use as a new ideotype in the future.
Beyond the relationship between different abiotic stresses and RSA, there are other factors that should be considered to allow for RSA ideotype breeding. High temperature stress often occurs simultaneously in environments where drought and salinity stresses are also present (Calleja-Cabrera et al. 2020). In such cases, it is necessary to select RSA profiles that show high resistance against multiple abiotic stresses. It is also possible that N and P deficiencies may occur simultaneously in some regions. In these cases, since the RSA ideotypes required for N and P deficiencies are completely different (Lynch 2019), breeders will have to determine whether deep, intermediate, or shallow root phenotypes would be prioritized. Thus, there is no universal RSA ideotype for multiple abiotic stresses (Lynch 2019, Schneider and Lynch 2020). When considering RSA ideotypes, the influence of cropping system, including factors such as planting density (Meister et al. 2014), cultivation period (Voss-Fels et al. 2018), water source (i.e., whether crops are irrigated or rainfed) (Schmidt and Gaudin 2017) and of soil physical properties such as soil compaction (Ramalingam et al. 2017) should also be considered. Data collection in all environments and for all cropping systems, is an unrealistic goal. For this reason, simulation studies that model RSA phenotypes can complement field-based RSA research (de Dorlodot et al. 2007, Postma et al. 2014, Rellán-Álvarez et al. 2016). With the accumulation of such data, accurate simulation-based design of RSA ideotypes will be possible in the future.
Materials: gene resources to enable design-oriented breeding
Breeding materials are essential for crop development. Plant crossbreeding involves selecting accessions with the desired characters and using them as breeding stock to produce new plants with novel phenotypes. On the other hand, recently developed breeding techniques, such as DNA marker-assisted selection and genome editing, cannot use directly such existing genetic resources as breeding material. For example, DNA marker-assisted selection requires the mapping of genes related to the target trait before existing genetic resources can be used as breeding materials. Moreover, the cloning and characterizing of genes responsible for target traits is required before genome editing can take place. So far, many promising RSA-related genes have been identified in crops. These discoveries have resulted from many different genetic approaches, including mutant assays, QTL analysis, and genome-wide association studies (Deja-Muylle et al. 2020, Meister et al. 2014, Rogers and Benfey 2015). Moreover, the molecular functions of RSA-related genes in maize and rice have been described in detail (Hochholdinger et al. 2018, Kitomi et al. 2018, Wachsman et al. 2015). Only a few RSA-related cloned genes have been shown to contribute to abiotic stress resistance under field conditions (Mickelbart et al. 2015). In this section, genes related to robust and plastic RSAs, which we could use as breeding materials, are introduced.
Robust RSA-related genes
Some robust RSA-related genes, including DRO1 and qSOR1, have been shown to contribute to resistance to abiotic stress in the field. DRO1 is the first cloned QTL for root growth angle in crops (Uga et al. 2013). This QTL was identified on rice chromosome 9 from a mapping population derived from a cross between ‘IR64’, lowland rice with a shallow root type, and ‘Kinandang Patong’, an upland rice with a deep root type (Uga et al. 2011). An near-isogenic line (NIL) of ‘IR64’ introducing a functional allele at DRO1 from ‘Kinandang Patong’ showed deeper rooting than ‘IR64’, resulting in higher yield under drought conditions in upland fields (Uga et al. 2013) and paddy fields (Uga et al. unpublished data). qSOR1 is the second cloned QTL for root growth angle in crops (Kitomi et al. 2020). This gene has been identified on rice chromosome 7 from a mapping population derived from a cross between ‘Gemdjah Beton’, a lowland rice with SOR, and ‘Sasanishiki’, a lowland rice without SOR (Uga et al. 2012). A NIL of ‘Sasanishiki’ containing a non-functional allele at qSOR1 from ‘Gemdjah Beton’ showed SOR in both upland and paddy fields (Kitomi et al. 2020). This NIL with SOR avoids the reducing stress found in saline paddy fields and results in reduced yield loss.
Homology searches based on the amino acid sequence of qSOR1 revealed that rice DRO1 was most closely related to rice qSOR1 (Kitomi et al. 2020). There are several regions of conserved sequence between these two genes, although the overall similarity of their amino acid sequences is low (i.e., less than 30%). Recent work has indicated that there are four DRO1 homologs in rice, which constitute three subgroups (Kitomi et al. 2020), named DRO1, qSOR1/DRL1 (DRO1-like 1), and DRL2, respectively. A study of DRL2 mutant lines made by genome editing demonstrated that DRL2 was also involved in root growth angle (Kitomi et al. 2020). Taken together, these data suggest that the three subgroups of the DRO1 family, have important roles in regulating RSA in rice.
Laboratory studies have shown that both DRO1 and qSOR1 are early auxin response genes involved in root gravitropism in rice (Fig. 3, Kitomi et al. 2020, Uga et al. 2013). In natural conditions, both genes show robust RSA phenotypes in both regular and paddy field environments but do not affect tiller angle (Kitomi et al. 2020). Furthermore, they have little effect on shoot and root morphology other than on root growth angle. Combining the effects of both genes may make it possible to produce a wide variation of RSAs, without considering investment trade-offs between shoots and roots. Root trait breeders have often assumed that it would be difficult to modify only target root traits without simultaneously affecting shoot characters because of this trade-off (Voss-Fels et al. 2017). However, our studies have demonstrated that it is possible to improve RSA without considering the trade-off between shoots and roots, although the effect of each gene must be carefully characterized in a field environment before being used for breeding.
Fig. 3.
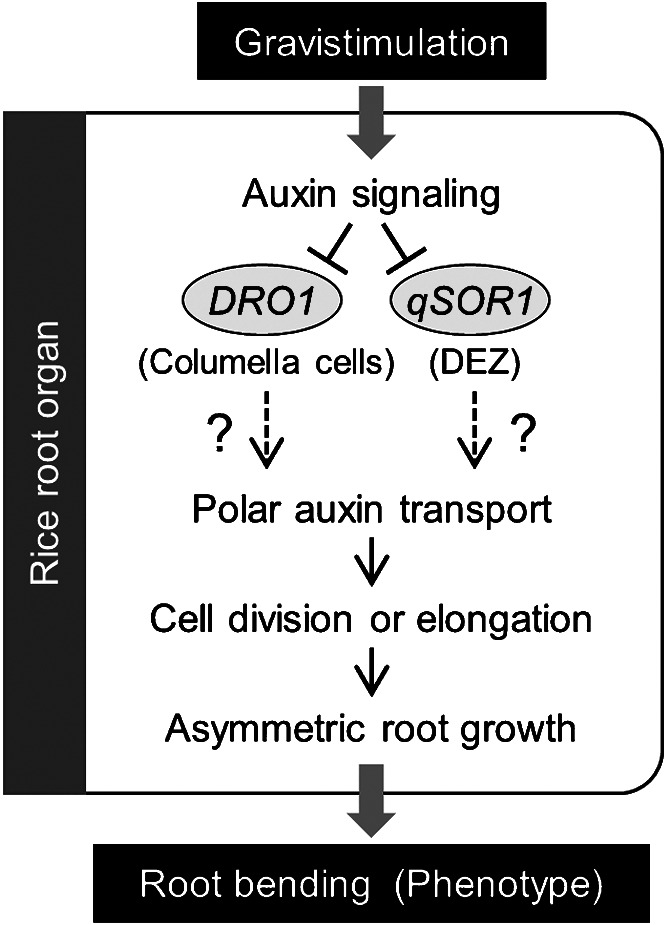
Proposed regulatory pathways of gravity-induced root bending via DRO1 homologs in rice. Term in parentheses indicates the major tissues expressing each gene. DEZ, distal elongation zone.
DRO1 homologs have also been reported to be involved in root gravitropism in dicots, including species in the genera Medicago, Arabidopsis, and Prunus (Ge and Chen 2016, Guseman et al. 2017, Taniguchi et al. 2017). This suggests that DRO1 homologs may form an important gene family regulating RSA across the angiosperms. If so, the DRO1 family is likely to be used as a gene resource for RSA breeding in crops other than rice. In what follows, I will summarize the DRO1 family members that have been identified thus far.
Among monocots other than rice, DRO1 homologs have also been identified in wheat, although whether they affect RSA is still unknown (Ashraf et al. 2019). In 2007, LAZY1, a gene involved in shoot gravitropism, was identified in a rice mutant (Yoshihara and Iino 2007). More recent studies have found that LAZY1 is highly similar to the DRO1 family in multiple domain sequences (Guseman et al. 2017), including a conserved C-terminal domain known as conserved C terminus in the LAZY1 family of proteins (CCL) (Taniguchi et al. 2017). Tiller Angle Control 1 (TAC1), a gene involved in tiller angle in rice (Yu et al. 2007), has also been found to have a high similarity to the DRO1 family in multiple domain sequences without a CCL domain (Nakamura et al. 2019). These data further suggest that DRO1 homologs form an extensive gene family associated with shoot and/or root gravitropism in monocots.
In dicots, several DRO1 homologs have been found in aberrant gravitropic mutants. In Medicago, a gene known as NEGATIVE GRAVITROPIC RESPONSE OF ROOTS (NGR) was isolated from mutants showing roots emerging above the soil surface (Ge and Chen 2016). In Arabidopsis, AtDRO1 mutants showed shallower lateral root angles compared to wild type plants (Guseman et al. 2017). LAZY1 homologs have also been found in Arabidopsis (Taniguchi et al. 2017), and each of these could be classified into one of three groups (Nakamura et al. 2019): (1) LZY1 (LAZY1-LIKE 1), a rice LAZY homolog, is involved in shoot gravitropism; (2) LZY4 is involved solely in root gravitropism; and (3) LZY2 and LZY3 are involved in both shoot and root gravitropism. Nakamura et al. (2019) mentioned that LZY3 is a rice DRO1 homolog. However, phylogenic analysis of the DRO1 gene family in both monocots and dicots revealed that many dicots showed more similar sequences to qSOR1 than to DRO1, suggesting that the qSOR1 sequence may be more common than the DRO1 sequence in angiosperms (Kitomi et al. 2020).
The homologs of DRO1 and LAZY1 are therefore present in both monocots and dicots, forming a large gene family that controls shoot and root gravitropism (Kitomi et al. 2020). In Arabidopsis, the CCL domain of the LZY protein binds to the Brevis radix (BRX) domains of the RCC-like domain (RLD) protein, thereby allowing LZY to transport RLD from the cytoplasm to the plasma membrane (Furutani et al. 2020). Furutani et al. (2020) showed that these genes affected gravitropism through their involvement in PIN-FORMED (PIN)-mediated auxin polarity transport. Recently, light signals were found to regulate LZY expression, resulting in change of response to the gravistimulus (Yang et al. 2020a). Light promotes accumulation of ELONGATED HYPOCOTYL5 (HY5) proteins, which directly bind and activate the expression of LAZY4/LZY3/NGR2/DRO1 in Arabidopsis roots. Thus, further clarification of the functions of individual genes in the DRO1/LAZY1 family is expected to enable more precise genetic control of the above- and belowground architecture of crops. In other words, DRO1/LAZY1 family genes have strong potential to be used as “materials” for design-oriented breeding of not only the RSA but of the architecture of the whole plant in many crops.
Plastic RSA-related genes
As for robust RSAs, few genes involved in plastic RSAs have been reported. However, genes involved in plastic RSAs may be just as important a breeding material as genes involved in robust RSAs (Dwivedi et al. 2020). In rice, Rice Morphology Determinant (RMD), a gene encoding an actin-binding protein, controls gravitropic responses to low P condition (Huang et al. 2018). Since RMD controls from deep to shallow rooting at low P conditions, RMD is expected to be a useful RSA gene, enhancing P capture under low P conditions. However, its usefulness has not been demonstrated in the field, so further studies designed to verify this effect are needed. The paucity of reports on the usability of genes for plastic RSAs may be due to the fact that it is difficult to evaluate them in the field, since it is difficult to control stress conditions underground. Another reason may be related to the crop domestication process. During domestication, humans may have selected mainly for yield-related traits, including seed size and number, as well as cultivation-related traits including seed shattering and dormancy. In cultivated fields where domesticated crops are more stable than they were in their natural habitat (Wallace et al. 2018), plastic RSA responses may have been unwanted and ultimately lost by the modern farming system (Calleja-Cabrera et al. 2020). Consequently, genetic diversity for RSAs in modern crops was likely reduced by domestication and breeding bottlenecks (Voss-Fels et al. 2018). Placido et al. (2020) reported that allele of Agropyron elongatum, a wild relative of wheat, at LATERAL ROOT DENSITY (LRD), a gene involved in lateral root formation, promoted lateral root growth under drought conditions. Crop wild species are a promising genetic resource for future breeding of improved root stress response (Calleja-Cabrera et al. 2020). Therefore, the discovery of useful alleles from genetic resources such as wild species is important for the design-oriented breeding of RSAs in the future.
Future perspective: is the design-oriented breeding of RSA possible?
In this review, I have described two elemental technologies (“design” and “materials”) for design-oriented breeding of RSA. Although RSA ideotypes have been proposed in many crops, few RSA ideotypes adapted to abiotic stresses have been established based on individual genes for RSA-related traits whose functions have been characterized in field studies (Arai-Sanoh et al. 2014, Kitomi et al. 2020, Uga et al. 2013, 2015). Bi-parental mapping populations, such as recombinant inbred lines, are not suitable for evaluating the effects of each trait because such populations include lines showing simultaneously varying phenotypic segregations (Yamamoto et al. 2014). However, plant materials with unified genetic backgrounds, such as NILs and gene pyramiding lines, may be better options to examine these traits. However, the isolation of genes or QTLs involved in target traits and the development of plant materials such as NILs are time-consuming. With respect to the “materials” available to study RSA ideotypes, a large number of RSA-related genes have been identified, but their effects on target traits in the field have not been adequately validated. This is also a time-consuming activity, and many individual genes isolated from different accessions should be evaluated in the same genetic background.
A common challenge with both elemental technologies is that the development of plant materials needed to evaluate target traits or genes is a significant time investment. Two techniques that have been developed in recent years that may help alleviate this problem. The first is genome editing. For example, multiple RSA-related genes found in different species or varieties can be easily introduced into a model or representative accession by genome editing techniques. The effect of each gene on the different environmental stresses (G × E) can then be evaluated in the same genetic background. By simultaneously editing multiple genes, we can also verify the effect of gene pyramiding on different environmental stresses (G × G × E). In addition, an allele gained by genome editing will allow us to identify the allelic variation involved in phenotypic variation. The second technique is speed breeding (Hickey et al. 2019, Watson et al. 2018). Speed breeding permits the shortening of plant generation time by controlling plant environments, resulting in rapid accumulation of genes or QTLs. Thus, both techniques can be used to collect information relevant to ideotype “design” and “materials” in a short period of time.
Another challenge with both elemental technologies is that high-throughput RSA phenotyping in the field is very difficult and labor-intensive (Yoshino et al. 2019). In particular, when evaluating root plasticity under abiotic stress conditions, it is necessary to observe RSA development continuously. However, this is still difficult to do in the field (Voss-Fels et al. 2018). Currently, phenotyping platforms permitting the controlling and monitoring of environmental conditions in greenhouses and/or growth chambers have been developing (Yang et al. 2020b). In addition, non-invasive RSA phenotyping techniques using X-ray computed tomography and magnetic resonance imaging are also being developed (Atkinson et al. 2019, Teramoto et al. 2020). Combining these technologies would permit the collection of highly reproducible RSA data non-destructively under reproducible environmental stress conditions and would be a viable alternative to evaluating plants in the field. I believe that progress in the research described above will make the design-oriented breeding of RSAs a reality.
Author Contribution Statement
Y.U. wrote the manuscript.
Acknowledgments
I thank Dr. Y. Tsujimoto (JIRCAS, Japan) for providing useful comments. This work was supported by JST CREST (Grant Number JPMJCR17O1), Japan.
Literature Cited
- Arai-Sanoh, Y., Takai T., Yoshinaga S., Nakano H., Kojima M., Sakakibara H., Kondo M. and Uga Y. (2014) Deep rooting conferred by DEEPER ROOTING 1 enhances rice yield in paddy fields. Sci. Rep. 4: 5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf, A., Rehman O.U., Muzammil S., Léon J., Naz A.A., Rasool F., Ali G.M., Zafar Y. and Khan M.R. (2019) Evolution of Deeper Rooting 1-like homoeologs in wheat entails the C-terminus mutations as well as gain and loss of auxin response elements. PLoS ONE 14: e0214145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, J.A., Pound M.P., Bennett M.J. and Wells D.M. (2019) Uncovering the hidden half of plants using new advances in root phenotyping. Curr. Opin. Biotechnol. 55: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlin, G.N., Lafitte H.R., Tao D., Laza M., Amante M. and Courtois B. (2006) Developing rice cultivars for high-fertility upland systems in the Asian tropics. Field Crops Res. 97: 43–52. [Google Scholar]
- Bailey-Serres, J., Parker J.E., Ainsworth E.A., Oldroyd G.E.D. and Schroeder J.I. (2019) Genetic strategies for improving crop yields. Nature 575: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher, K., Wick A.F., Desutter T., Chatterjee A. and Harmon J. (2016) Soil salinity: a threat to global food security. Agron. J. 108: 2189–2200. [Google Scholar]
- Calleja-Cabrera, J., Boter M., Oñate-Sánchez L. and Pernas M. (2020) Root growth adaptation to climate change in crops. Front. Plant Sci. 11: 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dorlodot, S., Forster B., Pagès L., Price A., Tuberosa R. and Draye X. (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 12: 474–481. [DOI] [PubMed] [Google Scholar]
- Deja-Muylle, A., Parizot B., Motte H. and Beeckman T. (2020) Exploiting natural variation in root system architecture via genome-wide association studies. J. Exp. Bot. 71: 2379–2389. [DOI] [PubMed] [Google Scholar]
- Dong, Z., Li W., Unger-Wallace E., Yang J., Vollbrecht E. and Chuck G. (2017) Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci. USA 114: E8656–E8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi, S.L., Stoddard F.L. and Ortiz R. (2020) Genomic-based root plasticity to enhance abiotic stress adaptation and edible yield in grain crops. Plant Sci. 295: 110365. [DOI] [PubMed] [Google Scholar]
- Fageria, N.K., Santos A.B., Barbosa Filho M.P. and Guimarães C.M. (2008) Iron toxicity in lowland rice. J. Plant Nutr. 31: 1676–1697. [Google Scholar]
- Fukai, S. and Cooper M. (1995) Development of drought-resistant cultivars using physiomorphological traits in rice. Field Crops Res. 40: 67–86. [Google Scholar]
- Furutani, M., Hirano Y., Nishimura T., Nakamura M., Taniguchi M., Suzuki K., Oshida R., Kondo C., Sun S., Kato K.et al. (2020) Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat. Commun. 11: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, L. and Chen R. (2016) Negative gravitropism in plant roots. Nat. Plants 2: 16155. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan, T., Hasan M.K., Haque A.T.M.S., Jayasinghe S.L. and Kumar L. (2019) Sustainability of coastal agriculture under climate change. Sustainability (Switzerland) 11: 1–24. [Google Scholar]
- Gupta, A., Rico-Medina A. and Caño-Delgado A.I. (2020) The physiology of plant responses to drought. Science 368: 266–269. [DOI] [PubMed] [Google Scholar]
- Guseman, J.M., Webb K., Srinivasan C. and Dardick C. (2017) DRO1 influences root system architecture in Arabidopsis and Prunus species. Plant J. 89: 1093–1105. [DOI] [PubMed] [Google Scholar]
- He, Y., Liao H. and Yan X. (2003) Localized supply of phosphorus induces root morphological and architectural changes of rice in split and stratified soil cultures. Plant Soil 248: 247–256. [Google Scholar]
- Herder, G.D., Van Isterdael G., Beeckman T. and De Smet I. (2010) The roots of a new green revolution. Trends Plant Sci. 15: 600–607. [DOI] [PubMed] [Google Scholar]
- Hickey, L.T., Hafeez A.N., Robinson H., Jackson S.A., Leal-Bertioli S.C.M., Tester M., Gao C., Godwin I.D., Hayes B.J. and Wulff B.B.H. (2019) Breeding crops to feed 10 billion. Nat. Biotechnol. 37: 744–754. [DOI] [PubMed] [Google Scholar]
- Hochholdinger, F., Yu P. and Marcon C. (2018) Genetic control of root system development in maize. Trends Plant Sci. 23: 79–88. [DOI] [PubMed] [Google Scholar]
- Huang, G., Liang W., Sturrock C.J., Pandey B.K., Giri J., Mairhofer S., Wang D., Muller L., Tan H., York L.M.et al. (2018) Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat. Commun. 9: 2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail, A.M., Heuer S., Thomson M.J. and Wissuwa M. (2007) Genetic and genomic approaches to develop rice germplasm for problem soils. Plant Mol. Biol. 65: 547–570. [DOI] [PubMed] [Google Scholar]
- Jiao, Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X.et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544. [DOI] [PubMed] [Google Scholar]
- Kano-Nakata, M., Gowda V.R.P., Henry A., Serraj R., Inukai Y., Fujita D., Kobayashi N., Suralta R.R. and Yamauchi A. (2013) Functional roles of the plasticity of root system development in biomass production and water uptake under rainfed lowland conditions. Field Crops Res. 144: 288–296. [Google Scholar]
- Khush, G.S. (2001) Green revolution: the way forward. Nat. Rev. Genet. 2: 815–822. [DOI] [PubMed] [Google Scholar]
- Kitomi, Y., J. Itoh and Y. Uga (2018) Genetic mechanisms involved in the formation of root system architecture. In: Sasaki, T. and M. Ashikari (eds.) Rice Genomics, Genetics and Breeding, Springer Nature, Singapore, pp. 241–274. [Google Scholar]
- Kitomi, Y., Hanzawa E., Kuya N., Inoue H., Hara N., Kawai S., Kanno N., Endo M., Sugimoto K., Yamazaki T.et al. (2020) Root angle modifications by the DRO1 homolog improve rice yields in saline paddy fields. Proc. Natl. Acad. Sci. USA 117: 21242–21250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafitte, H.R., Champoux M.C., McLaren G. and O’Toole J.C. (2001) Rice root morphological traits are related to isozyme group and adaptation. Field Crops Res. 71: 57–70. [Google Scholar]
- Lynch, J.P. and Brown K.M. (2001) Topsoil foraging—an architectural adaptation of plants to low phosphorus availability. Plant Soil 237: 225–237. [Google Scholar]
- Lynch, J.P. (2011) Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol. 156: 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J.P. (2013) Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 112: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J.P. and Wojciechowski T. (2015) Opportunities and challenges in the subsoil: pathways to deeper rooted crops. J. Exp. Bot. 66: 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J.P. (2019) Root phenotypes for improved nutrient capture: an underexploited opportunity for global agriculture. New Phytol. 223: 548–564. [DOI] [PubMed] [Google Scholar]
- Mano, Y. and Omori F. (2007) Breeding for flooding tolerant maize using “teosinte” as a germplasm resource. Plant Root 1: 17–21. [Google Scholar]
- Meister, R., Rajani M.S., Ruzicka D. and Schachtman D.P. (2014) Challenges of modifying root traits in crops for agriculture. Trends Plant Sci. 19: 779–788. [DOI] [PubMed] [Google Scholar]
- Mickelbart, M.V., Hasegawa P.M. and Bailey-Serres J. (2015) Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16: 237–251. [DOI] [PubMed] [Google Scholar]
- Munns, R., James R.A. and Läuchli A. (2006) Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 57: 1025–1043. [DOI] [PubMed] [Google Scholar]
- Nakamura, M., Nishimura T. and Morita M.T. (2019) Bridging the gap between amyloplasts and directional auxin transport in plant gravitropism. Curr. Opin. Plant Biol. 52: 54–60. [DOI] [PubMed] [Google Scholar]
- Oo, A.Z., Tsujimoto Y., Rakotoarisoa N.M., Kawamura K. and Nishigaki T. (2020) P-dipping of rice seedlings increases applied P use efficiency in high P-fixing soils. Sci. Rep. 10: 11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, O., Sauter M., Colmer T.D. and Nakazono M. (2021) Regulation of root adaptive anatomical and morphological traits during low soil oxygen. New Phytol. 229: 42–49. [DOI] [PubMed] [Google Scholar]
- Placido, D.F., Sandhu J., Sato S.J., Nersesian N., Quach T., Clemente T.E., Staswick P.E. and Walia H. (2020) The LATERAL ROOT DENSITY gene regulates root growth during water stress in wheat. Plant Biotechnol. J. 18: 1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma, J.A., Schurr U. and Fiorani F. (2014) Dynamic root growth and architecture responses to limiting nutrient availability: linking physiological models and experimentation. Biotechnol. Adv. 32: 53–65. [DOI] [PubMed] [Google Scholar]
- Qadir, M. and Schubert S. (2002) Degradation processes and nutrient constraints in sodic soils. Land Degrad. Dev. 13: 275–294. [Google Scholar]
- Ramalingam, P., Kamoshita A., Deshmukh V., Yaginuma S. and Uga Y. (2017) Association between root growth angle and root length density of a near-isogenic line of IR64 rice with DEEPER ROOTING 1 under different levels of soil compaction. Plant Prod. Sci. 20: 162–175. [Google Scholar]
- Rellán-Álvarez, R., Lobet G. and Dinneny J.R. (2016) Environmental control of root system biology. Annu. Rev. Plant Biol. 67: 619–642. [DOI] [PubMed] [Google Scholar]
- Rogers, E.D. and Benfey P.N. (2015) Regulation of plant root system architecture: implications for crop advancement. Curr. Opin. Biotechnol. 32: 93–98. [DOI] [PubMed] [Google Scholar]
- Schmidt, J.E. and Gaudin A.C.M. (2017) Toward an integrated root ideotype for irrigated systems. Trends Plant Sci. 22: 433–443. [DOI] [PubMed] [Google Scholar]
- Schneider, H.M. and Lynch J.P. (2020) Should root plasticity be a crop breeding target? Front. Plant Sci. 11: 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.P., Jain A., Anantha M.S., Tripathi S., Sharma S., Kumar S., Prasad A., Sharma B., Karmakar B., Bhattarai R.et al. (2017) Depth of soil compaction predominantly affects rice yield reduction by reproductive-stage drought at varietal screening sites in Bangladesh, India, and Nepal. Plant Soil 417: 377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, P.K., Gupta M., Pandey A., Pandey V., Singh N. and Tewari S.K. (2014) Effects of sodicity induced changes in soil physical properties on paddy root growth. Plant Soil Environ. 60: 165–169. [Google Scholar]
- Suralta, R.R., Niones J.M., Kano-Nakata M., Thi Tran T., Mitsuya S. and Yamauchi A. (2018) Plasticity in nodal root elongation through the hardpan triggered by rewatering during soil moisture fluctuation stress in rice. Sci. Rep. 8: 4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, Y. and Kamura T. (1966) The mechanism of reduction in waterlogged paddy soil. Folia Microbiol. (Praha) 11: 304–313. [Google Scholar]
- Taniguchi, M., Furutani M., Nishimura T., Nakamura M., Fushita T., Iijima K., Baba K., Tanaka H., Toyota M., Tasaka M.et al. (2017) The Arabidopsis LAZY1 family plays a key role in gravity signaling within statocytes and in branch angle control of roots and shoots. Plant Cell 29: 1984–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto, S., Kitomi Y., Nishijima R., Takayasu S., Maruyama N. and Uga Y. (2019) Backhoe-assisted monolith method for plant root phenotyping under upland conditions. Breed. Sci. 69: 508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto, S., Takayasu S., Kitomi Y., Arai-Sanoh Y., Tanabata T. and Uga Y. (2020) High-throughput three-dimensional visualization of root system architecture of rice using X-ray computed tomography. Plant Methods 16: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel, S., Kaeppler S.M., Brown K.M. and Lynch J.P. (2011) Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341: 75–87. [Google Scholar]
- Ueno, K. and Sato T. (1989) Aerial root formation in rice ecotype Bulu. Japan. J. Trop. Agr. 33: 173–175. [Google Scholar]
- Uga, Y., Okuno K. and Yano M. (2011) Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 62: 2485–2494. [DOI] [PubMed] [Google Scholar]
- Uga, Y., Hanzawa E., Nagai S., Sasaki K., Yano M. and Sato T. (2012) Identification of qSOR1, a major rice QTL involved in soil-surface rooting in paddy fields. Theor. Appl. Genet. 124: 75–86. [DOI] [PubMed] [Google Scholar]
- Uga, Y., Sugimoto K., Ogawa S., Rane J., Ishitani M., Hara N., Kitomi Y., Inukai Y., Ono K., Kanno N.et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45: 1097–1102. [DOI] [PubMed] [Google Scholar]
- Uga, Y., Kitomi Y., Ishikawa S. and Yano M. (2015) Genetic improvement for root growth angle to enhance crop production. Breed. Sci. 65: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zelm, E., Zhang Y. and Testerink C. (2020) Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71: 403–433. [DOI] [PubMed] [Google Scholar]
- Voss-Fels, K.P., Qian L., Parra-Londono S., Uptmoor R., Frisch M., Keeble-Gagnère G., Appels R. and Snowdon R.J. (2017) Linkage drag constrains the roots of modern wheat. Plant Cell Environ. 40: 717–725. [DOI] [PubMed] [Google Scholar]
- Voss-Fels, K.P., Snowdon R.J. and Hickey L.T. (2018) Designer roots for future crops. Trends Plant Sci. 23: 957–960. [DOI] [PubMed] [Google Scholar]
- Wachsman, G., Sparks E.E. and Benfey P.N. (2015) Genes and networks regulating root anatomy and architecture. New Phytol. 208: 26–38. [DOI] [PubMed] [Google Scholar]
- Wallace, J.G., Rodgers-Melnick E. and Buckler E.S. (2018) On the road to breeding 4.0: unraveling the good, the bad, and the boring of crop quantitative genomics. Annu. Rev. Genet. 52: 421–444. [DOI] [PubMed] [Google Scholar]
- Watson, A., Ghosh S., Williams M.J., Cuddy W.S., Simmonds J., Rey M.D., Asyraf Md Hatta M., Hinchliffe A., Steed A., Reynolds D.et al. (2018) Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4: 23–29. [DOI] [PubMed] [Google Scholar]
- Yamamoto, T., Y. Uga and M. Yano (2014) Genomics-assisted allele mining and its integration into rice breeding. In: Tuberosa, R., A. Graner and E. Frison (eds.) Genomics of Plant Genetic Resources, Springer, Germany, pp. 251–265. [Google Scholar]
- Yamamuro, S. (1986) Behavior of nitrogen in paddy soils. Jpn. Agric. Res. Q. 20: 100–107. [Google Scholar]
- Yang, P., Wen Q., Yu R., Han X., Deng X.W. and Chen H. (2020a) Light modulates the gravitropic responses through organ-specific PIFs and HY5 regulation of LAZY4 expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 117: 18840–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W., Feng H., Zhang X., Zhang J., Doonan J.H., Batchelor W.D., Xiong L. and Yan J. (2020b) Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives. Mol. Plant 13: 187–214. [DOI] [PubMed] [Google Scholar]
- Yoshihara, T. and Iino M. (2007) Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol. 48: 678–688. [DOI] [PubMed] [Google Scholar]
- Yoshino, K., Numajiri Y., Teramoto S., Kawachi N., Tanabata T., Tanaka T., Hayashi T., Kawakatsu T. and Uga Y. (2019) Towards a deeper integrated multi-omics approach in the root system to develop climate-resilient rice. Mol. Breed. 39: 165. [Google Scholar]
- Yu, B., Lin Z., Li H., Li X., Li J., Wang Y., Zhang X., Zhu Z., Zhai W., Wang X.et al. (2007) TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 52: 891–898. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Kaeppler S.M. and Lynch J.P. (2005) Topsoil foraging and phosphorus acquisition efficiency in maize (Zea mays). Funct. Plant Biol. 32: 749–762. [DOI] [PubMed] [Google Scholar]