Abstract
We identified the main changes in serum metabolites associated with severe (n = 46) and mild (n = 19) COVID-19 patients by gas chromatography coupled to mass spectrometry. The modified metabolic profiles were associated to an altered amino acid catabolism in hypoxic conditions. Noteworthy, three α-hydroxyl acids of amino acid origin increased with disease severity and correlated with altered oxygen saturation levels and clinical markers of lung damage. We hypothesize that the enzymatic conversion of α-keto-acids to α- hydroxyl-acids helps to maintain NAD recycling in patients with altered oxygen levels, highlighting the potential relevance of amino acid supplementation during SARS-CoV-2 infection.
Subject terms: Metabolomics, Viral infection, Hypoxia
Introduction
COVID-19 is an emerging infection caused by the severe acute respiratory syndrome (SARS)-CoV-2 virus, whose main clinical features are fever, fatigue, cough, lymphopenia and, in severe cases, pneumonia with SARS, which if uncontrolled leads to multi-organic failure and dead1. In severe cases, hypoxia seems to play a distinct role in the adverse clinical outcomes of COVID-19 patients. Therefore, several clinical and pharmacological approaches for restoring oxygen levels to viable parameters are currently under evaluation2.
The physiological changes related to the presence of hypoxia that occur as an attempt to normalize oxygen levels include increase in respiratory rate, vasodilation, and vascularization as mechanisms to fulfill energetic requirements until normoxia is attained3. At the molecular level, hypoxia inhibits oxidative phosphorylation in mitochondria, which is the main source of adenosine triphosphate (ATP) for critical and energetic expensive processes such as ionic equilibrium and protein synthesis3.
Hypoxia inducible factor 1-α (HIF 1-α) is the master regulator of oxygen homeostasis in mammals and has crucial roles on metabolism through up-regulating genes involved in the glycolytic pathway during hypoxic conditions4. The activity of this transcription factor favors anaerobic glycolysis over mitochondrial respiration as a source of ATP when environmental O2 falls4. Despite that anaerobic glycolysis produces only 2 ATP molecules per molecule of glucose, this metabolic shift is accompanied by an increased glucose influx as a compensatory mechanism to reach high levels of ATP4. However, a sustained glycolytic metabolism may potentially lead to reduced nicotinamide dinucleotide (NAD) pools, which must be preserved to keep the cytosolic synthesis of ATP during hypoxia5. Although pyruvate is considered the final electron acceptor in this pathway, oxidation of this α keto-acid (α-KA) by lactate dehydrogenase is not the only way to replenish NAD levels in hypoxia. Other α-keto-acid oxidases, such as malate dehydrogenase, are able to replenish the cytosolic NAD pools in hypoxic environments6.
In the present study, our metabolomics analysis disclosed dysregulation of pathways linked to energy production (Krebs cycle, Warburg effect) and amino acid catabolism [BCAA (branched chain amino acids)], threonine, glutamine and glutamate) in COVID-19 patients. An increment of three different α hydroxyl acids (α-HA) linked to valine and threonine catabolism in infected patients was prominent, leading to the proposal that in hypoxic patients the α-KA derived from these amino acids become the final electron acceptors through the activity of their corresponding α-keto-acid oxidases. Accordingly, the increment of serum glutamate in COVID-19 patients could be related with an increased transaminase activity, an enzymatic process that generates α-KA as reaction products5.
Our results are in line with other COVID-19 metabolomics analyses, with emphasis on the relevance of amino acid catabolism during hypoxic conditions7–9. Interestingly, these metabolic signatures may have implications on the appearance of adverse effects due to SARS-CoV-2 infection, such as diabetes and neurological disabilities.
Results
Demographic and clinical features of control donors and patients with COVID-19
Sixty-five COVID-19 patients were included, 19 with mild disease and 46 with severe disease. In addition, 27 subjects with a negative PCR test for SARS-CoV-2 infection were included. The main clinical characteristics and demographic features of the patients and controls are shown in Table 1.
Table 1.
Demographic, clinical, and laboratory parameters of the participants.
| Parameter | Control donors | Mild COVID–19 | Severe COVID-19 | P-value |
|---|---|---|---|---|
| Demographic features | ||||
| Sex | F:17–62.9% | F:9–47.4% | F:17–36.9% | 0.098 |
| F: Female | M:10– 37.1% | M:11–52.6% | M:29–63.0% | |
| M: Male | Total: 27 | Total: 19 | Total: 46 | |
| Age (Average-SD) | 35.4 ± 8.8 | 40.7 ± 10.7 | 49.6 ± 13.6 | 2.658 × 10–5 |
| BMI (Average SD) | 26.9 ± 4.3 | 26.9 ± 3.4 | 27.8 ± 3.8 | 0.011 |
| Comorbidities | ||||
| Overweight | 11/27—40.7% | 3/19—15% | 15/46—32% | 0.195 |
| Obesity | 5/27—18.5% | 3/19—15% | 20/46—42.6% | 0.014 |
| DM2 | 2/27– 7.4% | 2/19—10% | 12/46—25.5% | 0.085 |
| Hypertension | 1/27—3.7% | 1/19—5% | 17/46—36.2% | 5.697 × 10–4 |
| Cardiopathy | 0–0% | 0—0% | 5/46—10.6% | 0.1146 |
| Vital signs | ||||
| Temp (ºC) | – | 37.4 ± 0.7 | 37.2 ± 0.8 | 0.095 |
| Hearth rate (bpm) | – | 101.3 ± 19.3 | 103.6 ± 17.0 | 0.680 |
| Respiratory rate (bpm) | – | 20.1 ± 3.8 | 25.2 ± 6.5 | 0.002 |
| Mean arterial pressure (mmHg) | – | 95.1 ± 11.6 | 97.6 ± 14.9 | 0.982 |
| SO2 (%) | – | 94.8 ± 1.4 | 86.5 ± 9.2 | 4.889 × 10–8 |
| Symptoms | ||||
| Cough | – | 15/19—75% | 38/46—80.9% | 0.734 |
| Headache | – | 15/19—75% | 24/46—51.1% | 0.055 |
| Fever | – | 16/19—80% | 42/46—89.4% | 0.407 |
| Dyspnea | – | 5/19—25% | 33/46—70.2% | 0.001 |
| Arthralgia | – | 11/19—55% | 26/46—55.3% | 0.999 |
| Myalgia | – | 11/19—55% | 31/46—66% | 0.570 |
| Sore Throat | – | 5/19—25% | 15/46—31.9% | 0.770 |
| Nausea | – | 1/19—5% | 4/46—8.5% | 0.999 |
| Diarrhea | – | 2/19—10% | 10/46—21.3% | 0.483 |
| Fatigue | – | 9/19—45% | 20/46—42.5% | 0.790 |
| Arterial blood gases | ||||
| FiO2 (%) | – | – | 27.9 ± 12.6 | |
| pH art | – | – | 7.33 ± 0.2 | |
| Arterial PO2 (mmHg) | – | – | 83.7 ± 43.2 | |
| Arterial PCO2 (mmHg) | – | – | 28.9 ± 3.7 | |
| HCO3 (mmol/L) | – | – | 21.4 ± 2.8 | |
| Lactate (mmol/L) | – | – | 1.67 ± 0.9 | |
| PAFI | – | – | 265.7 ± 104.3 | |
| Anion gap (mmol/L) | – | – | 14.3 ± 2.4 | |
| Laboratory tests | ||||
| Glucose (mg/dL) | – | – | 134.04 ± 76.2 | |
| CRP (mg/dL) | – | – | 13.3 ± 8.4 | |
| Ferritin (ng/mL) | – | – | 555.3 ± 408.2 | |
| DHL (U/L) | – | – | 373.07 ± 162.8 | |
| ALT (U/L) | – | – | 39.06 ± 36.5 | |
| AST (U/L) | – | – | 41.6 ± 29.7 | |
| Creatinine (mg/dL) | – | – | 0.97 ± 0.3 | |
| Albumin (g/dL) | – | – | 3.99 ± 0.8 | |
P-value for Kruskal–Wallis, U, cχ2 or Fisher exact test.
Using an untargeted metabolomics approach, we determined 46 different metabolites ([< 30% relative standard deviation (RSD) in quality control (QC) samples] in serum samples from patients and controls. The main characteristics of these compounds are shown in Table S1. The method showed high reproducibility as observed by principal component analysis (PCA) of QC samples, which forms a well-defined compact cluster (Figure S1).
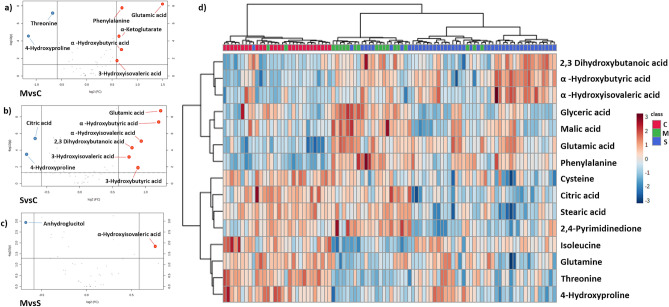
We performed volcano plots analysis to identify those metabolites with the highest changes between the groups (Fold change > 0.5 and p.-adjusted values < 0.05, Fig. 1). When compared to control, we detected lower levels of threonine and citrate in COVID-19 patients. Inversely, α-Ketoglutarate, phenylalanine, glutamic acid and several intermediaries of amino acid catabolism (3-Hydroxyisovaleric, 3-Hydroxybutyric acid, α-Hydroxybutyric acid, α-Hydroxyisovaleric, 2,3-Dihydroxybutanoic acid) increase with disease severity. Only α-Hydroxyisovaleric acid (a metabolite of valine degradation pathway) increases between mild and severe patients (Table 2).
Figure 1.
Changes in serum metabolites of controls vs COVID-19 patients. Volcano plots of metabolites that increase (red) or decrease (blue) in (a) mild COVID-19 versus controls, (b) severe COVID-19 vs controls and (c) severe versus mild COVID-19. Cut off fold change > 0.5, p value < 0.05. (d) Heatmap visualization and clustering analysis of differential metabolites among controls (C), mild (M) and severe (S) COVID-19 patients. Only the 15 metabolites with the lowest p values (by ANOVA) are shown.
Table 2.
Comparison of metabolites with fold changes > 0.5 according to disease severity.
| Combination | Metabolite | FC | log2(FC) | p-.adjusted | Kruskal–Wallis CvsMvsS p.-adjusted |
|---|---|---|---|---|---|
| M/C | Glutamic acid | 2.8108 | 1.491 | 5.89 × 10–9 |
M–C; S–C 2.21E−10–4.96E−09 |
| Phenylalanine | 1.616 | 0.69239 | 1.73 × 10–8 |
M–C; S–C 1.38E−10–4.96E−09 |
|
| α-Hydroxybutyric acid | 1.5999 | 0.67798 | 9.26 × 10–4 |
M–C; S–C; S–M 5.73E−09–8.59E−08 |
|
| α-Ketoglutarate | 1.5639 | 0.64511 | 3.04 × 10–5 |
M–C; S–C; M–S 0.00021638–0.00060858 |
|
| 3-Hydroxyisovaleric acid | 1.5081 | 0.59277 | 0.02247 |
M–C; S–C 0.00094552–0.0018499 |
|
| Threonine | 0.62777 | − 0.6717 | 6.68 × 10–8 |
C–M; C–S 5.73E−09–8.59E−08 |
|
| S/C | Glutamic acid | 2.3519 | 1.2338 | 1.45 × 10–9 |
M–C; S–C 2.21E−10–4.96E−09 |
| α-Hydroxybutyric acid | 2.3094 | 1.2075 | 4.04 × 10–8 |
M–C; S–C; S–M 5.73E−09–8.59E−08 |
|
| α-Hydroxyisovaleric acid | 1.9297 | 0.94837 | 7.55 × 10–6 |
S–C; S–M 3.04E−06–1.71E−05 |
|
| 3-Hydroxybutiric acid | 1.8519 | 0.88901 | 0.012145 |
S–C; S–M 0.008355–0.012532 |
|
| 2,3-Dihydroxybutanoic acid | 1.7324 | 0.79276 | 5.73 × 10–5 |
M–C; S–C 5.43E−05–0.00024421 |
|
| 3-Hydroxyisovaleric acid | 1.6932 | 0.75971 | 5.46 × 10–4 |
M–C; S–C 0.00094552–0.0018499 |
|
| Citric acid | 0.62276 | − 0.68326 | 3.21 × 10–6 |
C–S; M–S 9.24E−07–7.07E−06 |
|
| 4-Hydroxyproline | 0.57272 | − 0.80409 | 2.77 × 10–4 |
C–M; C–S 1.09E−05–5.47E−05 |
|
| M/S | α-Hydroxyisovaleric acid | 1.7032 | 0.76821 | 0.014705 |
S–C; S–M 3.04E−06–1.71E−05 |
| Anhydroglucitol | 0.62921 | − 0.66838 | 0.0011881 |
M–C; M–S 0.0004461–0.00095594 |
Fold-change was calculated using Metaboanalyst 5.0. p values were calculated using the Mann–Whitney test for MvsS and Kruskal Wallis test for multiple comparisons (CvsMvsS). Comparisons are included as follows: mild COVID-19 versus controls (MvsC); severe COVID-19 versus controls (SvsC) and severe versus mild COVID-19 (SvsM).
To obtain an overview of the main changes in our metabolomics data, we performed a heatmap and hierarchical clustering analysis using the 15 metabolites with the lowest p value (ANOVA p < 0.05) (Fig. 1). With this approach, we detected reduced levels of several amino acids (cysteine, isoleucine, glutamine, and threonine) and lower levels of glyceric and citric acid in COVID-19 patients. Conversely, increased levels of three different α-HA (α-hydroxyisovaleric acid, α-hydroxybutyric and 2,3 dihydroxybutanoic acid and malic acid) and two amino acids (glutamic acid and phenylalanine) were identified in infected patients. However, this analysis is not able to generate a perfect clustering between the analyzed groups and to discriminate for disease severity.
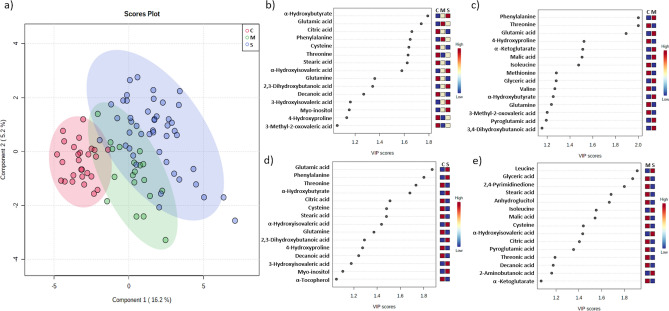
To further categorize the patients according to the metabolic profile, we performed a supervised multivariate partial least squares-discriminate analysis (PLS-DA). To assess the robustness of the model classification, cross validation and permutation analysis were performed for the following comparisons: Control versus severe COVID-19 (CvsS), control versus mild COVID-19 (CvsM), mild COVID-19 versus severe COVID-19 (MvsS), and control versus mild COVID-19 versus severe COVID-19 (CvsMvsS), obtaining good prediction parameters except for the MvsS groups, indicating a less metabolic variation in this set of subjects (Table 3). The Variable Importance to the Projection (VIP) metabolites relevant to group each cluster are depicted in Fig. 2; as shown, VIP values belong to amino acid, α-HA, fatty acids, and Krebs cycle intermediaries.
Table 3.
Predictability values of partial least squares-discriminate analysis (PLS-DA).
| Measure | C vs M | C vs S | M vs S | C vs M vs S |
|---|---|---|---|---|
| R22 | 0.836 | 0.845 | 0.568 | 0.797 |
| Q22 | 0.691 | 0.723 | 0.262 | 0.667 |
| Accuracy | 0.957 | 0.973 | 0.831 | 0.826 |
C versus M, Control versus Mild; C versus S, Control versus Severe; M versus S, Mild versus Severe; and C versus M versus S, Control versus Mild versus Severe. Q2, R2 and accuracy were calculated via double cross validation procedure with tenfold-CV, non-independent set was used.
Figure 2.
Partial least squares-discriminate analysis (PLS-DA) plot of differential metabolites from control and COVID-19 patients and their corresponding VIP values. (a) PLSDA score plots for control (H-red), mild (M-green) and severe (S-blue) patients .VIP schematic scores of PLS-DA analysis for CvsMvsS (b), CvsM (c), CvsS (d), and MvsS (e) groups.
Mitochondrial dysregulation as a feature of severe COVID-19
To identify the disturbed pathways between the study groups, enrichment analysis (Metaboanalyst v4.0) employing the metabolites with the highest VIP values from PLS-DA analysis was applied; pathways with p < 0.05 are shown in Table 4. Pathways associated with the metabolism of BCAA, glutamate and phenylalanine were differentially expressed in patients with mild and severe COVID-19 as well as in controls. Additionally, we observed a possible involvement of the Warburg effect, as this process is differentially regulated in COVID-19 patients bearing different disease severity in comparison with controls. Further, differential pathways analysis among COVID-19 patients identified a potential mitochondrial dysregulation in those with severe disease. We also found that BCAA metabolism was differentially expressed in the studied groups, and glutamine and glutamate metabolism exhibited a high impact on the metabolic profile of COVID-19 patients (Fig. S2).
Table 4.
Enriched pathway analysis identified in COVID19 patients.
| Groups | Enriched metabolite set | Hits | p value | Holm P | FDR |
|---|---|---|---|---|---|
| C versus M | Glycine and serine metabolism | 5 | 0.00103 | 0.101 | 0.0547 |
| Valine, leucine and isoleucine degradation | 5 | 0.00112 | 0.108 | 0.0547 | |
| Propanoate metabolism | 4 | 0.00241 | 0.231 | 0.0786 | |
| Phenylalanine and tyrosine metabolism | 3 | 0.00669 | 0.636 | 0.118 | |
| Urea cycle | 3 | 0.0074 | 0.695 | 0.118 | |
| Warburg effect | 4 | 0.00792 | 0.737 | 0.118 | |
| Malate-aspartate shuttle | 2 | 0.00843 | 0.775 | 0.118 | |
| C versus S | Glutamate metabolism | 3 | 0.0312 | 1.0 | 0.874 |
| Glutathione metabolism | 2 | 0.0358 | 1.0 | 0.874 | |
| Warburg effect | 3 | 0.0483 | 1.0 | 0.874 | |
| M versus S | Valine, leucine and isoleucine degradation | 4 | 0.00686 | 0.672 | 0.392 |
| Citric acid cycle | 3 | 0.00799 | 0.775 | 0.392 | |
| Glutathione metabolism | 2 | 0.0314 | 1.0 | 0.655 | |
| Transfer of acetyl groups into mitochondria | 2 | 0.0343 | 1.0 | 0.655 | |
| Warburg effect | 3 | 0.0402 | 1.0 | 0.655 | |
| Glycine and serine metabolism | 3 | 0.042 | 1.0 | 0.655 | |
| Cysteine metabolism | 2 | 0.0468 | 1.0 | 0.655 | |
| C versus M versus S | Glutamate metabolism | 3 | 0.0312 | 1.0 | 0.793 |
| Glutathione metabolism | 2 | 0.0358 | 1.0 | 0.793 | |
| Warburg effect | 3 | 0.0483 | 1.0 | 0.793 |
Results of the enrichment pathways according to Metaboanalyst (v4.0), employing the VIP values from PLS-DA analysis as the input metabolites.
Correlation of a distinct metabolic profile with ventilatory parameters and diverse clinical features
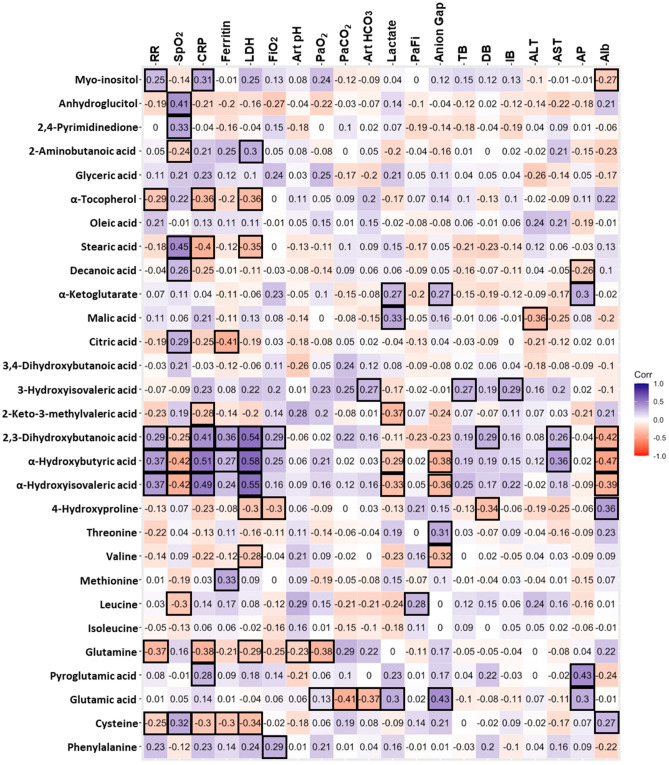
We performed a Spearman’s correlation analysis between VIP metabolites with routine clinical tests for oxygen homeostasis and clinical markers recently described for lung impairment and poor prognosis in SARS-CoV-2 infection. Figure 3 and Table S2 show significant correlations between metabolites and clinical variables. A generalized linear mixed models to examine the effect of patient condition (H, M or S) over VIP metabolites levels, including the presence of obesity and hypertension as random factors (Supplemental Table S3), revealed that only oleic acid did not show significant differences derived from the patients’ condition (HvsMvsS). For generalized linear models with age as a covariate, only 3-Hydroxyisovaleric acid, anhydroglucitol and threonic acid are influenced due to the patient age. Nonetheless, this variable does not replace the effect of COVID-19 (Supplemental Table 4). Furthermore, after adjustment, we also found that BMI influences the levels of 3-Hydroxyisovaleric and citric acid. However, these changes are explained based on the composite effect of BMI and COVID-19 status as well.
Figure 3.
Spearman’s correlation analysis. Correlation analysis of the modified metabolites in severe COVID-19 patients and the clinical parameters considered. Blue numbers indicate a positive association. Red numbers denote a negative association. Squared values denote associations with values p < 0.05. RR = Respiratory rate, SpO2 = Oxygen saturation, LDH = Lactate dehydrogenase, FiO2 = Fraction of inspired oxygen, Art pH = Arterial pH, PaO2 = Partial pressure of oxygen, PaCO2 = Partial pressure of CO2, Art HCO3 = Arterial bicarbonate, PaFi = PaO2/ FiO2, TB = Total bilirubin, DB = Direct bilirubin, IB = Indirect bilirubin, ALT = Alanine aminotransferase, AST = Aspartate aminotransferase, AP = Alkaline phosphatase, Alb = Albumin. Exact p values are referred in Supplemental Table 2.
Discussion
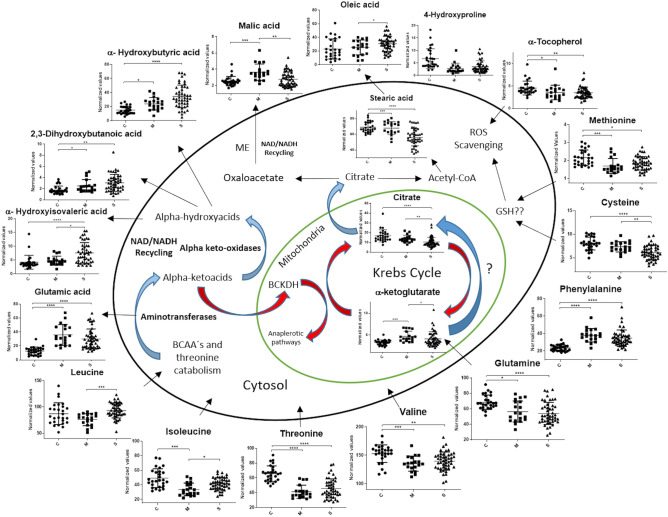
We identified altered levels of amino-acids, Krebs cycle intermediaries, and α-HA in serum samples from mild and severe COVID-19 patients. The analysis of these metabolic profiles revealed a relationship between an altered metabolism of BCAA, glutamate, glutamine, and Warburg effect with the severity of the disease. In particular, hypoxemia in COVID-19 patients could impair redox, energetic, and immune responses1. In this context, the term metabolic flexibility refers to the transcriptomic, proteomic and metabolic changes that need to be adapted to particular pathological conditions10. Globally, hypoxia favors anaerobic glycolysis (over beta oxidation, pentose phosphate pathway and cellular respiration) as a source of ATP to fulfill the energetic requirements of several critical processes such as RNA expression, and protein and lipid synthesis11.We detected modified levels of three intermediaries of the Krebs cycle. Citrate serum levels fell in severe COVID-19 patients and positively correlated with lower oxygen saturation levels. On the other hand, α-ketoglutarate and malate transiently increased in mild COVID-19 patients, showing a positive correlation with lactate and anion gap markers, both indicators of metabolic acidosis. Although it is unclear whether the degree of these changes were caused by the hypoxic conditions, the analysis of the differential metabolic signatures exhibited by mild and severe COVID-19 patients suggests that citric cycle and Warburg effect play a significant role for discriminating both groups. In normoxia, glucose conversion to acetyl Co-A and glutamine to α-ketoglutarate are the major sources of mitochondrial carbon intermediaries for the Krebs cycle12. However, in hypoxia, the flux of acetyl-coA is inhibited and glutamine-derived α-ketoglutarate can be reductively carboxylated to isocitrate by isocitrate dehydrogenase as a way to replenish mitochondrial NAD levels and increase citrate pools in the Krebs cycle12,13. This process has been extensively studied in cancer-derived hypoxia14. Thus, citrate may diffuse to cytosol where it could be further processed by the enzyme ATP citrate lyase to Acetyl-Co-A and oxaloacetate15. The cytosolic activity of malate dehydrogenase also might support an enhanced glycolysis through the conversion of oxaloacetate to malate, regenerating NAD6. In addition to the potential inhibition of the Krebs cycle, ATP citrate lyase activity could also explain the altered levels of citrate in mild COVID-19 patients.
We observed reduced levels of glutamine in severe and mild COVID-19 patients and this change negatively correlated with lactate dehydrogenase (LDH), C reactive protein (CRP), and PO2 levels, and positively with PCO2; these markers have been associated with lung damage and altered oxygen homeostasis in COVID-19 patients16,17. In vitro experiments also have demonstrated that hypoxia increases glutamine transport into the cells through HIF2-alpha upregulation of SLC1A5 genes18. These interconnected processes might potentially link glutamine hypoxic catabolism with NAD recycling and malate synthesis. According to our results, several reports detected reduced levels of this amino acid in COVID-19 cohorts, notwhitstanding, little is understood about this decrement and its potential therapeutic implications7,8,19. Further studies are needed to more precisely assess whether glutamine is linked to reductive carboxylation in hypoxic patients.
On the other hand, three different α-HA of amino acid origin (α-Hydroxyisovaleric acid, α-Hydroxybutyric, and 2,3-Dihydroxybutanoic acid) were significantly increased in samples from mild and severe COVID-19 patients and positively correlated with CRP and LDH and negatively with SO2 and serum albumin. Reduced levels of this protein have been associated with increased mortality in severe hypoxic hepatitis and COVID-1920–22. Similar to glutathione, albumin has an antioxidant activity over ROS control in ischemic and hypoxic liver23. The association between increased α-HA and lower albumin levels may be related to the decline in protein synthesis and modification of amino acid metabolism due to hypoxia24,25. In this regard, α-Hydroxyisovaleric acid is a product of valine catabolism and is used as a marker of maple syrup urine disease (MSUD), a clinical condition derived from inactivating mutations in branched-chain α-keto acid dehydrogenase complex (BCKDH), an enzyme essential for BCAA catabolism26. BCAA that are not required for protein synthesis are deaminated to α-KA (producing glutamate from α-ketoglutarate) by branched chain aminotransferases (BCAT) and funneled to mitochondrial BCKDH, where they are further metabolized in the Krebs cycle27. However, in hypoxic conditions, the halt of respiratory chain promotes an increase in NADH/NAD ratio, inactivating BCKDH complex and boosting α-KA levels28. Subsequently, cellular peroxidases oxidizes the resultant α-KA to α-HA in a NADH dependent manner29,30. As in MSUD, α-KA and α-HA are increased in patients with respiratory diseases that disturb oxygen homeostasis, and this increment is probably related to BCKDH inhibition and α-Keto oxidase activity28,31,32.
We identified as well modified serum valine, leucine, isoleucine, 2-keto-3-methylvaleric acid, and 3-Hydroxyisovaleric acid levels, which indeed are included in the BCAA metabolic pathway. These changes could suggest a modified BCAA metabolism in COVID-19 patients. Noteworthy, we also identified a trend towards restoration of the levels of these amino acids in patients with severe COVID-19 when compared to those with mild disease. In fact, 4-Hydroxyproline, a marker of amino acid mobilization from skeletal muscle to liver, displayed a trend towards higher levels in severe COVID-19 patients, suggesting that in severe disease, there is an increased requirement of tissue BCAA in order to replenish NAD.
Threonine levels decreased in severe and mild COVID-19 patients; this decrement could be potentially related to an enhanced catabolism as we also identified increased levels of α-Hydroxybutyric and 2,3-Dihydroxybutanoic acids, both oxidized metabolites of threonine conversion to α-HA. α-Hydroxybutyric acid is synthetized by the activity of lactate dehydrogenase over α-Ketobutyrate, a product of methionine/threonine catabolism and cysteine anabolism33. The increase of this metabolite correlated positively with higher levels of serum LDH. In this context, given that α-Ketobutyrate is a substrate of BCKDH, the increase of its corresponding α-HA could be related with the potential inactivity of the mitochondrial enzymatic machinery34,35. On the other hand, 2,3-Dihydroxybutanoic acid is a product of threonine catabolism probably generated by the activity of cytosolic transaminases and oxidases, a conversion that may potentially restore NAD during hypoxia36. All the enzymes involved in α-KA oxidation rely on NADH (or NADPH) as a co-substrate and their activities are important to maintain the energetic and redox states in health and disease. However, the mechanisms involved in BCAA and threonine intermediaries in hypoxia are still incompletely understood37.
A mechanism that links HIF1-alpha with the increase in BCAA transport and aminotransferase activity in hypoxic glioma cells was recently described38. In this cell context, the in vitro accumulation of BCAA in cerebral cortex of normoxic rats promotes glucose internalization and, interestingly, induces lower levels of CO2 release. These data suggest the presence of aerobic or anaerobic glycolysis regulated by the BCAA levels despite the presence of normoxia39. Interestingly, BCAT exhibits a CXXC motif that presumably acts as redox sensor and predisposes its aminotransferase activity by the cellular redox state40.
Although the effects of BCAA metabolism in hypoxia are still poorly understood, an evolutive advantage cannot be ruled out. In this regard, cells from the fungus Aspergillus nidulans increase the synthesis of BCAA as a mechanism to regenerate NAD and NADP during hypoxia41. Interestingly, the in vitro addition of BCAA, such as valine and leucine, rises the survival rate of mice infected with Klebsiella Pneumoniae42. Our present data suggest that hypoxia promotes the metabolic funneling of BCAA and threonine to α-KA synthesis and then to α-HA as a compensatory mechanism to replenish NAD levels in COVID-19 patients (Fig. 4).
Figure 4.
Proposed model for the serum metabolic changes observed in COVID-19 patients. Red arrows represent metabolic flux inhibited during hypoxia. Blue arrows represent increased flux in hypoxia. ME Malic enzyme. The graphs were constructed with the normalized values from control subjects (C), mild (M), and severe (S) patients. Asterisks indicate statistical significance according to the Dunn test. p* < 0.05 ** < 0.01, *** < 0.001, **** < 0.0001.
In support to this hypothesis, we observed decreased serum levels of BCAA and threonine in COVID 19 patients, an effect that has been previously identified in several studies of chronic obstructive pulmonary disease37. It has been shown that BCAA mobilization from skeletal muscle to liver is essential for human adaptability to lower oxygen levels43, which is in agreement with our findings. Recent data highlight the relevance of BCAA administration as an strategy to attenuate protein muscle loss in COVID-19 critical patients44.
An scenario with high amino transferase activity requires a constant flux of α-Ketoglutarate, a nitrogen acceptor, which could be achieved through an enhanced glutamine catabolism12. In this regard, α-KA synthesis mediated by aminotransferases generates glutamate as a byproduct. Accordingly, we observed an increase in glutamic acid levels, which correlated positively with anion gap values in severe COVID-19. Glutamate is an important neurotransmitter that, if unbalanced, promotes several neurological abnormalities45,46. In fact, patients with hypoxia may exhibit impaired activity of glutamate and several other neurotransmitters involved in homeostasis of the central nervous system, which may explain some of the neurological features found in COVID-19 patients47.
On the other hand, the appearance of new onset or worsening of pre-existing diabetes has been identified in COVID-19 patients48. α-Hydroxybutyric acid is an early marker of insulin resistance in the non-diabetic population49,50. Additionally, high levels of 2,3- Dihydroxybutanoic acid and α-Hydroxybutyric acid were detected in type I diabetes patients36. Gall et al54 proposes that individuals with an augmented fatty acid metabolism display NADH/NAD imbalance resembling hypoxic conditions that unleash a change in BCAA metabolic fate, predisposing to diabetes development. Although there are still missing paths connecting BCAA metabolism with diabetes, a plausible explanation for the presence of insulin resistance in COVID-19 patients may be partly connected to BCKDH state51.
Conclusion
Three different α-hydroxyl-acids are increased in severe COVID-19. The underlying biochemical traits may involve a modified amino acid metabolism derived from lung damage and hypoxic conditions. It is tempting to propose that the enzymatic conversion of α-keto-acids to α-hydroxyl-acids helps to maintain NAD recycling in patients with altered oxygen levels, highlighting the potential relevance of amino acid supplementation during SARS-CoV-2 virus infection.
Methods
Subjects
Sixty-five COVID-19 patients, confirmed by a positive RT-PCR test for SARS-CoV-2 on a nasopharyngeal swab, and 27 control donors with a negative PCR test for SARS-CoV-2 were recruited at a third level referral center in Mexico City (Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán) from March to June, 2020.
Upon patient assessment, general laboratory tests were performed (complete blood count, glucose, blood urea nitrogen (BUN), creatinine (Cr), liver function tests, C-reactive protein (CRP), lactate dehydrogenase (LDH), creatinine kinase (CPK), fibrinogen, D-dimer, coagulation tests and ferritin). The severity of the disease was classified as follows: Mild/Moderate illness: Fever, signs of airway disease, with or without a tomographic image indicating pneumonia. Severe illness, any of the following: respiratory failure, respiratory rate > 30 bpm, O2 saturation < 92% at rest, PaO2/FiO2 < 300 mmHg52 . Once obtained, the serum was processed immediately and frozen to − 80 °C until GC/MS analysis, which was performed following a randomized scheme in the same way as COVID-19 samples. All samples from subjects included were drawn upon admission to the triage at the emergency department for the patients and at the epidemiology unit where the hospital staff attended for randomized nasopharyngeal PCR testing, during the morning shift and in a fasting state (minimum 8 h). The study was approved by the institutional ethics and research committees of Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán (Ref. 3341). In those subjects that met the inclusion criteria, serum samples were collected from an antecubital vein and stored at − 80 °C until the day of metabolomics processing. Written informed consent to participate in the study was obtained from all participants. All analytical methods were carried out in accordance with relevant guidelines and regulations.
Gas chromatography/mass spectrometry (GC/MS) analysis
Forty-five microliters of serum sample and 10 µL of internal standard (IS—tetradecanoic acid, methyl tricosanoate, 5α-cholestane—0.36 mg/mL) were mixed in 150 µL of 1:3 chloroform–methanol and thoroughly mixed for 2 min. Samples were incubated at − 20 °C for 20 min and then centrifuged at 14,000 rpm for 15 min. One-hundred and fifty microliters of recovered supernatants were dried under nitrogen flow. Each precipitate was redissolved in 20 uL of methoxyamine and incubated for 90 min at 37 °C in a shaking incubator. Thereafter, 40 µL of MBSTFA + 1% TMCS was added to each sample and incubated for 30 min at 37 °C. One microliter was applied to a GC/MS system (Agilent 5977A/7890B, Santa Clara, CA, USA) with an automatic autosampler (G4513A, Agilent) and run under the following conditions: splitless column flow 1 ml/min, inlet temperature 200 °C, EI source temperature 200 °C, and interface temperature 250 °C. A column HP5ms (30 m × 250 µm × 0.25 µm, Agilent) with helium 99.9999% as a mobile phase was employed. The running method consisted of 1 min hold at 60 °C with an increased ramp of 10 °C/min to 325 °C, with a final held time of 10 min.
Briefly, to assess the system suitability along all the analysis we employed three different internal standards (IS). To identify any issues in the derivatization process, we employed tridecanoic acid as a metabolite prone to derivatization. To check shifts over the retention times we included additionally 5-alpha cholestane and tricosanoic acid methyl ester. The RSD values for the IS over all the experimental process (encompassing 104 samples in 12 batches = 12 days of analysis) were 12.89, 10.78 and 11.57% respectively (before the normalization process). For the intraday variation, we injected a QC1 (consisting of equal volumes of all the samples of the batch) every 5 samples along the entire batch. We observed an average %RSD value of 17.7% (the average of all metabolite RSD values (triplicate) in each batch [12 batches]). For the inter-day variation, we generated a QC2 consisting of ten aleatory selected samples. We observed an inter-day RSD value of 17.5% (average of RSD values for each metabolite in 12 batches). The %RSD values for each metabolite are shown in supplementary Table 1. These results are in accordance with Fiehn Laboratory recommendations for metabolomic analysis (< 30% RSD)53.
Deconvolution and identification
GC/MS data was transformed to .mzdata using the Agilent Chemstation software (Agilent). Feature detection, spectral deconvolution and peak alignment were performed using the Mzmine2 software54. The parameters used were: RT range, 5.5–27.5 min; m/z range, 50–500; m/z tolerance, 0.5; noise level, 1 × 103; and peak duration range, 0.01–0.2 min. Further metabolite selection was performed according to the rule of 80%. Identifications were determined using the National Institute of Standards and Technology (NIST) 2.0 spectral library. Results higher than 70% (R > 700), were accepted as correct, while values below this limit were marked as unknown and omitted. Only identified metabolites with %RSD lower than 30% were selected to further uni- or multivariate statistical analysis.
Statistical analysis
Peak heights raw data was normalized by sum, log-transformed and autoscaled (mean-centered and divided by the standard deviation) for statistical multivariate analysis. PLS-DA and hierarchical cluster analysis were performed by the Ward’s method using Metaboanalyst 4.055. For univariate analysis, data were normalized by sum and then the Kruskal–Wallis test was applied to examine the three levels of patient condition (control, moderate, and severe), followed by the Dunn’s post hoc test and unpaired Mann–Whitney test to examine differences between mild and severe conditions. For these analyses, the GraphPad Prism version 6 software (GraphPad Software Inc., San Diego, CA) was employed. p values < 0.05 were considered significant. Spearman’s correlation analysis, generalized linear models and generalized linear mixed models were performed in R version 4.0.2 (R Foundation for Statistical Computing).
Metabolic pathway analysis: Enrichment analysis
The VIP values from PLS-DA analysis were selected to implement the enrichment pathway analysis in Metaboanalyst (v.4.0). In the case of α-Hydroxyisovaleric acid, the corresponding α-KA was used as the input metabolite. KEGG database does not have the correspondent α-HA derived of BCAA’s catabolism in its library. Only the enrichment analysis with p values < 0.05 are presented.
Supplementary Information
Acknowledgements
This study was supported by a grant from CONACyT (F0005-2020-01-313252). We would like to thank the technical support from the Bioinformatics, Biostatistics, and Computational Biology Unit of the Red de Apoyo a la Investigación of the National University of Mexico and the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán.
Author contributions
J.C.P.F. and J.J.T.R. contributed equally in the design and performance of experiments, analysis, and wrote the manuscript. V.A.S.H., R.C.D. and S.R.R. performed experiments and analyzed data. D.E.M.S. assisted in processing and preservation of patient samples. A.P.F. collect patient data, generated and organize the clinical database. J.M.G.A., J.L.M.M., A.P.L. and A.U.A. assisted in writing and editing the manuscript. N.R.M.D. performed bioinformatics and statistical analyses. D.G.M., L.G.L.P. designed the experiments, supervised general work and edited the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: José C. Páez-Franco and Jiram Torres-Ruiz.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-85788-0.
References
- 1.Lake MA. What we know so far: COVID-19 current clinical knowledge and research. Clin. Med. 2020;20:124–127. doi: 10.7861/clinmed.2019-coron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geier MR, Geier DA. Respiratory conditions in coronavirus disease 2019 (COVID-19): Important considerations regarding novel treatment strategies to reduce mortality. Med. Hypotheses. 2020;140:109760. doi: 10.1016/j.mehy.2020.109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michiels C. Physiological and pathological responses to hypoxia. Am. J. Pathol. 2004;164:1875–1882. doi: 10.1016/S0002-9440(10)63747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray LR, Tompkins SC, Taylor EB. Regulation of pyruvate metabolism and human disease. Cell. Mol. Life Sci. 2014;71:2577–2604. doi: 10.1007/s00018-013-1539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everaers RA. 乳鼠心肌提取 HHS Public Access. Physiol. Behav. 2017;176:139–148. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen B, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. SSRN Electron. J. 2020 doi: 10.1101/2020.04.07.20054585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas T, et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020 doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Troisi J, et al. Serum metabolomic prole of symptomatic and asymptomatic SARS-CoV-2 infected patients. Cell. 2020;1:21. doi: 10.21203/rs.3.rs-34085/v1. [DOI] [Google Scholar]
- 10.Goodpaster BH, Sparks LM. Metabolic flexibility in health and disease. Cell Metab. 2017;25:1027–1036. doi: 10.1016/j.cmet.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheaton WW, Chandel NS. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am. J. Physiol. Cell Physiol. 2011;300:C385–C393. doi: 10.1152/ajpcell.00485.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenasedependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. USA. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun RC, Denko NC. Growth. 2015;19:285–292. doi: 10.1016/j.cmet.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Icard P, Poulain L, Lincet H. Understanding the central role of citrate in the metabolism of cancer cells. Biochim. Biophys. Acta Rev. Cancer. 2012;1825:111–116. doi: 10.1016/j.bbcan.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Han Y, et al. Lactate dehydrogenase, an independent risk factor of severe COVID-19 patients: A retrospective and observational study. Aging (Albany. NY) 2020;12:11245–11258. doi: 10.18632/aging.103372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L. C-reactive protein levels in the early stage of COVID-19. Med. Mal. Infect. 2020;50:332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo HC, et al. A variant of SLC1A5 is a mitochondrial glutamine transporter for metabolic reprogramming in cancer cells. Cell Metab. 2020;31:267–283.e12. doi: 10.1016/j.cmet.2019.11.020. [DOI] [PubMed] [Google Scholar]
- 19.Barberis E, et al. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to sars-cov-2. Int. J. Mol. Sci. 2020;21:1–25. doi: 10.3390/ijms21228623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Rica R, et al. Low albumin levels are associated with poorer outcomes in a case series of COVID-19 patients in Spain: a retrospective cohort study. Microorganisms. 2020;8:1–13. doi: 10.3390/microorganisms8081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Violi F, et al. Is Albumin predictor of mortality in COVID-19? Antioxid. Redox Signal. 2020;00:1–4. doi: 10.1089/ars.2020.8142. [DOI] [PubMed] [Google Scholar]
- 22.Chang P, et al. Low serum albumin predicts early mortality in patients with severe hypoxic hepatitis. World J Hepatol. 2017;9:959–966. doi: 10.4254/wjh.v9.i22.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strubelt O, Younes M, Li Y. Protection by albumin against ischaemia- and hypoxia-induced hepatic injury. Pharmacol. Toxicol. 1994;75:280–284. doi: 10.1111/j.1600-0773.1994.tb00361.x. [DOI] [PubMed] [Google Scholar]
- 24.Preedy VR, Smith DM, Sugden PH. The effects of 6 hours of hypoxia on protein synthesis in rat tissues in vivo and in vitro. Biochem. J. 1985;228:179–185. doi: 10.1042/bj2280179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chee NT, Lohse I, Brothers SP. MRNA-to-protein translation in hypoxia. Mol. Cancer. 2019;18:1–13. doi: 10.1186/s12943-019-0968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burrage LC, Nagamani SCS, Campeau PM, Lee BH. Branched-chain amino acid metabolism: from rare Mendelian diseases to more common disorders. Hum. Mol. Genet. 2014;23:1–8. doi: 10.1093/hmg/ddu123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neinast M, Murashige D, Arany Z. Branched chain amino acids. Annu. Rev. Physiol. 2019;81:139–164. doi: 10.1146/annurev-physiol-020518-114455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker V, Gentry AJ, Green LR, Hanson MA, Bennet L. Effects of hypoxia on plasma amino acids of fetal sheep. Amino Acids. 2000;18:147–156. doi: 10.1007/s007260050013. [DOI] [PubMed] [Google Scholar]
- 29.Jones JM, Morrell JC, Gould SJ. Three human peroxisomal 2-hydroxy acid oxidases. Biochemistry. 2000;275:12590–12597. doi: 10.1074/jbc.275.17.12590. [DOI] [PubMed] [Google Scholar]
- 30.Liebich HM, Först C. Hydroxycarboxylic and oxocarboxylic acids in urine:products from branched-chain amino acid degradation and from ketogenesis. J. Chromatogr. B Biomed. Sci. Appl. 1984;309:225–242. doi: 10.1016/0378-4347(84)80031-6. [DOI] [PubMed] [Google Scholar]
- 31.Bakkeren JAJM, Sengers RCA, Trijbels JMF. Simulating an inborn error of metabolism. Eur. J. Pediatr. 1977;47:41–47. doi: 10.1007/BF00465564. [DOI] [PubMed] [Google Scholar]
- 32.Cooper AJL, Ginos JZ, Meister A. Synthesis and properties of the α-keto acids. Chem. Rev. 1983;83:321–358. doi: 10.1021/cr00055a004. [DOI] [Google Scholar]
- 33.Ballèvre O, Prugnaud J, Houlier ML, Arnal M. Assessment of threonine metabolism in vivo by gas chromatography/mass spectrometry and stable isotope infusion. Anal. Biochem. 1991;193:212–219. doi: 10.1016/0003-2697(91)90011-H. [DOI] [PubMed] [Google Scholar]
- 34.Paxton R, Scislowski PWD, Davis EJ, Harris RA. Role of branched-chain 2-oxo acid dehydrogenase and pyruvate dehydrogenase in 2-oxobutyrate metabolism. Biochem. J. 1986;234:295–303. doi: 10.1042/bj2340295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettit FH, Yeaman SJ, Reed LJ. Purification and characterization of branched chain a-keto acid dehydrogenase complex of bovine kidney (multienzyme complex/subunit composition/structural organization) Biochemistry. 1978;75:4881–4885. doi: 10.1073/pnas.75.10.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kassel DB, Martin M, Schall W, Sweeley CC. Urinary metabolites of L-threonine in type 1 diabetes determined by combined gas chromatography/chemical ionization mass spectrometry. Biol. Mass Spectrom. 1986;13:535–540. doi: 10.1002/bms.1200131004. [DOI] [PubMed] [Google Scholar]
- 37.Engelen MPKJ, Schols AMWJ. Altered amino acid metabolism in chronic obstructive pulmonary disease: new therapeutic perspective? Curr. Opin. Clin. Nutr. Metab. Care. 2003;6:73–78. doi: 10.1097/00075197-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 38.Zhang B, et al. Regulation of branched-chain amino acid metabolism by hypoxia-inducible factor in glioblastoma. Cell. Mol. Life Sci. 2020 doi: 10.1007/s00018-020-03483-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sgaravatti MÆ, et al. Inhibition of brain energy metabolism by the branched-chain amino acids accumulating in maple syrup urine disease. Neurochem. Res. 2008;33:114–124. doi: 10.1007/s11064-007-9423-9. [DOI] [PubMed] [Google Scholar]
- 40.Conway ME. Emerging moonlighting functions of the branched-chain aminotransferase (BCAT) proteins. Antioxid. Redox Signal. 2020 doi: 10.1089/ars.2020.8118. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu M, Fujii T, Masuo S, Takaya N. Mechanism of de novo branched-chain amino acid synthesis as an alternative electron sink in hypoxic Aspergillus nidulans cells. Appl. Environ. Microbiol. 2010;76:1507–1515. doi: 10.1128/AEM.02135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S, et al. Metabolic regulation protects mice against Klebsiella pneumoniae lung infection. Exp. Lung Res. 2018;44:302–311. doi: 10.1080/01902148.2018.1538396. [DOI] [PubMed] [Google Scholar]
- 43.Chicco AJ, et al. Adaptive remodeling of skeletal muscle energy metabolism in high-altitude hypoxia: lessons from AltitudeOmics. J. Biol. Chem. 2018;293:6659–6671. doi: 10.1074/jbc.RA117.000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romano L, et al. Short report—medical nutrition therapy for critically ill patients with COVID-19. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4035–4039. doi: 10.26355/eurrev_202004_20874. [DOI] [PubMed] [Google Scholar]
- 45.Yudkoff M. Interactions in the metabolism of glutamate and the branched-chain amino acids and ketoacids in the CNS. Neurochem. Res. 2017;42:10–18. doi: 10.1007/s11064-016-2057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sperringer JE, Addington A, Hutson SM. Branched-chain amino acids and brain metabolism. Neurochem. Res. 2017;42:1697–1709. doi: 10.1007/s11064-017-2261-5. [DOI] [PubMed] [Google Scholar]
- 47.Payus AO, Lin CLS, Noh MM, Jeffree MS, Ali RA. SARS-CoV-2 infection of the nervous system: A review of the literature on neurological involvement in novel coronavirus disease-(COVID-19) Bosn. J. Basic Med. Sci. 2020;20:283–292. doi: 10.17305/bjbms.2020.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubino F, et al. New-onset diabetes in Covid-19. N. Engl. J. Med. 2020;383:789–791. doi: 10.1056/NEJMc2018688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tricò D, et al. Elevated a-hydroxybutyrate and branched-chain amino acid levels predict deterioration of glycemic control in adolescents. J. Clin. Endocrinol. Metab. 2017;102:2473–2481. doi: 10.1210/jc.2017-00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gall WE, et al. Α-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS ONE. 2010;5:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gentile S, Strollo F, Mambro A, Ceriello A. COVID-19, ketoacidosis and new-onset diabetes: are there possible cause and effect relationships among them? Diabetes Obes. Metab. 2020;22:2507–2508. doi: 10.1111/dom.14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu W, et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin. Med. J. (Engl.) 2020;133:1032–1038. doi: 10.1097/CM9.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fiehn O. Metabolomics by gas chromatography-mass spectrometry: combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2016;20:16. doi: 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pluskal T, Castillo S, Villar-Briones A, Orešič M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010;11:1–11. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chong J, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.