Abstract
Background
When grown in human serum, laboratory isolates of Pseudomonas aeruginosa exhibit tolerance to antibiotics at inhibitory concentrations. This phenomenon, known as serum-associated antibiotic tolerance (SAT), could lead to clinical treatment failure of pseudomonal infections. Our purpose in this study was to determine the prevalence and clinical impact of SAT in Pseudomonas isolates in hospitalized children.
Methods
The SAT phenotype was assessed in patients aged <18 years admitted with respiratory or blood cultures positive for P. aeruginosa. The SAT phenotype was a priori defined as a ≥2-log increase in colony-forming units when grown in human serum compared with Luria-Bertani medium in the presence of minocycline or tobramycin.
Results
SAT was detected in 29 (64%) patients. Fourteen patients each (34%) had cystic fibrosis (CF) and tracheostomies. Patient demographics and comorbidities did not differ by SAT status. Among CF patients, SAT was associated with longer duration of intravenous antibiotics (10 days vs 5 days; P < .01).
Conclusions
This study establishes that SAT exists in P. aeruginosa from human serum and may be a novel factor that contributes to differences in clinical outcomes. Future research should investigate the mechanisms that contribute to SAT in order to identify novel targets for adjunctive antimicrobial therapies.
Keywords: antibiotic resistance, antibiotic tolerance, cystic fibrosis, drug efflux, tracheitis
When grown in human serum, laboratory isolates of Pseudomonas aeruginosa exhibit tolerance to antibiotics (serum-associated antibiotic tolerance [SAT]). SAT was detected in two-thirds of P. aeruginosa from pediatric patients and may be a novel factor that contributes to differences in clinical outcomes.
Morbidity and mortality from infectious diseases continue to rise in both adult and pediatric hospitals throughout the United States [1]. Complicating the treatment of bacterial infectious diseases is clinical failure of antimicrobial therapy against strains shown to be susceptible in clinical laboratory testing (antibiotic treatment failure [ATF]). Observations in multiple clinical settings have shown that ATF leads to prolonged hospital lengths of stay, use of multiple antibiotics, and increased morbidity and mortality [2, 3]. The chronic persistence of bacteria (colonization) that can become pathogenic in susceptible hosts is an additional cause of morbidity related to infectious diseases. Individuals with chronic respiratory disease such as cystic fibrosis (CF) and chronic respiratory failure that requires a tracheostomy are particularly susceptible to colonization by the gram-negative bacterial pathogen Pseudomonas aeruginosa. In CF, the acquisition of P. aeruginosa is associated with more frequent pulmonary exacerbations and hospitalizations as well as decreased survival [4–7]. Similarly, nearly 90% of children with tracheostomies will have respiratory cultures positive for P. aeruginosa [8], and acquisition of P. aeruginosa post-tracheostomy is independently associated with increased odds of bacterial tracheostomy-associated respiratory tract infections [9]. Both populations, although heterogeneous in underlying pathophysiology, are frequently treated with anti-pseudomonal antibiotics [9, 10]. Nevertheless, a substantial subpopulation of these children, for example, 13% of those with CF, develop chronic pseudomonal infections despite early and aggressive antibiotic therapy [11].
The pathophysiology and molecular basis of ATF in P. aeruginosa infection remain mostly undefined. Prior work has shown that ATF in patients with P. aeruginosa ventilator-associated pneumonia is not associated with antibiotic class or the resistance profile of the organisms, suggesting that additional factors of the pathogen itself may contribute to ATF [12]. Pseudomonas aeruginosa has been demonstrated to exhibit adaptive tolerance to antibiotics when grown in biologically relevant media such as human serum, a phenotype known as serum-associated antibiotic tolerance (SAT) [13]. In contrast to static determinants of resistance where drug-binding sites are altered or enzymes degrade the antibiotic, SAT does not confer a change in the clinical minimum inhibitory concentration (MIC). Laboratory strains of P. aeruginosa are susceptible to ciprofloxacin and tobramycin in broth cultures but become tolerant to the antibiotics’ MICs when exposed to human serum [13]. The SAT phenotype has been hypothesized to allow P. aeruginosa to persist in the host environment despite exposure to otherwise appropriate antimicrobial treatment. Furthermore, susceptibility of P. aeruginosa to ciprofloxacin but not tobramycin can be restored with treatment with the known efflux pump inhibitor phenylalanine-β-naphthylamide (PAβN), suggesting efflux- and nonefflux-mediated mechanisms for this tolerance [13].
Our purpose in this study was to determine the prevalence of SAT among P. aeruginosa isolates obtained from pediatric patients, characterize the population with P. aeruginosa isolates that exhibit SAT, and investigate the association between SAT and relevant clinical outcomes.
METHODS
Study Design and Population
Children aged <18 years hospitalized between 1 July 2016 and 28 February 2017 at Johns Hopkins All Children’s Hospital with cultures positive for P. aeruginosa obtained from blood, endotracheal tube aspirate, tracheostomy aspirate, or expectorated sputum in the emergency department (during the encounter that immediately preceded admission), general pediatric inpatient ward, or pediatric/cardiovascular intensive care units were included in the study. An individual patient could contribute multiple specimens during the study period. Specimens were excluded if they could not be processed within 5 days of collection or were resistant to minocycline or tobramycin according to MICs in broth medium. Specimens from patients hospitalized in the neonatal intensive care unit were also excluded in order to reduce the potential influence of longer neonatal hospitalizations on clinically relevant outcomes. Patient data were obtained from the electronic health information system by 2 investigators and were further reviewed by a pediatric infectious diseases specialist (D. B.). The Johns Hopkins All Children’s Hospital Institutional Review Board approved the study, with waiver of informed consent granted.
Bacterial Strains and Growth Conditions
Pseudomonas aeruginosa laboratory strain PAO1 was provided by Barbara Iglewski (University of Rochester, Rochester, NY). All clinical specimens were collected according to institutional standards of care and grown on sheep’s blood agar plates. A single colony of P. aeruginosa was subcultured onto agar slants and transferred to the laboratory at the University of Rochester. All strains were grown in either Luria-Bertani (LB) medium (VWR, Radnor, PA) or 100% human serum (Corning, Corning, NY). Where indicated, LB or serum was supplemented with the indicated concentration of tobramycin (Abraxis Pharmaceutical Products, Wilmington, DE) or minocycline (MP Biomedicals, Solon, OH). Minocycline was chosen as a previously studied surrogate for efflux-mediated SAT in our laboratory [13, 14]. Tobramycin was chosen as a clinically relevant antibiotic for the treatment of pseudomonal infections.
Antibiotic Susceptibility Assays
Pseudomonas aeruginosa was grown overnight from the agar slants in LB medium, diluted into fresh medium (1:100), and grown to the mid-exponential phase (optical density at 600 nm [OD600], 0.4) at 37°C with aeration. A total of 1 × 104 colony-forming units (CFUs) were transferred to individual wells of a 96-well round bottom plate containing 100 µL of LB supplemented with 2-fold increasing concentrations of tobramycin or minocycline (0.25 µg/mL to 256 µg/mL). The MIC was considered to be the lowest concentration of antibiotic in which no bacterial growth could be detected by eye. Once the MIC was determined, a fresh colony from the same frozen stock of the indicated bacterial strain was grown overnight in LB medium, diluted into fresh medium (1:100), and grown to OD600 = 0.4 at 37°C with aeration. The susceptibility assays were performed in triplicate by transferring 1 × 104 CFU to individual wells of a 96-well round bottom plate containing 100 µL of LB or 100% human serum. LB medium was utilized for the SAT assays due to the decreased variability in MIC with differing cation concentrations in comparison to that seen with Mueller-Hinton broth [15]. Cells were incubated in the plates for 2 hours at 37°C without shaking and then supplemented with the indicated MIC concentrations of tobramycin or minocycline and then incubated at 37°C for 48 hours. The contents of each well were serially diluted in sterile 0.8% sodium chloride and plated on LB agar to quantitate CFU/mL. SAT was defined a priori as a ≥2-log increase in CFU/mL grown in serum + antibiotic compared to LB + antibiotic. This definition was based on prior observations that P. aeruginosa laboratory strain PAO1 exhibited this level of growth in serum + antibiotic [13]. Growth controls for both LB and serum were performed without the addition of antibiotic; isolates that exhibited a difference in growth potential at the 48-hour time point were excluded. The increase in growth noted in the presence of serum + antibiotic was further assessed as resulting from inducible tolerance vs a conferred genetic mutation in PAO1 and a subset of clinical isolates. Single colonies obtained after exposure to serum + antibiotic (at the maximum concentration with observable visible growth) were subsequently subjected to MIC determination in LB + antibiotic as described.
Clinical Data Collection
Patient demographic information including age, sex, chronic diagnoses, and primary admitting diagnosis was collected. Chronic diagnoses were categorized into the following organ systems: respiratory, neurologic, gastrointestinal, cardiac, renal, and hepatic disease and immunosuppression status. Patient home respiratory support including tracheostomy status, noninvasive positive-pressure ventilation requirement, and/or ventilator settings were recorded. The number of hospitalizations, the admitting diagnosis for these hospitalizations, and the number of respiratory and blood cultures in the 12 months preceding enrollment were collected. Additionally, we examined clinical outcomes including total length of stay (LOS), duration of intravenous (IV) antibiotic use, duration of fever (>38°C), number of critical care days, duration of intensity of respiratory support exceeding baseline requirements, and mortality during the study period. For patients with positive cultures from repeated hospitalizations within the study period, data from the first (“index”) hospitalization were used in the analysis of clinical outcomes described above.
Statistical Analyses
Continuous variables were reported as either medians (with interquartile range [IQR]) or means (standard deviation) dependent on whether the continuous variable was normally distributed based on visual and statistical assessment. Categorical variables were reported as frequency counts (with percentages). Differences in nonnormally distributed continuous variables between a priori specified groups were assessed using the Wilcoxon rank sum test, whereas the independent sample t test was used to determine whether differences in normally distributed continuous variables existed. The χ2 test was used to determine whether categorical variables differed between a priori specified groups. Due to the small group sizes, exact methods were used for all calculations. Statistical significance was defined as a P value < .05. Statistical analysis was performed using SAS software (v. 9.4).
RESULTS
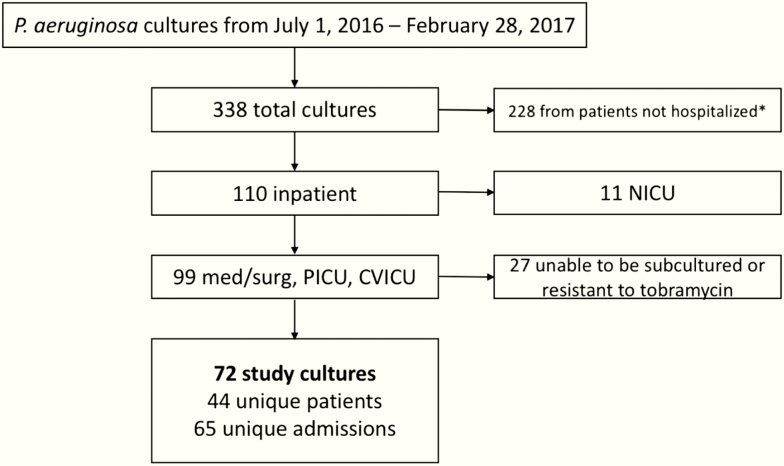
During the study period, 338 cultures were positive for P. aeruginosa, 32.5% (110/338) of which were obtained on hospitalized patients (Figure 1). SAT assays were performed on 72 cultures from 44 unique patients during 65 unique hospital admissions. Individual MIC values for each strain tested are described in Supplementary Table 1.
Figure 1.
Determination of eligible cultures for inclusion in the serum-associated antibiotic tolerance assay. Cultures obtained from patients in the emergency department who were not admitted were considered outpatient cultures and not included in the study (*). Abbreviations: CVICU, cardiovascular intensive care unit; NICU, neonatal intensive care unit; med, general pediatric medicine unit; PICU, pediatric intensive care unit; surg, pediatric surgery unit.
Patient Population
Comparisons of clinical characteristics and outcomes by SAT status were performed in 44 patients. Thirty-eight (86%) patients had at least 1 medical comorbidity identified (Table 1). The most common comorbidities were attributed to the respiratory, neurologic, and gastrointestinal systems. Equal proportions of patients had either CF (32%; n = 14) or were tracheostomy-dependent (32%; n = 14). The majority of patients (84%; n = 37) were hospitalized for respiratory-related conditions. The most common admitting diagnoses were respiratory distress (n = 14), exacerbation of CF disease (n = 11), and tracheitis (n = 6). Patients were previously admitted a median (IQR) of 1.5 (1.0–5.0) times in the 12 months prior to enrollment and had a median (IQR) of 4 (1.0–5.0) respiratory cultures obtained during the same time frame. Thirty-two (73%) of the patients had a respiratory culture positive for P. aeruginosa within 12 months prior to the study period.
Table 1.
Baseline Characteristics of the Study Cohort (n = 44)
| Baseline Characteristic | Total |
|---|---|
| Patient age at admission, median (IQR), y | 12.2 (4.9, 16.5) |
| Sex, n (%) | |
| Male | 20 (45.4) |
| Female | 24 (54.6) |
| Admitting diagnoses, n (%)a | |
| Cough | 8 (18.2) |
| Fever | 6 (13.6) |
| Hypoxia | 10 (22.7) |
| Pneumonia | 11 (25.0) |
| Respiratory distress | 14 (31.8) |
| Tachypnea | 1 (2.3) |
| Cystic fibrosis/exacerbation | 14 (31.8) |
| Tracheitis | 6 (13.6) |
| Other | 30 (68.2) |
| Chronic diagnoses, n (%)a | |
| Respiratory diagnoses | |
| Other respiratoryb | 17 (38.6) |
| Cystic fibrosis | 14 (31.8) |
| Tracheostomy dependence | 14 (31.8) |
| Neurologic diagnoses c | 11 (75.0) |
| Cardiac diagnoses d | 4 (9.1) |
| Gastrointestinal diagnoses e | 38 (86.4) |
| Immunosuppressed f | 3 (6.8) |
| Renal diagnoses | 2 (4.6) |
| Hepatic diagnoses | 3 (6.8) |
a Patients could have had multiple admitting or chronic diagnoses.
b Asthma, bronchopulmonary dysplasia (BPD), chronic lung disease (CLD), home respiratory support.
c Cerebral palsy, developmental delay, hydrocephalus, intellectual disability, nonambulatory, and/or ventriculoperitoneal shunt.
d Congenital heart disease, pulmonary hypertension.
e Aspiration, feeding-tube dependency, gastric fundoplication, supplemental nutrition requirement.
f Immunosuppressed due to medication, genetic, or both medication and genetic.
Prevalence of SAT in P. aeruginosa Isolates
Specimen sources were as follows: sputum (n = 19; 43%), tracheal aspirate (n = 15; 34%), endotracheal tube aspirate (n = 8; 18%), and blood (n = 2; 5%). Forty-five (63%) cultures exhibited the SAT phenotype that involved at least minocycline or tobramycin (Table 2). Tobramycin tolerance alone was observed in 11 (15%) of the isolates, whereas tolerance to minocycline alone was observed in 22 (31%). Tolerance to both antibiotics was observed in 12 (17%) of the isolates. When the analysis was limited to cultures from the index hospitalization (ie, each patient represented once), the frequency of SAT was similar to that for all isolates at 66% (n = 29). Six (14%) patients at the index hospitalization exhibited tolerance to tobramycin alone and 14 (34%) exhibited tolerance to minocycline alone. Overall, 9 (20%) exhibited tolerance to both minocycline and tobramycin.
Table 2.
Prevalence of Serum-Associated Antibiotic Tolerance Phenotypes Among Total Study Cultures (n = 72) and Index Hospitalizations (n = 44)
| Antibiotic Serum-Associated Antibiotic Tolerance Phenotype | Total Cultures, n (%) | Index Hospitalization, n (%) |
|---|---|---|
| No | 27 (37) | 14 (32) |
| Yes | 45 (63) | 29 (66) |
| Tobramycin alone | 11 (15) | 6 (14) |
| Minocycline alone | 22 (31) | 14 (24) |
| Tobramycin + minocycline | 12 (17) | 9 (20) |
Determination of SAT as an Adaptive Phenotype
The SAT phenotypes observed in laboratory strain PAO1 and a subset of clinical isolates (JHACHPA15, JHACHPA37, and JHACHPA39) were assessed for an inducible response (hypothesized mechanism) vs static resistance phenotypes conferred from a genetic mutation resulting from antibiotic exposure. For all isolates tested, the measured MIC for cells subcultured after exposure to serum + antibiotic was equivalent to that for cells not previously exposed to serum or antibiotic (Table 3).
Table 3.
Determination of Minimum Inhibitory Concentrations for Laboratory Strain PAO1 and Clinical Isolates Exhibiting the Serum-Associated Antibiotic Tolerance Phenotype Before and After Exposure to Serum + Antibiotic
| Strain | Antibiotica | MIC LB-pre Serum (µg/mL) | MIC Serum (µg/mL) | MIC LB-post Serum (µg/mL) |
|---|---|---|---|---|
| PAO1 | Minocycline | 8 | >32 | 8 |
| JHACHPA15 | Minocycline | 8 | >32 | 8 |
| Tobramycin | 2 | 8 | 2 | |
| JHACHPA37 | Minocycline | 16 | >32 | 16 |
| Tobramycin | 1 | >1 | 1 | |
| JHACHPA39 | Minocycline | 16 | >32 | 16 |
Abbreviation: LB, Luria-Bertani; MIC, minimum inhibitory concentration.
a Minimum inhibitory concentrations determined only for antibiotics to which the strain exhibited serum-associated antibiotic tolerance.
Relationship Between SAT and Patient Characteristics
Medical comorbidities did not differ by SAT status (Table 4). Among patients with SAT-negative P. aeruginosa isolates, 4 (27%) had a diagnosis of CF and 4 (27%) were tracheostomy-dependent. Similarly, 11 (38%) and 10 (35%) of SAT-positive patients had a diagnosis of CF or were tracheostomy-dependent, respectively. The majority of patients, regardless of SAT group, were hospitalized for respiratory conditions. The median (IQR) number of prior hospitalizations for SAT-negative patients (1.0 [1.0–5.0]) and SAT-positive patients (2.0 [0.0–3.0]) was not significantly different (P = .93).
Table 4.
Baseline Patient Characteristics, Admitting Diagnoses, and Chronic Diagnoses by Serum-Associated Antibiotic Tolerance Status
| Baseline Characteristic | SAT-Negative (n = 15) | SAT-Positive (n = 29) | P Value |
|---|---|---|---|
| Patient age at admission, median (IQR), y | 9.8 (2.6,16.7) | 12.6 (8.0,16.4) | .34 |
| Sex, n (%) | .34 | ||
| Male | 5 (33.3) | 15 (51.7) | … |
| Female | 10 (66.7) | 14 (48.3) | … |
| Admitting diagnoses, n (%)a | |||
| Cough | 1 (6.7) | 7 (24.1) | .23 |
| Fever | 2 (13.3) | 4 (13.8) | 1.0 |
| Hypoxia | 4 (26.7) | 6 (20.7) | .71 |
| Pneumonia | 3 (20.0) | 8 (27.6) | .72 |
| Respiratory distress | 5 (33.3) | 9 (31.0) | 1.0 |
| Tachypnea | 0 (0.0) | 1 (3.5) | 1.0 |
| Cystic fibrosis/exacerbation | 4 (26.7) | 10 (34.5) | .74 |
| Tracheitis | 1 (6.7) | 5 (17.2) | .65 |
| Other | 8 (53.3) | 22 (75.9) | .18 |
| Chronic diagnoses, n (%) | |||
| Respiratory diagnoses | |||
| Other respiratoryb | 13 (86.7) | 21 (72.4) | .45 |
| Cystic fibrosis | 4 (26.7) | 10 (34.5) | .74 |
| Tracheostomy dependence | 4 (26.7) | 10 (34.5) | .74 |
| Neurologic diagnosesc | 11 (73.3) | 22 (75.9) | 1.0 |
| Cardiac diagnosesd | 2 (13.3) | 2 (6.9) | .60 |
| Gastrointestinal diagnosese | 13 (86.7) | 25 (86.2) | 1.0 |
| Immunosuppressedf | 0 (0.0) | 3 (10.3) | .54 |
| Renal diagnoses | 0 (0.0) | 2 (6.9) | .54 |
| Hepatic diagnoses | 1 (6.7) | 2 (6.9) | 1.0 |
Abbreviation: SAT, serum-associated antibiotic tolerance.
a Patients could have had multiple admitting or chronic diagnoses.
b Asthma, bronchopulmonary dysplasia (BPD), chronic lung disease (CLD), home respiratory support.
c Cerebral palsy, developmental delay, hydrocephalus, intellectual disability, nonambulatory, and/or ventriculoperitoneal shunt.
d Congenital heart disease, pulmonary hypertension.
e Aspiration, feeding-tube dependency, gastric fundoplication, supplemental nutrition requirement.
f Immunosuppressed due to medication, genetic, or both medication and genetic.
Relationship Between SAT and Clinical Outcomes
Clinical outcomes are summarized by SAT status in Table 5. Overall, median (IQR) total hospital LOS was 9 days (5.5–16). There was no significant difference in median LOS between SAT-positive and SAT-negative patients in the overall study population (P = .53). Similarly, mean duration of antibiotic administration did not differ by SAT status (P = .23). However, in the study subpopulation of CF patients (Table 6), those with SAT-positive isolates at the index hospitalization had a significantly longer median duration of IV antibiotic administration compared with SAT-negative isolates (10 vs 5 days, respectively; P < .01). Median LOS was also longer in SAT-positive CF patients whose isolates at the index hospitalization were SAT-positive when compared with those whose isolates were SAT-negative (12 vs 5 days; P = .07), although this difference did not meet the threshold for statistical significance. By contrast, in the tracheostomy subpopulation, there was a lack of information to detect a difference in duration of IV antibiotic use (P = .56) or LOS (P = .73) between patients whose isolates were SAT-positive and SAT-negative at the index hospitalization (Table 7). Patients with tracheostomy with the tobramycin SAT-positive phenotype had a longer median duration of IV antibiotic administration and LOS, although these results did not reach the threshold for statistical significance (P = .72 and P = .90, respectively; Table 7).
Table 5.
Clinical Outcomes for Entire Study Cohort by Overall Serum-Associated Antibiotic Tolerance (SAT) Status and SAT Pharmacologic Phenotype
| Outcome | SAT-Negative (n = 15) | SAT-Positive (n = 29) | P Value | Minocycline SAT-Positive (n = 23) | P Value | Tobramycin SAT-Positive (n = 13) | P Value |
|---|---|---|---|---|---|---|---|
| Duration with fever, median (IQR), d | 1.0 (0.0, 1.0) | 0.0 (0.0, 1.0) | .16 | 0.0 (0.0, 1.0) | .22 | 0.0 (0.0, 1.0) | .45 |
| Duration in pediatric intensive care unit, median (IQR), d | 1.0 (0.0, 5.0) | 0.0 (0.0, 7.0) | .87 | 0.0 (0.0, 10.0) | .90 | 0.0 (0.0, 11.0) | .87 |
| Hospital length of stay, median (IQR), d | 7.0 (5.0, 14.0) | 9.0 (6.0, 18.0) | .53 | 9.0 (6.0, 18.0) | .52 | 11.0 (7.0, 20.0) | .28 |
| Duration of antibiotics, median (IQR), d | 5.0 (5.0, 8.0) | 8.0 (5.0, 10.0) | .23 | 8.0 (3.0, 11.0) | .25 | 8.0 (6.0, 10.0) | .14 |
| Duration of oxygen therapy (>home requirement), median (IQR), d | 5.0 (0.0, 11.0) | 4.0 (0.0, 8.0) | .66 | 5.0 (0.0, 9.0) | .80 | 3.0 (0.0, 8.0) | .72 |
| Mortality, n (%) | 0 (0.0) | 2 (6.9) | .54 | 2 (8.7) | .51 | 2 (15.4) | .21 |
Abbreviation: IQR, interquartile range; SAT, serum-associated antibiotic tolerance.
Table 6.
Clinical Outcomes for Cystic Fibrosis Patient Subpopulation by Overall Serum-Associated Antibiotic Tolerance (SAT) Status and SAT Pharmacologic Phenotype
| Outcome | SAT-Negative (n = 4) | SAT-Positive (n = 10) | P Value | Minocycline SAT-Positive (n = 7) | P Value | Tobramycin SAT-Positive (n = 5) | P Value |
|---|---|---|---|---|---|---|---|
| Duration with fever, median (IQR), d | 0.0 (0.0, 0.0) | 0.0 (0.0, 0.0) | 1.0 | 0.0 (0.0, 0.0) | 1.0 | 0.0 (0.0, 0.0) | 1.0 |
| Duration in pediatric intensive care unit, median (IQR), d | 0.0 (0.0, 0.0) | 0.0 (0.0, 0.0) | 1.0 | 0.0 (0.0, 0.0) | 1.0 | 0.0 (0.0, 0.0) | 1.0 |
| Hospital length of stay, median (IQR), d | 5.0 (4.0, 10.0) | 12.0 (9.0, 18.0) | .07 | 11.0 (9.0, 18.0) | .09 | 14.0 (11.0, 21.0) | .08 |
| Duration of antibiotics, median (IQR), d | 5.0 (4.0, 5.5) | 10.0 (8.0, 15.0) | <.01 | 10.0 (9.0, 17.0) | <.01 | 10.0 (8.0, 15.0) | .02 |
| Duration of oxygen therapy (>home requirement), median (IQR), d | 0.0 (0.0, 0.) | 0.0 (0.0, 0.0) | 1.0 | 0.0 (0.0, 0.0) | 1.0 | 0.0 (0.0, 0.0) | 1.0 |
| Mortality, n (%) | 0 (0.0) | 0 (0.0) | 1.0 | 0 (0.0) | 1.0 | 0 (0) | 1.0 |
Abbreviation: IQR, interquartile range; SAT, serum-associated antibiotic tolerance.
Table 7.
Clinical Outcomes for Tracheostomy Patient Subpopulation by Overall Serum-Associated Antibiotic Tolerance (SAT) Status and SAT Pharmacologic Phenotype
| Outcome | SAT-Negative (n = 4) | SAT-Positive (n = 10) | P Value | Minocycline SAT-Positive (n = 7) | P Value | Tobramycin SAT-Positive (n = 5) | P Value |
|---|---|---|---|---|---|---|---|
| Duration with fever, median (IQR), d | 1.0 (0.5, 4.0) | 0.0 (0.0, 0.0) | .21 | 0.0 (0.0, 0.0) | .21 | 0.0 (0.0, 1.0) | .63 |
| Duration in pediatric intensive care unit, median (IQR), d | 1.0 (0.0, 2.0) | 0.5 (0.0, 3.0) | .83 | 0.0 (0.0, 3.0) | .82 | 1.0 (0.0, 19.0) | .73 |
| Hospital length of stay, median (IQR), d | 6.0 (4.5, 39.0) | 5.5 (3.0, 11.0) | .73 | 6.0 (5.0, 11.0) | .94 | 11.0 (2.0, 18.0) | .90 |
| Duration of antibiotics, median (IQR), d | 6.0 (4.0, 8.0) | 3.0 (0.0, 9.0) | .56 | 3.0 (3.0, 9.0) | .74 | 9.0 (3.0, 10.0) | .72 |
| Duration of oxygen therapy (>home requirement), median (IQR), d | 6.5 (4.5, 10.0) | 6.0 (4.0, 9.0) | 1.0 | 6.0 (6.0, 9.0) | .85 | 9.0 (3.0, 11.0) | 1.0 |
| Mortality, n (%) | 0 (0.0) | 1 (10.0) | 1.0 | 1 (11.1) | 1.0 | 1 (20) | 1.0 |
Abbreviation: IQR, interquartile range; SAT, serum-associated antibiotic tolerance.
Discussion
In this study, we are the first to report the existence of SAT among clinical isolates of P. aeruginosa obtained from pediatric patients. We demonstrated the SAT phenotype in more than two-thirds of all clinical isolates collected in this cohort, the majority of which was comprised of children with CF or tracheostomy who had 2 or more comorbidities. Furthermore, although there were no statistically significant differences in outcomes of the index hospitalization between SAT-positive and SAT-negative groups for the overall study population, we found a statistically significant increase in duration of IV antibiotics and a clinically significant (albeit not statistically significant) increase in hospital LOS in the CF subpopulation.
The aforementioned findings are important for better understanding the mechanisms by which P. aeruginosa evades eradication in its host, particularly those individuals affected by chronic and recurrent pseudomonal infections. The colonization of patients with CF and patients with tracheostomy by P. aeruginosa has been associated with poorer clinical outcomes [5, 9]. Treatment approaches aimed at eradicating P. aeruginosa from the airways of patients with CF have limited long-term success [4, 11, 16]. Similarly, children with tracheostomy have high readmission rates of approximately 30% for respiratory illnesses despite frequent administration of systemic anti-pseudomonal antibiotics [17]. The reasons for this microbial persistence are characterized best in patients with CF and are multifactorial [18–20]. Antibiotic extrusion through multidrug efflux pumps is one mechanism by which P. aeruginosa may persist in its host, conferring resistance to multiple antibiotics [21–28] and fitness advantages for the organism [29, 30]. Further elucidation of the mechanisms that underlie SAT could therefore have important implications for enhanced therapeutic strategies in patients at risk for recurrent pseudomonal infection and/or colonization.
Within our clinical cohort, we determined that the SAT phenotype was present in 63% of total isolates and 68% of unique patients. Because this has not been previously established, it is difficult to determine whether a similar prevalence would be found in populations external to our institution. Multidrug-resistant P. aeruginosa, which may share common molecular mechanisms to SAT, has been estimated to affect up to 45% of patients with CF [31–33]. The prevalence of multidrug-resistance in non-CF patients is not as well characterized. The influence of SAT on clinical outcomes warrants an understanding of the drug-targetable mechanisms that could inhibit this phenomenon. Multiple mechanisms for antibiotic resistance in P. aeruginosa have been previously described [19]. While intrinsic and acquired mechanisms for resistance have been well characterized in P. aeruginosa, adaptive mechanisms by which the pathogen responds to an environmental stimulus to alter its tolerance to antibiotics are a more recent discovery [13, 34–38]. Previously, we showed that the laboratory strain of P. aeruginosa, PAO1, becomes tolerant to ciprofloxacin and tobramycin when exposed to human serum and does so in both an efflux-dependent and -independent manner [13]. The present work confirms this in vitro observation from a laboratory strain via ex vivo findings from patient-derived isolates in a cohort of children with CF or tracheostomy.
The mechanisms for the SAT identified here are not entirely clear. Previous studies in the gram-negative bacterium Acinetobacter baumannii have shown that SAT is the result of the overexpression of a wide repertoire of efflux pumps when exposed to human serum [14] and physiological concentrations of sodium chloride [39]. Treatment with the known efflux pump inhibitor PAβN restored the susceptibility of A. baumannii to antibiotic exposure. Similar to A. baumannii, P. aeruginosa harbors numerous known and putative efflux pumps [40]. Most notable are the resistance-nodulation-division (RND) efflux systems that are capable of extruding antibiotic and nonantibiotic substrates [41]. Specifically, the MexXY and MexAB-OprM efflux pumps are capable of extruding aminoglycosides such as tobramycin [42]. While constitutive overexpression of these pumps is known to contribute to multidrug-resistance in P. aeruginosa [43], adaptive mechanisms for SAT are possible in response to bacterial exposure to human plasma in vivo and serum ex vivo. Induction of RND-type efflux pumps has been described in P. aeruginosa after exposure to electrophilic stressors [44] and nitrosative stresses [45]; human serum as a growth medium may represent conditions that elicit a similar physiologic response. Albumin, a prominent component of human serum, has also been linked to regulation of quorum-sensing apparati [46], the function of which is affected by drug efflux pump expression [47–50]. Although not directly evaluated in our study, it is possible that serum-specific components increase the expression of drug efflux pumps and thereby result in the SAT phenotype. The relevance of the SAT phenotype within the lung environment and whether exposure to alveolar epithelial lining fluid (aELF) induces a similar tolerance to antibiotic challenge were not definitively addressed in this study. While it was determined in previous studies that the composition of aELF protein [51–54] and elemental compositions in healthy volunteers [55, 56] are similar to those in serum, further investigation to elucidate the host factors responsible for SAT and whether these factors are specific to serum or ubiquitous among biologically relevant growth media is warranted.
In addition, nonefflux-mediated mechanisms may be contributing to the SAT phenotype. For instance, human serum may be a relatively nutrient-limited environment and thus induce the bacterium’s stringent response with formation of persister cells known to be more tolerant to antibiotics [38]. One alternative explanation for the increased growth in human serum includes decreased effective antibiotic concentration due to significant protein-binding of the antibiotic itself. This has been shown to be negligible for aminoglycoside antibiotics such as tobramycin [57] but could be relevant for minocycline, which is known to exhibit significant binding [58]. However, this mechanism alone would not explain the variability of the minocycline SAT phenotype observed in this study, particularly for isolates with equivalent minocycline MICs. Furthermore, the restoration of susceptibility to minocycline by efflux pump inhibition suggests that efflux contributes, at least in strong part, to the SAT phenotype [13]. Additional studies into the mechanism(s) that underlies the SAT phenotype will provide more insight into the feasibility of this phenomenon as a druggable target for the treatment of pseudomonal infections.
There are several limitations to this study. First, as the inclusion criteria for the study was pathogen-based and not disease-based, the study population was heterogeneous with respect to baseline clinical characteristics and the clinical management that may have affected the results of our secondary clinical outcomes. The resultant limitation of the between-group comparisons for clinical outcomes may have led to an underestimation of the relationship between SAT and clinical outcomes. Second, the nonprospective design of our study engenders heterogeneity in patient management, which historically tends to regress putative associations toward the null of no association. A prospective study will help not only to validate our cross-sectional study findings but also to generate more reliable estimates of the strength of the association between SAT phenotype and clinical outcomes. Third, given its sample size, the study may have been underpowered to definitively perform our secondary objective of investigating the relationship between SAT and clinical outcomes. Nevertheless, we were able to demonstrate that duration of IV antibiotic use in the CF population was significantly increased for patients with SAT-positive isolates compared with those with SAT-negative isolates at the index hospitalization. In addition, our study did not collect aELF from patients in whom our isolates were obtained. Thus, we cannot address whether aELF alone would induce the antibiotic tolerance observed in this study. Identification of additional media in which the SAT phenotype is observed is an important next step for future research efforts. Last, we were unable to accurately measure the influence of antibiotic exposure preceding inclusion in this cross-sectional study. Future studies are warranted to investigate the influence of a patient’s longitudinal exposure to antibiotics on the prevalence of SAT and clinically relevant outcomes associated with SAT. Notwithstanding these limitations, our findings warrant further adequately powered research in expanded pediatric populations with infectious disease or heightened infectious disease risk.
In conclusion, this study establishes that clinical isolates of P. aeruginosa harbor the SAT phenotype and that this phenotype is found in multiple pediatric populations at risk for chronic and acute-on-chronic pulmonary infections. Our results further show that SAT is a novel factor associated with differences in clinical outcomes of patients affected by P. aeruginosa. Future research should investigate the mechanisms that contribute to SAT in pediatric populations, such as those with CF or tracheostomy dependence, toward the identification of novel targets for future adjunctive antimicrobial therapies.
Supplementary Material
Notes
Acknowledgments. We are grateful for the efforts of the faculty and staff in the Johns Hopkins All Children’s Hospital Clinical and Translational Research Track. We give special thanks to Ernest Amankwah, PhD, for assistance with study design.
Financial support. This work was supported by an institutional grant from the Johns Hopkins All Children’s Foundation.
Potential conflicts of interest. D. B. is a clinical consultant to Teleflex Inc. All remaining authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 2009; 48:1–12. [DOI] [PubMed] [Google Scholar]
- 2. Fox MP, Thea DM, Sadruddin S, et al. ; Pneumonia Studies Group . Low rates of treatment failure in children aged 2-59 months treated for severe pneumonia: a multisite pooled analysis. Clin Infect Dis 2013; 56:978–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sánchez García M. Early antibiotic treatment failure. Int J Antimicrob Agents 2009; 34(Suppl 3):S14–9. [DOI] [PubMed] [Google Scholar]
- 4. Konstan MW, Morgan WJ, Butler SM, et al. ; Scientific Advisory Group and the Investigators and Coordinators of the Epidemiologic Study of Cystic Fibrosis . Risk factors for rate of decline in forced expiratory volume in one second in children and adolescents with cystic fibrosis. J Pediatr 2007; 151:134–9, 139.e1. [DOI] [PubMed] [Google Scholar]
- 5. Zemanick ET, Emerson J, Thompson V, et al. ; EPIC Study Group . Clinical outcomes after initial pseudomonas acquisition in cystic fibrosis. Pediatr Pulmonol 2015; 50:42–8. [DOI] [PubMed] [Google Scholar]
- 6. Henry RL, Mellis CM, Petrovic L. Mucoid Pseudomonas aeruginosa is a marker of poor survival in cystic fibrosis. Pediatr Pulmonol 1992; 12:158–61. [DOI] [PubMed] [Google Scholar]
- 7. Michael K, Lan Z, Susan W, et al. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol 2003; 32:277–87. [DOI] [PubMed] [Google Scholar]
- 8. McCaleb R, Warren RH, Willis D, Maples HD, Bai S, O’Brien CE. Description of respiratory microbiology of children with long-term tracheostomies. Respir Care 2016; 61:447–52. [DOI] [PubMed] [Google Scholar]
- 9. Russell CJ, Mack WJ, Schrager SM, Wu S. Care variations and outcomes for children hospitalized with bacterial tracheostomy-associated respiratory infections. Hosp Pediatr 2017; 7:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cogen JD, Oron AP, Gibson RL, et al. Characterization of inpatient cystic fibrosis pulmonary exacerbations. Pediatrics 2017; 139:e20162642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heltshe SL, Khan U, Beckett V, et al. Longitudinal development of initial, chronic and mucoid Pseudomonas aeruginosa infection in young children with cystic fibrosis. J Cyst Fibros 2018; 17:341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Planquette B, Timsit JF, Misset BY, et al. ; OUTCOMEREA Study Group . Pseudomonas aeruginosa ventilator-associated pneumonia. Predictive factors of treatment failure. Am J Respir Crit Care Med 2013; 188:69–76. [DOI] [PubMed] [Google Scholar]
- 13. Blanchard C, Barnett P, Perlmutter J, Dunman PM. Identification of Acinetobacter baumannii serum-associated antibiotic efflux pump inhibitors. Antimicrob Agents Chemother 2014; 58:6360–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jacobs AC, Sayood K, Olmsted SB, et al. Characterization of the Acinetobacter baumannii growth phase-dependent and serum responsive transcriptomes. FEMS Immunol Med Microbiol 2012; 64:403–12. [DOI] [PubMed] [Google Scholar]
- 15. Reller LB, Schoenknecht FD, Kenny MA, Sherris JC. Antibiotic susceptibility testing of Pseudomonas aeruginosa: selection of a control strain and criteria for magnesium and calcium content in media. J Infect Dis 1974; 130:454–63. [DOI] [PubMed] [Google Scholar]
- 16. Cohen-Cymberknoh M, Gilead N, Gartner S, et al. Eradication failure of newly acquired Pseudomonas aeruginosa isolates in cystic fibrosis. J Cyst Fibros 2016; 15:776–82. [DOI] [PubMed] [Google Scholar]
- 17. Russell CJ, Mamey MR, Koh JY, Schrager SM, Neely MN, Wu S. Length of stay and hospital revisit after bacterial tracheostomy–associated respiratory tract infection hospitalizations. Hosp Pediatr 2018; 8:72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Winstanley C, O’Brien S, Brockhurst MA. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol 2016; 24:327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv 2019; 37:177–92. [DOI] [PubMed] [Google Scholar]
- 20. Stefani S, Campana S, Cariani L, et al. Relevance of multidrug-resistant Pseudomonas aeruginosa infections in cystic fibrosis. Int J Med Microbiol 2017; 307:353–62. [DOI] [PubMed] [Google Scholar]
- 21. Llanes C, Hocquet D, Vogne C, Benali-Baitich D, Neuwirth C, Plésiat P. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob Agents Chemother 2004; 48:1797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Köhler T, Kok M, Michea-Hamzehpour M, et al. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1996; 40:2288–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li XZ, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1995; 39:1948–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okamoto K, Gotoh N, Nishino T. Pseudomonas aeruginosa reveals high intrinsic resistance to penem antibiotics: penem resistance mechanisms and their interplay. Antimicrob Agents Chemother 2001; 45:1964–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao Q, Li XZ, Mistry A, et al. Influence of the tonB energy-coupling protein on efflux-mediated multidrug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1998; 42:2225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aires JR, Köhler T, Nikaido H, Plésiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 1999; 43:2624–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vogne C, Aires JR, Bailly C, Hocquet D,Plésiat P. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob Agents Chemother 2004; 48:1676–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Westbrock-Wadman S, Sherman DR, Hickey MJ, et al. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother 1999; 43:2975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olivares J, Álvarez-Ortega C, Martinez JL. Metabolic compensation of fitness costs associated with overexpression of the multidrug efflux pump MexEF-OprN in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2014; 58:3904–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sánchez P, Linares JF, Ruiz-Díez B, et al. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J Antimicrob Chemother 2002; 50:657–64. [DOI] [PubMed] [Google Scholar]
- 31. Cystic fibrosis patient registry: 2017 annual data report. Bethesda, Maryland: Cystic Fibrosis Foundation; 2018. [Google Scholar]
- 32. Lechtzin N, John M, Irizarry R, Merlo C, Diette GB, Boyle MP. Outcomes of adults with cystic fibrosis infected with antibiotic-resistant Pseudomonas aeruginosa. Respiration 2006; 73:27–33. [DOI] [PubMed] [Google Scholar]
- 33. Tam VH, Chang K-T, Abdelraouf K, et al. Prevalence, resistance mechanisms, and susceptibility of multidrug-resistant bloodstream isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2010; 54:1160–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Currie CJ, Berni E, Jenkins-Jones S, et al. Antibiotic treatment failure in four common infections in UK primary care 1991–2012: longitudinal analysis. BMJ 2014; 349:g5493. [DOI] [PubMed] [Google Scholar]
- 35. Fournier PE, Vallenet D, Barbe V, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2006; 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khanam S, Guragain M, Lenaburg DL, et al. Calcium induces tobramycin resistance in Pseudomonas aeruginosa by regulating RND efflux pumps. Cell Calcium 2016; 61:32–43. [DOI] [PubMed] [Google Scholar]
- 37. Skiada A, Markogiannakis A, Plachouras D, Daikos GL. Adaptive resistance to cationic compounds in Pseudomonas aeruginosa. Int J Antimicrob Agents 2011; 37:187–93. [DOI] [PubMed] [Google Scholar]
- 38. Nguyen D, Joshi-Datar A, Lepine F, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 2011; 334:982–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hood MI, Jacobs AC, Sayood K, Dunman PM, Skaar EP. Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob Agents Chemother 2010; 54:1029–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y, Venter H, Ma S. Efflux pump inhibitors: a novel approach to combat efflux-mediated drug resistance in bacteria. Curr Drug Targets 2016; 17:702–19. [DOI] [PubMed] [Google Scholar]
- 41. Fernando DM, Kumar A. Resistance-nodulation-division multidrug efflux pumps in gram-negative bacteria: role in virulence. Antibiotics 2013; 2:163–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morita Y, Tomida J, Kawamura Y. MexXY multidrug efflux system of Pseudomonas aeruginosa. Front Microbiol 2012; 3:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grkovic S, Brown MH, Skurray RA. Transcriptional regulation of multidrug efflux pumps in bacteria. Semin Cell Dev Biol 2001; 12:225–37. [DOI] [PubMed] [Google Scholar]
- 44. Juarez P, Jeannot K, Plésiat P, Llanes C. Toxic electrophiles induce expression of the multidrug efflux pump MexEF-OprN in Pseudomonas aeruginosa through a novel transcriptional regulator, CmrA. Antimicrob Agents Chemother 2017; 61:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fetar H, Gilmour C, Klinoski R, et al. mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob Agents Chemother 2011; 55:508–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith AC, Rice A, Sutton B, et al. Albumin inhibits Pseudomonas aeruginosa quorum sensing and alters polymicrobial interactions. Infect Immun 2017; 85:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alcalde-Rico M, Olivares-Pacheco J, Alvarez-Ortega C, et al. Role of the multidrug resistance efflux pump MexCD-OprJ in the Pseudomonas aeruginosa quorum sensing response. Front Microbiol 2018; 9:2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Evans K, Passador L, Srikumar R, et al. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J Bacteriol 1998; 180:5443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blanco P, Hernando-Amado S, Reales-Calderon J, et al. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 2016; 4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rampioni G, Pillai CR, Longo F, et al. Effect of efflux pump inhibition on Pseudomonas aeruginosa transcriptome and virulence. Sci Rep 2017; 7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu J, Kobayashi M, Sousa EA, et al. Differential proteomic analysis of bronchoalveolar lavage fluid in asthmatics following segmental antigen challenge. Mol Cell Proteomics 2005; 4:1251–64. [DOI] [PubMed] [Google Scholar]
- 52. Bowler RP, Duda B, Chan ED, et al. Proteomic analysis of pulmonary edema fluid and plasma in patients with acute lung injury. Am J Physiol Lung Cell Mol Physiol 2004; 286:L1095–104. [DOI] [PubMed] [Google Scholar]
- 53. Bell DY, Haseman JA, Spock A, McLennan G, Hook GE. Plasma proteins of the bronchoalveolar surface of the lungs of smokers and nonsmokers. Am Rev Respir Dis 1981; 124:72–9. [DOI] [PubMed] [Google Scholar]
- 54. Müller B, von Wichert P. Bronchoalveolar lavage proteins. Klin Wochenschr 1985; 63:781–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Joris L, Dab I, Quinton PM. Elemental composition of human airway surface fluid in healthy and diseased airways. Am Rev Respir Dis 1993; 148(6 Pt 1):1633–7. [DOI] [PubMed] [Google Scholar]
- 56. Hull J, Skinner W, Robertson C, Phelan P. Elemental content of airway surface liquid from infants with cystic fibrosis. Am J Respir Crit Care Med 1998; 157:10–4. [DOI] [PubMed] [Google Scholar]
- 57. Myers DR, DeFehr J, Bennet WM, et al. Gentamicin binding to serum and plasma proteins. Clin Pharmacol Ther 1978; 23:356–60. [DOI] [PubMed] [Google Scholar]
- 58. Zhou J, Tran BT, Tam VH. The complexity of minocycline serum protein binding. J Antimicrob Chemother 2017; 72:1632–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.