Abstract
Data sharing anchors reproducible science, but expectations and best practices are often nebulous. Communities of funders, researchers and publishers continue to grapple with what should be required or encouraged. To illuminate the rationales for sharing data, the technical challenges, and the social and cultural challenges, we consider the stakeholders in the scientific enterprise. In biomedical research, participants are key among those stakeholders. Ethical sharing requires considering both the value of research efforts and the privacy costs for participants. We discuss current best practices for various types of data, as well as opportunities to promote ethical data sharing that accelerates science by aligning incentives.
Graphical Abstract
Data sharing can maximize the benefit and reach of genomics research. However, sharing must occur in a responsible manner, particularly when there are privacy risks to human participants. In this article, the authors discuss the principles of data sharing, strategies for assessing and mitigating privacy risks, as well as practical guidelines for researchers and wider stakeholders.
Introduction
Genomics has a robust culture of data sharing. We are now nearing the two-decade mark of strong expectations for sharing genome-wide transcriptomic assays and associated metadata [G] 1. This wealth of data has enabled new approaches that rely on the analysis of very large collections of public data by investigators who were not involved in the original data collection2-7. It is also possible to assay genotypes8,9, methylation10, and many other features of a sample at a genome-wide level, which presents considerable opportunities for secondary analysis.
With proof-of-concept studies showing the potential to uniquely identify an individual in ever-widening types of detailed datasets, the sharing process has become murkier11,12. As expression profiling has switched from array-based profiling to sequencing-based profiling, the re-identification risk from human-derived samples has also increased13-15. For genetic data, the risk of re-identification has led to controlled-access sharing, which is mediated via services such as the database of Genotypes and Phenotypes (dbGaP)16. However, genotype-related data that contains aggregated estimates, such as variant-level association statistics, pose some risk that individuals could be re-identified17.
Investigators, funders, and other stakeholders supporting responsible data sharing must consider both the risks and benefits to participants as well as other individuals who could be affected positively or negatively by sharing a research dataset in different ways. In addition to ethical concerns, it is important to consider the impact of data sharing practices on the overall research ecosystem. Genomic profiling technologies are now ubiquitously available and are becoming widely used in fields with different cultures of sharing. Funders and publishers must balance multiple considerations to develop appropriate policies. For example, adding data sharing requirements, particularly as unfunded mandates, could hamper the establishment of a pro-sharing culture by creating resentment around re-use18. However, early genomic scientists recognized the potential for high-dimensional profiling [G] to lead to irreproducible results and spurious findings if source data were not shared19. Funders and publishers ultimately must take steps to foster a robust, responsible data sharing culture to support rigorous research with high-dimensional genomic profiling technologies.
Investigators who have shared well increase the impact of their research: publications linked to a data repository or persistent identifier are more cited20. In this Review, we first outline types of data, metadata and frameworks for sharing. We next describe the steps that researchers can take to assess risks and responsibly share data derived through genome-wide profiling technologies. We discuss the rationale for specific data sharing practices. For some data types, there are not widely recognized single point-of-truth repositories [G] , and these principles can guide researchers’ current decision making. For data types with widely used repositories, we provide more detailed guidance. We extensively cover privacy challenges posed by individual-level data derived from human samples because these data pose the most substantial challenges, but we recognize that many types of genomic data such as those derived from model organisms pose little to no risk and should be publicly shared in appropriate repositories. Although we focus on genomic profiling, the underlying principles apply to other data-intensive research projects as well. We also note the roles that other stakeholders including funders and publishers can play in the process to enhance the pace of discovery, ultimately helping patients. We identify practical changes that could better align researcher incentives and support the efficient enforcement of sharing for valuable research products.
What are research data?
Research data in genomics are of many different classes and types. We can divide data by the types of biomolecules that they represent. For example, certain assays measure RNA in a sample21, and others DNA22, protein23, or metabolite24 content. We can also divide data by the type of measurement technology used to gather them. For example, RNA assays could be based on microarray or sequencing profiling25. A sample itself could be derived from a single organism or many26: it could be a cell line with a treatment27, a human tissue sample28, or a population of organisms gathered from an ocean location29. For the purposes of this Review, we consider genomic data to be those that include the potential to profile the genes or gene products of most of an organism’s or collection of organisms’ genes.
We also consider derived data that are intermediate between the raw data produced by an instrument and a finding to be research data. In the terminology used in this manuscript, we consider read files produced by an RNA sequencing (RNA-seq) experiment and represented in FASTQ format30 to be raw data, we consider gene expression estimates to be intermediate data [G] , and we consider the findings to be plots, figures, and underlying statistics produced by analysis of the gene expression data. There could be multiple intermediate data representations between raw data and a finding. Researchers sequencing paired tumour and normal samples to identify somatic and germline variants would be likely to produce FASTQ files for each tumour and normal sample, variant call format (VCF) files for each sample, separate mutation annotation format (MAF) files for the germline and somatic variants, and finally summary results and figures. In this case there could be hundreds of intermediate VCF files and two separate MAF files between the raw data and the findings. We provide recommendations for how investigators can select which items from raw data to findings should be archived and how they can best be shared.
An increasingly common type of derived data is a model produced by machine learning methods applied to genomic data. Researchers can download publicly available data or process data associated with their study, analyze those data with neural networks31,32 or other approaches33,34, and then use those models to either infer something about the biological system that generated the data6, to better understand the methods themselves35, or to develop a deeper understanding of a related disease or process7. Machine-learning models can often be repurposed in much the same way as underlying data. For example, Gulshan et al.36 took a model trained on generic images and fine-tuned it to detect diabetic retinopathy. In genomics, Kelley et al.37 demonstrated that a model trained on a collection of data from certain cell types could quickly and accurately be adapted to a new cell type. Because machine-learning models are executable, they can also be automatically tested38. New repositories, such as Kipoi, have been designed to support and automatically test such models39, providing downstream researchers with a library of working models.
Throughout this Review we maintain these distinctions between raw data, intermediate data, and findings and provide specific sharing recommendations for each. We also discuss why certain data are more or less likely to identify a study participant and how sharing is controlled for certain high-risk data. In the interests of providing a review that is as broadly applicable as possible, we also describe the principles that underlie specific recommendations. For data modalities that are either not discussed within this Review or that are developed in the future, we expect that these principles can be applied to develop an appropriate sharing plan.
What are research metadata?
Research metadata are the data that describe research data. If a biospecimen’s genomic sequence data are represented in raw form by a FASTQ file, information about the biospecimen is metadata. This could include a coded identifier, the tissue from which a biospecimen was taken, information about the handling of the biospecimen, information extracted from an electronic health record describing the individual from which the biospecimen was taken, and more.
We divide our consideration of research metadata into information about the subject of study, which we term ‘sample metadata’, and information about a sample’s handling and processing, which we term ‘handling metadata’. This framing is aligned with how the influential minimum information about a microarray experiment1 (MIAME) recommendations can be applied to non-microarray settings. It is also aligned with how these types of resources are represented in major databases: for example in the BioSample database, frequently reused biospecimens such as cell lines or references are designated a single, reusable identifier with additional sample metadata40. For derived data, the sample’s metadata would often remain unchanged while the handling metadata would differ based on the computational processing steps; however, this distinction begins to blur for intermediate forms that integrate multiple samples, such as machine-learning models.
Metadata are provided with a level of detail that can be high or low. The fields that are included as metadata can enhance or reduce the level of detail. For example, a hypothetical sample41 could be described as “tumour” or as “tumour from an 18-month-old male”. The latter has additional age and gender information, which are akin to additional fields. The level of specificity for each field also affects the level of detail of the metadata: the same sample could be described as “malignant peripheral nerve sheath tumour from an 18-month-old male”.
Metadata can be structured or unstructured. Structured metadata could be represented as a tab-delimited text file containing a unique identifier and experiment factor ontology42 (EFO) terms relevant to a sample or its handling. Unstructured metadata could be a paragraph in a manuscript describing the experiment. In our example above, derived from Kudesia et al.41, “malignant peripheral nerve sheath tumour from an 18-month-old male” is an unstructured description of a sample. Databases designed to store research data often include fields that allow highly structured information, such as ontology terms that apply to a sample, to be provided alongside fields that are relatively unstructured. For example, the EFO term for malignant peripheral nerve sheath tumour is EFO:0000760, age is EFO:0000246, and male, which is included in EFO from the phenotype and trait ontology (PATO), is term PATO:0000384. The metadata that describe most repository-stored genomic data are available with some structured and other relatively unstructured elements.
How are data shared?
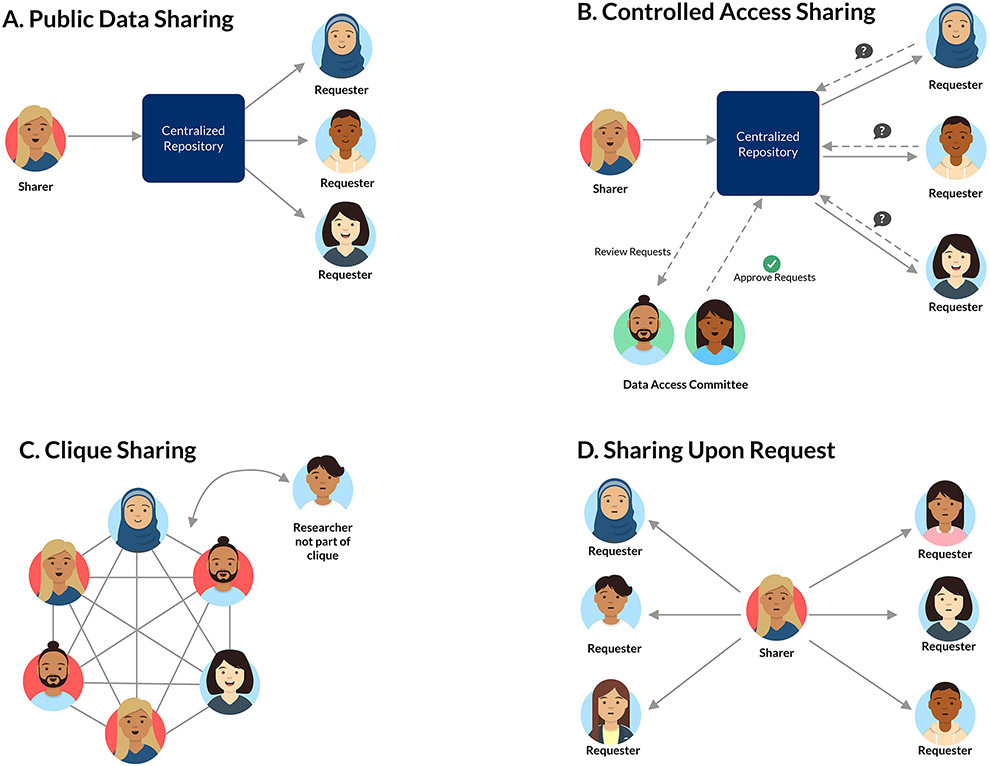
Genomic data are shared in many ways. We distinguish between public, controlled-access, clique, and upon-request sharing approaches (Figure 1). Data are also shared on many different platforms, from those purpose-built for a data type to general-purpose repositories that support many data types to investigator-specific solutions.
Figure 1 ∣. Diverse types of data sharing.
a ∣ In public sharing, researchers make data broadly available without restrictions on use. b ∣ In controlled-access data sharing, researchers place some conditions on access and re-use but ideally do not discriminate on the basis of individual projects or proposers. c ∣ Clique sharing occurs when researchers form consortia and share within the consortia but have very restrictive policies for external sharing, which hampers engagement and impact. d ∣ Sharing upon request offloads the burden for negotiating sharing procedures until there is demand, but when there are multiple requests this approach can become time consuming to manage.
Public data sharing (Figure 1a) occurs when data are released for reuse without barriers (beyond any applicable ethical considerations and laws, with which the user is expected to be familiar). This level provides the lowest barrier to entry for reuse as researchers can probe the data to gain an understanding of its characteristics. Public data sharing combined with detailed sample and handling metadata can allow researchers to answer numerous questions. The Cancer Genome Atlas (TCGA) dataset provides somatic mutation, gene expression estimates, a limited set of clinical metadata, and certain other profiling information, which were made available in a fully-open form and available for publication by anyone after an embargo period43. It has become a remarkably successful example of a public, reusable data resource laying the groundwork for numerous discoveries44. At a smaller scale individually — although covering more biological samples — microarray gene expression datasets are also publicly shared in data-type-specific repositories such as ArrayExpress45 and the Gene Expression Omnibus (GEO)46.
Controlled-access sharing (Figure 1b) occurs when data are available for reuse if some fixed criteria are met. These criteria may include a review of protocols, a commitment to use data only for health-related research, or other elements that affect how one obtains and uses the data but that are not applied differently to different requestors. This level usually provides a modest barrier to entry for reuse efforts and is currently the favoured approach for de-identified genomics data that pose significant re-identification concerns. We discuss such datasets as high-risk. The UK Biobank47 is an example of a resource that is made available under such criteria. A similar effort is underway in the United States via the All of Us project48. Making datasets available in this way allows dataset developers to confirm that adequate oversight structures are in place for research that could potentially lead to re-identification of a study participant.
Clique sharing (Figure 1c) and sharing upon request (Figure 1d) occur when investigators join a consortium or make individual arrangements to share data. These mechanisms place substantial burdens on data requesters, and those within the clique or who hold datasets can select which requesters will be disadvantaged. Data ostensibly made available upon request are not widely shared in practice49,50. In these cases, the data sharing decisions at each point come down to individual scientists. There may be a mismatch between researchers’ perceptions of their own sharing behaviour and their practices. Even when the commitment to share is strong, failure to quickly deposit data in a repository may degrade the investigator’s ability to share as personnel come and go from the lab, as data are likely to be managed less reliably than they would be in established repositories. Earlier-career scientists report being the most enthusiastic about sharing and senior researchers report the most reticence51. In the same survey, early-career researchers report worse sharing behaviours than more senior ones51; however, Campbell et al.52 made data requests and found better sharing behaviours among early-career scientists. These seemingly contradictory results suggest that early-career researchers may hold themselves to a higher standard for sharing. For the purposes of this Review on behaviours supporting an ecosystem that accelerates discovery, we focus on public or controlled-access sharing because of the considerable limitations of clique-based and request-based approaches.
Although the type of sharing influences the extent to which sharing efforts will enhance the impact of the work, it is not the sole factor. For example, Learned et al.53 describe efforts to access and compile publicly available genomic data into a reusable resource for the paediatric cancer community. Even among public data, the authors found barriers to using some of the data: samples that were mislabelled, purportedly uploaded data that were missing, or in certain cases a requirement that they would have to use a proprietary cloud platform for analysis at a substantial cost. In subsequent sections we describe potential risks as well as principles and practices that can help investigators maximize the impact of their data through effective sharing.
Data have variable levels of risk
Although we focus a considerable amount of attention in this Review on the risks associated with sharing certain data, in many cases sharing data poses little to no risk. Many experiments involve genomic assays of model organisms, cell lines, environmental samples, or agricultural subjects. In other cases, the measurement technology may not be capable of revealing individual characteristics or the assay may provide information that is transient and thus poses little risk. Other data clearly identify the individual from which the data were derived, either through the data themselves or the metadata that describe them.
Data that accurately describes a person for long periods of time typically carries a greater privacy risk compared with information that is only transiently true. For example, the sequence of our genome is with us for our lifetime while triglyceride levels may fluctuate with fasting. The risk of re-identification is also related to the extent to which the data modality uniquely identifies individuals. The idea of an equivalence class can help to develop an intuitive understanding of risk: consider an equivalence class to be the number of people for whom a set of values would be true. A measure of the risk of re-identification, given those values, can be considered to be 1/[the number of people in that equivalence class]54,55. In general, the richer the data elements, the smaller the equivalence classes. Transformations of the data can alter the size of equivalence classes; using decade of life, rather than age increases the size of many equivalence classes. However, the effect is not uniform across the dataset: equivalence classes can remain very small for those at the extremes of age. Although it is not possible to exhaustively enumerate data types and their associated risk levels, we provide certain examples (Table 1) and a fuller discussion of risk levels in the following subsections.
Table 1 ∣.
Genomic data types and levels of risk
| Data type | Usual risk level | Sharing with less risk |
|---|---|---|
| RNA-seq reads of model organisms | None | NA |
| Whole-genome sequencing reads of endangered species | Usually none, although location metadata could put species at risk | Public data but controlled-access metadata |
| RNA-seq reads of human tissue samples | High | Public gene expression estimates Controlled-access for sequencing reads |
| Whole-exome sequencing reads of cancer biopsies | High | Public access for somatic-variant data, but controlled access for germline-variant data Potential summary-level queries of germline variants |
| Exome sequencing of human tissue samples | High | Public summary-level information aggregated across many individuals |
| High-density DNA methylation array of human tissue | High | Remove data from probes that contain common variants before public sharing Controlled access for full dataset |
NA, not applicable; RNA-seq, RNA sequencing.
Other types of data encountered in genomic research could also pose risks when shared for reasons different than identification. Certain data, such as the genome sequences of particular pathogens, could pose biosafety concerns. Data that inadvertently discloses the location of endangered species could facilitate poaching. We expect these cases to be rare. In the absence of a clear overriding concern of this type, data not derived from participants should be considered low risk.
Genomic variants are one path to risk.
Certain types of genomic data, such as those directly assaying numerous variants across the genome, cannot be de-identified. For other data types, de-identification can be attempted but may not succeed, and as with other data types the key points to consider are the duration and uniqueness.
Certain types of genomic data are designed to reveal many of an individual’s genetic variants: whole-genome sequencing, high-density genotyping array profiling, and whole-exome sequencing. Germline genetic variants accurately describe a person for long periods of time and, with modest numbers of variants, produce very small equivalence classes. For genomic data sharing beacons, which were an attempt to share only limited, summary-level genomic information to control risks, on the order of hundreds to thousands of variants was often sufficient to reidentify an individual as a member of a beacon56. The clearest avenue to risk is with high-density germline variant calls57. Even noisy variant information can be readily cross-referenced with study participants to re-identify an individual58. In addition, systems for storing genomic data have at times permitted queries of the database using uploaded sequences. Such systems make it possible to find individuals related to an unknown person, given that unknown person’s DNA sequence. Law enforcement entities have used these systems to solve previously unsolved cases, including that of the Golden State Killer59. One database has sought to use an opt-in preference from data contributors to control what can be searched; however, a court has recently ruled that with a search warrant, a police agency can search that database without regard for the opt-in preference of the data contributors60. The extent to which data can be accessed and obtained in this manner depends greatly on the legal jurisdictions that apply.
Sequencing-based assays can reveal the genetic variants that characterize an individual, even if that was not an intended portion of the experiment. Sequencing cancer genomes with the goal of identifying somatic variants reveals both germline and somatic variants. Sequencing-based assays are one avenue of risk. Even if the goal is to simply measure gene expression with RNA-seq, an experiment of normal human tissue that captures a large fraction of messenger RNA and long non-coding RNAs with high sequencing depth is likely to contain sufficient sequencing depth to call genetic variants13-15. The RNA isolation strategy and sequencing depth will affect the windows of the genome in which variants could be revealed. For certain body sites, a substantial fraction of metagenomic reads intended to measure our microbiomes align to a human reference61. On the other hand, highly targeted sequencing technologies may assay only small portions of the genome. The key question in each case is whether or not the technology reveals enough variants to identify an individual12.
Array-based assays can also reveal genetic variants. This can occur intentionally: SNP genotyping arrays are specifically designed to capture differences depending on the allele present at a locus. It can also occur unintentionally: methylation profiling with dense arrays can reveal genotypes at roughly one thousand loci62, those for which some people have genetic variants that directly overlap with the profiled positions. In many cases, data from microarray-based transcriptomic profiling technologies is currently considered to be low risk. For any array-based technology, the more of the genome that is assayed and the more sensitive probes are to short mismatches, the more risk there is of revealing genomic variants.
Especially in the case of genomic data, the probability of re-identification is not static over time and changes based on what other resources are available. Genetic measurements of many individuals provide sufficient information to design artificial queries against data resources that could reveal alleles of interest63. As our understanding of the interrelatedness of genotype and molecular phenotypes grows, it will become easier to identify alleles that underlie high-dimensional data that do not directly measure genotypes64. As more data is made available it becomes easier to find individuals who are closely enough related to a target individual to identify that participant. The observation that certain genetic variants affect gene expression has led to reports of a related risk for gene expression microarray data, but the accuracy of the imputed genotypes is currently relatively low64. We find the considerations in NOT-OD-19-023 from the US National Institutes of Health (NIH) for genomic summary results (GSR) to be particularly helpful for data with theoretical risks but limited current danger65. This policy favours broad sharing except in the case of “studies for which there are particular sensitivities, such as studies including potentially stigmatizing traits, or with identifiable or isolated study populations.”
Metadata can confer risk.
Submitters should supply metadata at the highest level of detail that is ethically and legally feasible. Certain identifiers are direct identifiers [G]. Others may not be direct identifiers but may produce small enough equivalence classes to make reidentification possible. Although defining which entities or research projects are covered by the Health Insurance Portability and Accountability Act (HIPAA) of 1996 is beyond the scope of this paper, the law defines useful concepts regarding data sharing and privacy, particularly as it relates to metadata. The HIPAA privacy rule [G] provides two approaches for de-identification [G] of a dataset: expert determination and the ‘safe harbor [G] ’ method66. The expert determination method requires that a person with appropriate knowledge certify the risk of re-identifying an individual as ‘very small’. The safe harbor method requires removal of 18 HIPAA-specified potentially identifying pieces of information from the database. These types of identifiers pose avenue of risk and include specific geographic locations tied to an individual, absolute dates and times, and other elements. In these cases, it can be helpful to remove absolute date and times and replace date and time fields with intervals. In any case where certain metadata fields introduce risk, we recommend that these fields be separated and low-risk elements be shared openly while high-risk fields be shared only via controlled access in accordance with legal and ethical guidelines.
Machine learning models can confer risk.
Machine-learning models are an emerging form of derived research data that often poses little to no risk. Models trained on publicly available data do not pose a risk above and beyond the data themselves. Models with few parameters relative to the number of subjects also pose less risk. However, models with many parameters that are trained on individual-level genomic data or metadata could reveal detailed information about study participants. Certain attacks have been described that are capable of extracting substantial information about training examples from models or, in certain cases, even the predictions from models67. In some cases, models can be trained using techniques such as differential privacy that allow investigators to manage this risk68,69. Such techniques should be considered if sensitive data from human study participants is used during model training. We recommend that high-dimensional models trained on sensitive data without any form of protection be treated as high-risk.
The principles that guide best practices
Data sharing is simply a means to an end. The goal of research with genomic data is often to improve human health or to better understand a biological process. For such research, stakeholders often include foundations and their donors, taxpayers, study participants who are each dedicating personal or financial resources to these ends, and patients who could someday benefit from the research. Participants in clinical trials overwhelmingly want their data to be shared with other academic researchers70. Researchers generating genomic data should be driven to responsibly advance the aims of these stakeholders as well as their own. We begin from the premise that the goal of sharing is to enhance the overall pace of research in an ethical manner.
Where feasible, data should be shared through data-type-specific repositories that are widely used within a field. Existing data-type-specific repositories are ideal data warehouses because they have the following four properties. First, they support publicly available or controlled-access sharing, thus increasing the speed at which data can be requested and obtained. Second, they provide long-term access to the data through provision of a persistent ID, such as a digital object identifier (DOI), and archiving. Third, they lower costs of research by making large collections of similar data available in a consistent place, which can reduce redundant work and encourage the generation of new hypotheses from secondary analyses. Finally, they allow data to be cited, which lets scientists generating data accrue credit for sharing data sets71. For controlled-access datasets, these repositories provide a consistent request approach. In certain circumstances, particularly early in the development of a data modality, there may be no such repository. In these cases, investigators should choose the last-resort option of placing data in general purpose archiving platforms such as Figshare (https://figshare.com/) or Zenodo (https://zenodo.org/) along with metadata that precisely describe the included files and their format. For data that cannot be publicly shared due to privacy concerns, Synapse (https://synapse.org) provides a similar general-purpose archiving platform that supports controlled access sharing.
Principles that should guide sharing of data with reduced risk.
The lowest risk data, including those derived from model organisms or experiments not involving humans, should be maximally shared with minimal restrictions. Investigators should apply a license to public datasets to provide certainty that they can be reused: Creative Commons Public Domain Dedication [G] (CC0) allows for data to be freely used, and Creative Commons Attribution [G] (CC BY) allows re-use as long as the data sources are attributed. Failure to apply a license can create substantial barriers to reuse for other researchers72,73. In countries that separate the copyright status of facts from those on creative works, it is possible that much genomic data already falls into the public domain but applying a CC0 license makes the intent to promote reuse clear. We recommend CC0 for all public data. Certain licenses create particular challenges for re-use efforts74. Additionally, academic norms require attribution, so CC BY adds barriers but is unlikely to change behaviour. Finally, in the event that someone violates a CC BY license it seems unlikely that investigators would pursue legal action to enforce a citation requirement. For these reasons, we suggest that CC0 is the most appropriate choice for genomic data that are intended to be public.
Sample metadata should be provided in as structured a form as possible. Unstructured text elements should be used only when a structured representation is not supported by the database. Well-structured metadata maximizes the value for downstream use and also make it easier to verify that metadata do not inadvertently reveal a participant’s identity. In data-type-specific databases (Box 1), structured fields are often in place for metadata related to sample handling. Some, such as ArrayExpress, provide entries for commonly used protocols that can be re-used45. Using existing entries makes it easier to add new experiments and allows subsequent users to select all experiments that follow a specific protocol. For handling metadata, unstructured fields should be used sparingly and may not need to be used at all for very common analytical strategies.
Box 1 ∣. Repositories for sharing genomic data.
Repository selection process
There are numerous data-type-specific repositories for sharing genomic data. Investigators should prioritize repositories that are likely to be maintained: these include those built by the National Center for Biotechnology Information (NCBI) in the United States and the European Bioinformatics Institute (EBI) in Europe. We discuss major NCBI and EBI repositories below. For data modalities for which no data-type-specific repositories are maintained by these or similar organizations, investigators should prioritize repositories that are well-adopted within the focused research community. In such cases and also in cases for which no such repositories exist, investigators should archive their data in general purpose repositories.
NCBI’s SRA
The Sequence Read Archive (SRA) supports read-level sequencing data. This includes RNA sequencing (RNA-seq), chromatin immunoprecipitation followed by sequencing (ChIP-seq), assay for transposase-accessible chromatin sequencing (ATAC-seq), whole-exome sequencing, whole-genome sequencing and the results of other such assays. The repository is primarily intended to support US National Institutes of Health (NIH)-funded research; however, datasets of under 1TB in size may be uploaded without cost. Only data that are intended to be public should be uploaded directly to the SRA submission system. This repository still holds controlled-access data, but the upload process is managed via dbGaP, which is discussed below.
EBI’s ENA
The European Nucleotide Archive (ENA) also supports public read-level sequencing data and the same data types as SRA. This database shares a data model with SRA, including the BioProject and BioSample concepts. Uploaded public data are mirrored across both systems. We recommend that investigators in Europe or those without NIH funding use ENA as a mechanism to publicly disseminate sequencing data. Controlled-access data should be uploaded to EGA.
NCBI’s dbGaP
The database of Genotypes and Phenotypes (dbGaP) is designed to support the controlled-access sharing of genomic data for NIH-funded projects. Access is managed through a Data Access Committee (DAC). dbGaP shares data from genetic association studies, studies of methylation, and other individual-level data that are high-risk. In the case of raw sequencing data, the data are housed at NCBI’s SRA but access and upload are managed through dbGaP.
EBI’s EGA
The European Genome–phenome Archive (EGA) holds individual-level genetic data similarly to dbGaP. Access is also managed by a DAC. EGA also holds controlled-access sequencing data. We recommend it as the primary choice for investigators working in Europe.
NCBI’s GEO
The Gene Expression Omnibus (GEO) holds array-based profiling data intended for public release as well as summary-level data from sequencing experiments. Essentially, if the results of an assay can be expressed as a level of observation for a gene, probe, or other such entity it can be housed here. For microarray-based experiments, raw data should be uploaded directly to GEO. For sequencing data, raw data should be uploaded to SRA and summary-level data should be supplied to GEO.
EBI’s ArrayExpress
ArrayExpress is the EBI repository that parallels GEO. For many years, ArrayExpress also imported nearly all GEO data, making it the easiest way to get access to high-throughput profiling data. Datasets that were imported in the past still exist and have E-GEOD- prefixes on their identifiers. However, ArrayExpress no longer imports GEO data so investigators seeking to query these resources will need to query both or a meta-repository that contains the contents of both.
Kipoi
The Kipoi model repository is a recently developed repository for storing machine-learning models that operate over genomic sequences. The models are regularly tested and paired with an application program interface (API) that facilitates their reuse. We recommend that investigators developing models compatible with Kipoi upload them there. For models not yet compatible with Kipoi, we recommend that investigators use general purpose repositories.
Principles that should guide sharing of data with elevated risk.
The vast majority of clinical trial participants favour data sharing despite potential privacy risks70. These privacy risks of data sharing scale according to: first, the chance of one or more parties re-identifying a person in the dataset, and second, the potential consequences of re-identification. Successful de-identification is key to reducing the chance of re-identification, and investigators should take care to avoid identifier leakage, which is a particular risk with metadata elements.
The 2015 Institute of Medicine consensus report entitled “Sharing Clinical Trial Data: Maximizing Benefits, Minimizing Risk”54 is a uniquely comprehensive discussion of the risks of data sharing, and steps that can be taken to mitigate those risks. Although not specific to genomic data, much of the report applies to genomic data. Among the principles identified in the consensus report is that context must be considered when thinking about risks of data sharing. If data sharing is made controlled access, then the risks of data sharing are mitigated to some extent. The privacy risks of sharing data sets that focus on rare diseases are generally greater than for common diseases, but not necessarily too great to undertake. To identify risks that could be deemed acceptable requires selecting a way to measure re-identification risk and selecting an appropriate threshold of risk, and finally measuring the risk in the actual data to be shared. The report encourages investigators to consider the maximum risk to an individual when calculating the risks of publicly shared data sets, and the average risk to individuals for controlled-access datasets.
The risk-benefit ratio of data sharing will look different to different study participants because of varying levels of tolerance for risk and individual reasons for participating in the study. Consent to share de-identified data for secondary analyses can be obtained by design. This approach demonstrates the highest regard for study participants’ interest in the issue of data sharing54. However, other, less clear forms of consent language have also been used, with varying degrees of consideration for the privacy of the participants. The approach that is most invasive of participants’ privacy is neither to obtain consent for data sharing up front, nor to notify the participants that the de-identified data are being shared. Researchers owe it to their participants to make sure that the impact of the data is maximized within ethical and legal constraints. We recommend that researchers ensure that informed consent language explicitly allows for data to be shared and to “promote research initiatives at other institutions” to maximize the impact of participants’ data75.
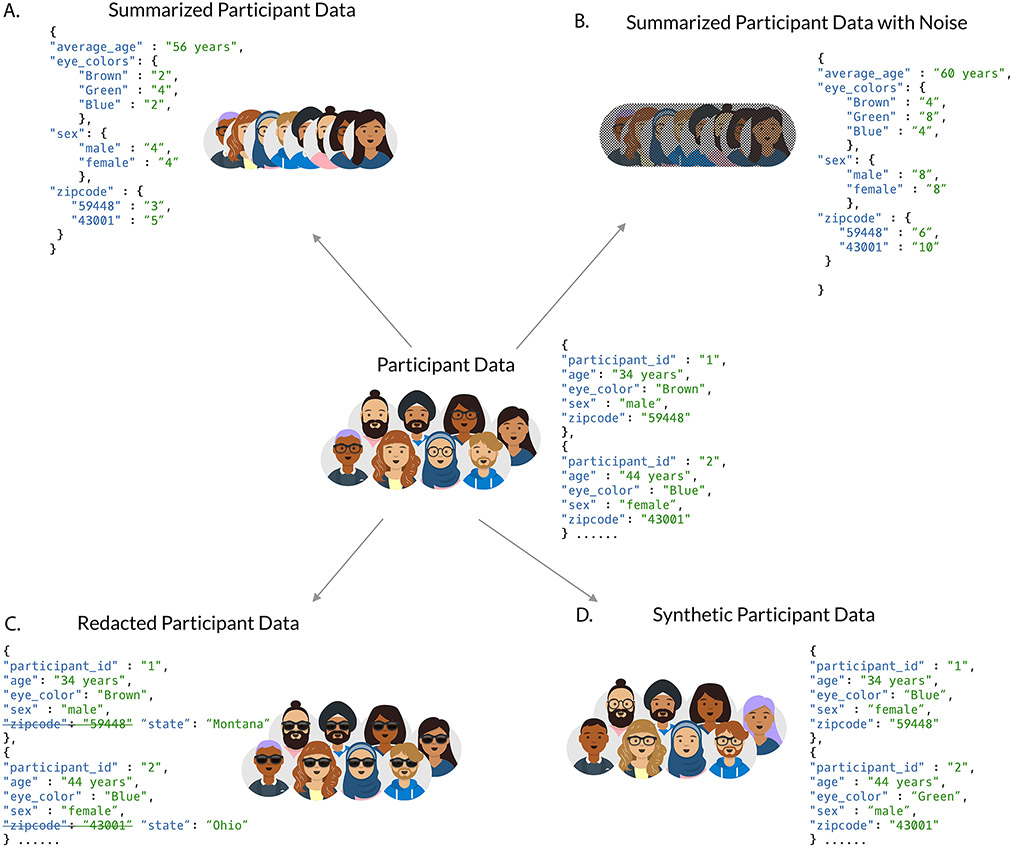
In many cases it is possible to produce low-risk derivatives or views of high-risk data that retain much of the utility while mitigating much of the risk (Figure 2). Methods include presenting only summary-level data (Figure 2a), potentially adding noise (Figure 2b). The Exome Aggregation Consortium (ExAC) and Genome Aggregation Database (gnomAD) browsers focus on germline exome and whole-genome sequencing data, and yet are relatively low-risk to participants, even though the underlying data are not, by providing summary information and limiting the complexity of queries76,77. Other methods of risk mitigation include redacting data (Figure 2c) or generating synthetic data that preserve certain statistical properties (Figure 2d). Given that participants often want their data shared, researchers should aim to identify methods to share valuable derivatives while guarding participant privacy, such as the step of removing human reads performed by the Human Microbiome Project before public sharing.
Figure 2 ∣. Strategies for de-risking data.
Participant data (centre) can be modified or reported in certain ways to minimize risk. It can be reported only at the summary level (a). Those summaries can include added noise to make it difficult or impossible to determine the membership of an individual in an aggregate membership (b). It can have identifying fields redacted (c). In certain cases, the data can be replaced with entirely synthetic data that have many of the same statistical properties but none of the original individuals (d).
Investigators who wish to maximize the impact of their research projects should always share findings-level data publicly unless they pose some risk. In many cases it is also possible to responsibly share intermediate-level data publicly as well. Public sharing reduces the barrier to entry for reanalysis and reduces the chance that a request for data will be received years after the work is done: such requests can be time consuming to answer, and the risk of data being irretrievable increases over time. Finally, data that cannot be responsibly shared in a public manner should be shared through a controlled-access repository.
Privacy is a non-renewable resource
Data has been said to be the new oil, the fuel that will power the economic engine of the 21st century78-80. However, the metaphor is imperfect; in stark contrast to oil, data are not lost when shared and are not destroyed when used. On the other hand, privacy is a resource that can be lost, and once it is lost, it cannot be regained. Although fully open data sharing would be ideal from the perspective of the pace of scientific discovery, it is important to consider the privacy costs of sharing study participants’ data.
In general, measurements that are transient are of a lower risk than information that rarely or never changes. For example, sharing metadata that reveals participants’ white blood cell count — which can transiently increase for many reasons — would impose less of a privacy risk than sharing participants’ HIV status. Certain measurements also pose additional concerns: HIV infection has unfortunately been the focus of stigmatization. Information associated with social stigma has a greater risk when sharing data.
The potential for someone to cross-reference information in a de-identified database with other data sources expands the possible threats to privacy81. Suppose a person tweets that she is proud to have volunteered for a clinical study at a medical school on a particular date, while choosing not to disclose which study. A data analyst may accidentally or intentionally become aware that a particular row of data fits that person’s data due to the date of the tweet. Cross-referencing risk and rare observation risk can be interactive. In a different example, if the date of a visit to the research facility is shared, and the research community is aware that only two families in the US have a particular disorder, the participant’s home state and decade of life could be sufficient to identify the participant. In general: the rarer a measurement, the more risk it poses for privacy.
For newly designed studies, researchers should plan for sharing at the outset. Ultimately, we need to be guided by the feelings of participants, and most participants do want to see data sharing among academic scientists70. Still, careful consideration should be given to what certain technologies, especially sequencing-based technologies, can reveal. In many cases it may not currently be possible to re-identify individuals from a certain data type, but this is a function of other data available for cross referencing, the computational methods and hardware available, and other factors: future risks for re-identification are difficult or impossible to predict. Consent forms should clearly discuss how data will be shared and the known associated risks, including the caveat that for genomic data there is significant risk that data that are not currently identifiable can become so in the future. Mechanisms for dynamic consent may be helpful in some regards82, but the control that they promise must be carefully considered alongside the potential future risk of re-identification due to new analytical methods.
Making repositories the single point of truth
In software engineering, the concept of a single point of truth can reduce errors83, and similar considerations emerge for research involving genomic data. Data and metadata accumulate during the course of a study, and ideally, they are stored in one place with one set of metadata descriptors. At this stage it is particularly important for scientists to have procedures in place to track the single point of truth for the data and metadata.
Depositing data into a repository as soon as possible offloads responsibility to the repository and prevents knowledge about the data, including metadata, from atrophying84. Depositing data in an accepted repository during a study reduces the risk of turnover leading to lost critical knowledge: with the passage of time, scientists generating data may not remember where the data sets are located and the details describing the data75. The repositories also frequently support versioning, allowing researchers to track the state of data over time. Repositories typically do not require that data be made public immediately after it is added: most allow investigators to deposit the data and release it once it is suitably complete and validated for public use.
The concept of a single point of truth also has implications for efforts to construct study-specific data portals or ‘data commons’. For such efforts it is helpful to first deposit data and metadata in data-type-specific repositories that are widely used by the biomedical community and then to construct the metadata summaries and derivative files made available on a data commons from these single points of truth.
Repositories for sharing high-risk data.
For genomic data, the primary repositories for sharing high-risk data support controlled access. Genetic data, raw RNA-seq reads from human samples, and other related data types can often be shared through the same repositories as low-risk data, but with an access control mechanism. As an example consider NCBI’s sequence read archive (SRA): access for certain datasets is controlled by dbGaP. For this database, access is controlled by a data access committee (DAC). Investigators who wish to use such data submit a project description, and the request is submitted by an institutional signing official. This confirms that the host institution is aware of the research and has given ethical approval. The DAC examines the project description and assesses the extent to which the described analysis aligns with the consent that was granted. If an investigator’s access is approved, they are then able to access the data.
What if there are no standard repositories?
In some cases, there will be no standard repository for the data type. For example, there is not currently a controlled-access repository for machine-learning models trained on clinical data that may leak information about individuals. If there are no standard repositories for the data type, investigators may consider a controlled-access general purpose repository: one of the primary such repositories is Synapse (https://www.synapse.org/), produced by Sage Bionetworks. As with all general-purpose repositories, there are certain limitations to such sharing. It is harder for users to perform consistent analyses across the contents of the repository, and more onus is on the uploaders to fully document their data formats, metadata, and other elements. Because this form of sharing requires more effort on both sharers and requesters, it should be only used in the case of last resort.
Benefits that accrue to good sharers
Sharing research outputs benefits the scientific community and increases transparency with the public, who predominantly fund the work, through taxpayer dollars as well as charitable giving to non-profit funders85. Sharing research outputs promotes reproducible science with fewer unintentionally duplicative studies allowing research dollars to be put to maximal use. Effective sharing should also accelerate the pace of discovery. Even though sharing benefits the community, it is not necessarily apparent to scientists generating data how sharing can benefit them and their careers directly, and this, in particular, is crucially important to address in order to increase their willingness to share high-quality data.
Science progresses by building upon the work of others. Sharing outputs openly leads to better utility and visibility of the research, which leads to more citations of that work71,86. For example, publications with preprints are more cited than those without preprints87 and publications with data in openly accessible repositories are more cited when compared to those without accessible data88.
To empower the sharing ecosystem, researchers recently created awards to recognize those who share data as well as those who re-analyze publicly available data. The Research Symbiont Awards founded by J.B.B. are given annually to researchers who share data beyond the expectation of their field89. The companion award to the Research Symbiont is the Research Parasite Award, founded by C.S.G., which honours those who conduct rigorous secondary analysis of existing data90. The goal of these awards is to publicly celebrate those who are committed to sharing and re-using data in a way that contributes to a greater understanding of the world around us.
Open data empowers researchers with the ability to pool data, effectively increasing sample size for appropriately powered studies91. Furthermore, open data facilitates linking, for example, genomic and epigenomic data with clinical and environmental exposure data for a greater understanding of disease biology92. To further illustrate the power of open data, Milham et al.91 recently compared the publications resulting from the use of the International Neuroimaging Data-sharing Initiative (INDI) repository by those who contributed data versus those that did not. They found that 90.3% of publications resulting from reanalysis of the data in the repository were authored by teams without any data contributions, suggesting that clique/consortium models that only allow access to the data to those that contribute are missing out on bringing new expertise and collaborators into their field who are able to re-analyze the data with fresh perspectives91.
Funding practices that support sharing
Funders of biomedical research can play a large role in shifting scientific sharing practices. In the absence of sharing requirements, researchers can be reticent to share, but sharing mandates can increase data sharing prevalence93-95. Funders should promote a culture of sharing, and in particular a data sharing culture that builds upon FAIR data [G] standards: ensuring that data are Findable, Accessible, Interoperable and Reproducible96. Barriers to sharing among researchers are multifactorial. Some barriers are practical: researchers may lack the time, funding, or understanding of how and where to share. Others are cultural and may include the lack of adoption in a field, concerns with data misuse or reproducibility and disincentives for sharing related to the potential loss of future publications derived from the dataset97,98. We strongly recommend that funders require that data are deposited into standard repositories that provide identifiers to enable output tracking. However, we expect that this alone will not be enough because the quality of data sharing can vary widely53. Hence, there must also be practical ways that funders can incentivize greater researcher-focus on effective sharing, which we describe next.
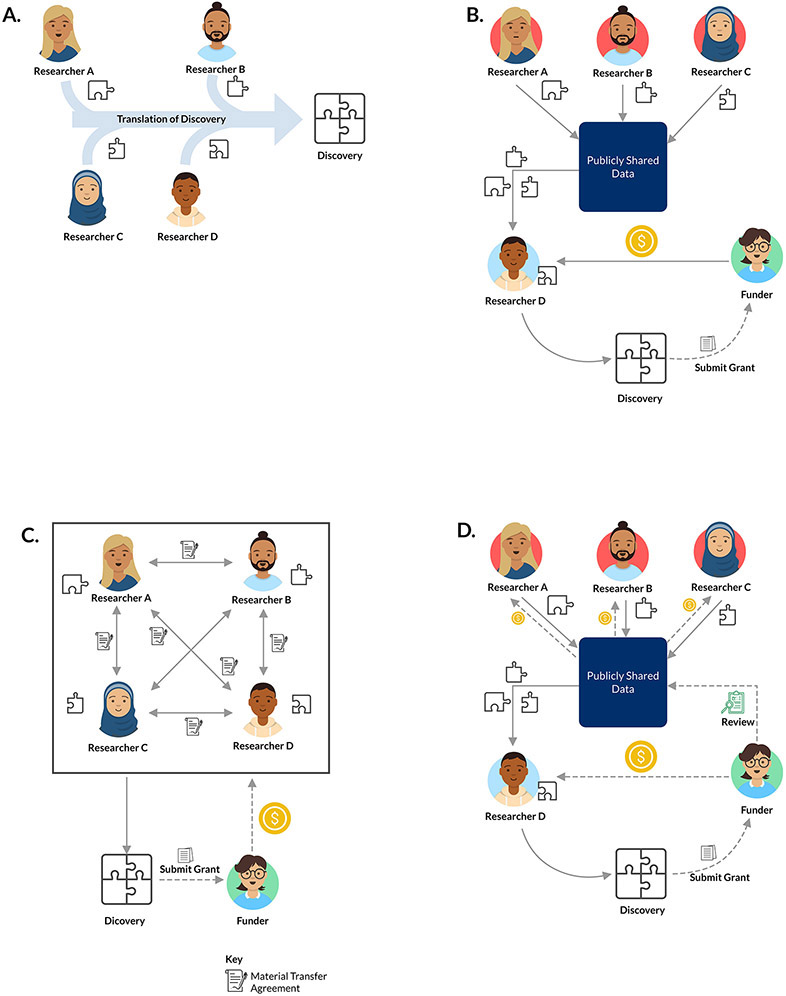
A fundamental challenge with incentivizing greater sharing is that resources, including data, may not be obviously valuable until a major discovery is made from them (Figure 3a). However, once a discovery is made, credit for the discovery accrues to the researchers who made that discovery and not necessarily to those who built and publicly shared the resources that enabled it (Figure 3b). This practice disadvantages sharers: those who share well would do better to hold on to resources and only trade them in the context of a negotiated contract that provides a part of the share of the discovery credit (Figure 3c). Funders have the ability to break this state of poor incentives by considering applicants’ track record of sharing by asking reviewers to consider the evidence of prior sharing. In particular, manuscripts written by unrelated groups using the shared dataset can provide primae facie evidence of the sharing reputation of the researcher under consideration for funding. If funding decisions are positively influenced by a strong track record, the reputational benefit for sharing can have concrete value that supersedes the value of refusing to share (Figure 3d). Rewarding open sharing by assessing sharing reputations in funding decisions has the potential to reduce the friction of contract negotiation and accelerate the pace of discovery. Alex’s Lemonade Stand Foundation, a leading funder of paediatric cancer research in the U.S., is one of the few funders requiring and reviewing prior sharing histories as part of resource sharing plans for all grant applicants, where resource sharing is inclusive of all research outputs including data90,99.
Figure 3 ∣. How funders consider resources can affect sharing cultures.
a ∣ Researcher work products from multiple groups need to be combined to produce a discovery that improves human health. b ∣ In a system of open sharing, if funders allocate credit without considering sharing behaviour, much of the credit and funding can accrue to the researcher who brings the final component that enables translation. c ∣ Researchers can restrict sharing and negotiate agreements through cliques to enhance the equity of credit distribution, but negotiating agreements is time consuming and may delay or prevent advances. d ∣ Funders who consider the value of shared resources when assessing impact provide a benefit not only to the researcher bringing the final component but also all others on the value chain.
When funders collectively require and review sharing plans, they provide an amplified voice to this issue which helps to shape sharing practices in the long term. To increase transparency and compliance in data sharing, funders should consider releasing the sharing plans to the public so that the scientific and lay community knows what was promised to be shared, especially when the projects are publicly funded, such as work supported by the NIH97. Funders should also require clear statements of when data will be made available.
Although it is important for funders to ask for resource sharing plans, it is also equally important that funders support the budgeting of reasonable costs for sharing. Sharing effectively requires knowledge, time and money, and funders must be willing to support these costs in order to ensure compliance with sharing policies. For example, Couture et al.84 found that compliance with data sharing mandates, despite being higher than without sharing requirements, is still low: 26% of data was recovered even when required to be shared by funder-mandate. Funders must provide monetary support for high-quality data deposition so that the community does not end up with ‘data dumpsters’ containing data that are difficult to use due to lack of metadata or meaningful documentation100.
Funders should also promote the use of university libraries as a resource for the development and implementation of data sharing plans and may consider supporting infrastructure grants that allow for the hiring of personnel devoted to data management or, where needed, support repository formation and/or maintenance101,102. Funders may also consider offering or funding research data management training workshops101. Funders should consider supporting the use of existing tools for the creation of data management plans, including California Digital Library’s DMPTool103 and Digital Curation Centre’s DMPonline104 which provide templates for data sharing plans75.
In summary, funder policies and practices have the potential to dramatically shift the data sharing landscape. Funders should make clear through their actions and funding decisions that they value all research outputs, including data sets, as important scientific contributions105,106. For this to be feasible, unique research outputs should have persistent identifiers that allow them to be cited, highlighting the key importance of sharing via repositories that we emphasize in this Review. Additional open science practices, such as research output sharing, open access publishing and preprinting105, can help to support this transition. Ultimately, funders should move to establish funding policies based in part on a past track record of effective sharing: this promotes the proactive sharing of high-quality outputs to create an ecosystem where researchers compete to share the highest quality data possible by the most effective method possible.
Publishing practices that support sharing
Journals played a key role in requiring microarray-based gene expression data to be made available at the time of publication107. Publishers must similarly require that data that are described in publications are made available. Reviewers should be asked specifically if any data or datasets should be made available. Before an article is published, journal staff should check not only that an accession number is present but also that the accession number resolves to a resource that contains the data described in the published work53. This would avoid certain cases where data that are shared are not as they are described53.
The complement to requiring data availability is ensuring that usage is responsible. Investigators have published research108 using controlled-access data resources such as the UK Biobank where the research questions were at best tangentially related to the underlying data access request109. Journals should require investigators using controlled-access data resources to provide the description of the proposed work as supplementary materials. Reviewers should be asked if the study in question aligns with the proposed work. Editors should also use their expert judgement during the editorial review process to assess the extent to which the work described in the manuscript aligns with the underlying request. Journals should refuse to publish work if the data were obtained under pretenses that do not match the results.
Perspectives
Investigators must simultaneously balance the wishes of participants to participate in impactful research with the informed risks that participants take in doing so. For genomic data in particular, the risks of participation are not static over time. Our understanding of underlying biological mechanisms, the presence of other complementary data types, and the power of our analytical approaches all affect the risk of re-identification. Research is needed on processes that can generate derivatives that maximize reuse value while mitigating the re-identification risk for as long as is possible. Still, because perfect risk reduction is likely to be impossible, researchers should not consent participants under promises that genomic data will be made de-identifiable. Certain efforts are underway to make computing environments that expose data for analysis but that limit risk, but guidance from the trajectory of beacons110,111 to reidentification56 suggests that technical solutions may be insufficient. In an era where we can expect those interested in reusing data to aim to train high-parameter machine learning models, investigators should take guidance in designing consent processes from the limited number of efforts that intended to publicly release variant-level data. For the 1000 Genomes Project112,113 and Harvard Personal Genome Project114 participants consented to have their germline genetic data openly shared. In a pilot program in Texas, many patients with cancer elected to have both germline and somatic variants shared openly115. It is clear that at least some are willing to participate in research, even if it leads to the public release of their germline genetic variants. Even for projects where the primary sharing mechanism is intended to be controlled access, investigators may wish to offer participants the opportunity to become ‘data donors’ whose data would be publicly shared.
Researchers recruiting participants must also make every effort to make sure that data sharing and consent processes do not marginalize certain participants or groups of individuals. The overwhelming presence of individuals of European descent in genetic databases has been widely documented116,117. A fuller communication of the potential risks of participating could discourage individuals from certain groups, particularly those who have been minoritized, from participating. Researchers have a responsibility to make sure that benefits of research accrue broadly to society: an increased proportion of individuals who decline to participate in genomic research should not be an acceptable excuse for disparities in the extent to which research benefits the members of that group.
Researchers who generate genomic data can take certain steps to make those data as impactful as possible: adding key metadata elements, sharing the data with the fewest restrictions possible, and putting data in data-type-specific repositories. However, creating a responsible culture of data sharing that accelerates research is more than just the responsibility of those who generate data in the course of their research. For controlled-access human study participants’ data, those analysing the data have a responsibility to do so in accordance with the consent of participants and supplied study plans. Journals have a responsibility to decline to publish analyses that are not conducted in accordance with ethical research practices. Funders have a responsibility to support ethical research in diverse populations while preferentially supporting those who have established exemplary records of generating widely reused resources.
Acknowledgements
This work was funded in part by a grant from Alex’s Lemonade Stand Foundation (CCDL), grants from the National Institutes of Health, K23 HL128909, R01 CA237170 and R01 HG010067, and a grant from the Gordon and Betty Moore Foundation (GBMF 4552).
Glossary
- Metadata
This term refers to data that describe the data. For genomic samples, this could be how the sample was processed, the platform that was used to assay it, characteristics about the conditions in which the sample was obtained, or any other elements that provide context to the genomic data in question
- High-dimensional profiling
Assays of samples that produce many measurements for each sample. Genomic profiling technologies are high-dimensional ones. For example, assaying the expression level of all protein-coding genes in the genome characterizes each sample in approximately 20,000 dimensions. Genotyping of single-nucleotide polymorphisms can produce more than one million dimensions for each human sample
- Single point-of-truth repositories
Repositories that are designed to store the archival form of a dataset and assign a unique identifier. Investigators are responsible for all aspects of data provenance until data are put into a single point-of-truth repository, at which point the repository becomes responsible for these
- Intermediate data
A term that refers to results between raw data and the desired final representation for reporting. For example, in an analysis to identify differentially expressed pathways from RNA sequencing (RNA-seq) reads, gene expression estimates and differential expression p-values could both be considered intermediate results
- HIPAA privacy rule
These are standards for privacy of individually identifiable health information introduced in the Health Insurance Portability and Accountability Act (HIPAA) of 1996. The rule introduces the concepts of expert determination and ‘safe harbor’ as means of de-identifying data
- De-identification
As defined by the the Health Insurance Portability and Accountability Act (HIPAA), de-identified data has been processed by the expert determination method or the ‘safe harbor’ method
- Safe harbor
A Health Insurance Portability and Accountability Act (HIPAA)-designated method of de-identification that relies on the removal of identifiers of the individual, or of relatives, employers, or household members of the individual. To achieve this method of de-identification, 18 different types of identifiers including email addresses, social security numbers, all elements of dates directly related to an individual except year for individuals 89 and younger, and many other elements must be removed
- Creative Commons Public Domain Dedication
(CC0). The Creative Commons Public Domain Dedication licence is designed to allow a data generator to waive all rights to the extent allowable by law, enabling any receipient to reuse the content to which it is applied without asking permission or meeting other terms. The current version of the licence is 1.0 and it is sometimes referred to as CC0 1.0
- Creative Commons Attribution
(CC BY).The Creative Commons Attribution licence is designed to enable reuse and sharing as long as the person sharing provides appropriate credit, a link to the licence, and a notice of whether or not any changes were made. The current version of the licence is 4.0, and it is sometimes referred to as CC BY 4.0.
- FAIR data
Data that are findable, accessible, interoperable, or resusable are considered to be FAIR; however, there is not a precise definition for each of these criteria, so this is an aspirational goal as opposed to a specific standard
- Direct Identifiers
Information that is replicable, distinguishable, and knowable, and that can identify individuals uniquely
Footnotes
Competing interests
A.C.G. is an employee of a funder, Alex's Lemonade Stand Foundation. As an author, A.C.G. participated in all aspects of conceptualization, design, preparation of the manuscript, and the decision to publish. D.V.P. is an employee of a funder, Alex's Lemonade Stand Foundation. As an author, D.V.P. participated in preparation of the manuscript and the decision to publish. C.S.G. is the Director of the Alex’s Lemonade Stand Foundation’s Childhood Cancer Data Lab. As an author, C.S.G. participated in all aspects of conceptualization, design, preparation of the manuscript, and the decision to publish.
Peer review information
Nature Reviews Genetics thanks O. Hofmann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links:
ArrayExpress: https://www.ebi.ac.uk/arrayexpress/
Database of Genotypes and Phenotypes (dbGaP): https://www.ncbi.nlm.nih.gov/gap/
European Genome–phenome Archive (EGA): https://www.ebi.ac.uk/ega/home
European Nucleotide Archive (ENA): https://www.ebi.ac.uk/ena
Figshare: https://figshare.com/
Gene Expression Omnibus (GEO): https://www.ncbi.nlm.nih.gov/geo/
Kipoi: https://kipoi.org/
NOT-OD-19-023 notice: https://grants.nih.gov/grants/guide/notice-files/NOT-OD-19-023.html
Sequence Read Archive (SRA): https://www.ncbi.nlm.nih.gov/sra
Synapse: https://synapse.org
Zenodo: https://zenodo.org/
References
- 1.Brazma A et al. Minimum information about a microarray experiment (MIAME) - Toward standards for microarray data. Nature Genetics vol. 29 365–371 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Myers CL et al. Discovery of biological networks from diverse functional genomic data. Genome Biol. 6, R114 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mostafavi S, Ray D, Warde-Farley D, Grouios C & Morris Q GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9 Suppl 1, S4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huttenhower C et al. Exploring the human genome with functional maps. Genome Res. 19, 1093–106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee I et al. Predicting genetic modifier loci using functional gene networks. Genome Res. 20, 1143–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan J et al. Unsupervised Extraction of Stable Expression Signatures from Public Compendia with an Ensemble of Neural Networks. Cell Syst. 5, 63–71.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taroni JN et al. MultiPLIER: A Transfer Learning Framework for Transcriptomics Reveals Systemic Features of Rare Disease. Cell Syst. 8, 380–394.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ragoussis J Genotyping Technologies for Genetic Research. Annu. Rev. Genomics Hum. Genet 10, 117–133 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Ng PC & Kirkness EF Whole Genome Sequencing BT - Genetic Variation: Methods and Protocols. in (eds. Barnes RM & Breen G) 215–226 (Humana Press, 2010). doi: 10.1007/978-1-60327-367-1_12. [DOI] [PubMed] [Google Scholar]
- 10.Beck S & Rakyan VK The methylome: approaches for global DNA methylation profiling. Trends in Genetics vol. 24 231–237 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Harmanci A & Gerstein M Analysis of sensitive information leakage in functional genomics signal profiles through genomic deletions. Nat. Commun 9, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gürsoy G, Brannon CM, Navarro FCP & Gerstein M FANCY: Fast Estimation of Privacy Risk in Functional Genomics Data. bioRxiv 775338 (2020) doi: 10.1101/775338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piskol R, Ramaswami G & Li JB Reliable identification of genomic variants from RNA-seq data. Am. J. Hum. Genet 93, 641–651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brouard JS, Schenkel F, Marete A & Bissonnette N The GATK joint genotyping workflow is appropriate for calling variants in RNA-seq experiments. J. Anim. Sci. Biotechnol 10, 44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deelen P et al. Calling genotypes from public RNA-sequencing data enables identification of genetic variants that affect gene-expression levels. Genome Med. 7, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mailman MD et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet 39, 1181–1186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homer N et al. Resolving Individuals Contributing Trace Amounts of DNA to Highly Complex Mixtures Using High-Density SNP Genotyping Microarrays. PLoS Genet. 4, e1000167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longo DL & Drazen JM Data Sharing. N. Engl. J. Med 374, 276–277 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Perou CM Show me the data! Nat. Genet. 29, 373–373 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Colavizza G, Hrynaszkiewicz I, Staden I, Whitaker K & McGillivray B The citation advantage of linking publications to research data. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perou CM et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Clarke L et al. The 1000 Genomes Project: Data management and community access. Nature Methods vol. 9 1–4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aebersold R & Mann M Mass spectrometry-based proteomics. Nature vol. 422 198–207 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Trivedi DK, Hollywood KA & Goodacre R Metabolomics for the masses: The future of metabolomics in a personalized world. New Horizons in Translational Medicine vol. 3 294–305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marioni JC, Mason CE, Mane SM, Stephens M & Gilad Y RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18, 1509–1517 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handelsman J Metagenomics: Application of Genomics to Uncultured Microorganisms. Microbiol. Mol. Biol. Rev 68, 669–685 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437–1452.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konecny GE et al. Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J. Natl. Cancer Inst 106, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zinger L et al. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS One 6, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cock PJA, Fields CJ, Goto N, Heuer ML & Rice PM The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 38, 1767–1771 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan J, Hammond JH, Hogan DA & Greene CS ADAGE-Based Integration of Publicly Available Pseudomonas aeruginosa Gene Expression Data with Denoising Autoencoders Illuminates Microbe-Host Interactions. mSystems 1, e00025–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J & Troyanskaya OG Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12, 931–934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W & Altman RB Data-driven human transcriptomic modules determined by independent component analysis. BMC Bioinformatics 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein-O’Brien GL et al. Decomposing Cell Identity for Transfer Learning across Cellular Measurements, Platforms, Tissues, and Species. Cell Syst. 8, 395–411.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Way GP, Zietz M, Rubinetti V, Himmelstein DS & Greene CS Sequential compression of gene expression across dimensionalities and methods reveals no single best method or dimensionality. bioRxiv 573782 (2019) doi: 10.1101/573782. [DOI] [Google Scholar]
- 36.Gulshan V et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA - J. Am. Med. Assoc 316, 2402–2410 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Kelley DR, Snoek J & Rinn JL Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 26, 990–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaulieu-Jones BK & Greene CS Development and application of continuous analysis enables reproducible computational workflows. Nat. Biotechnol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avsec Ž et al. The Kipoi repository accelerates community exchange and reuse of predictive models for genomics. Nature Biotechnology vol. 37 592–600 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett T et al. BioProject and BioSample databases at NCBI: Facilitating capture and organization of metadata. Nucleic Acids Res. 40, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kudesia S Primary MPNST in Childhood- A Rare Case Report. J. Clin. DIAGNOSTIC Res (2014) doi: 10.7860/jcdr/2014/9380.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malone J et al. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics 26, 1112–1118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, Jensen MA & Zenklusen JC A practical guide to The Cancer Genome Atlas (TCGA). in Methods in Molecular Biology vol. 1418 111–141 (Humana Press Inc., 2016). [DOI] [PubMed] [Google Scholar]
- 44.Park Y & Greene CS A parasite’s perspective on data sharing. Gigascience 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rustici G et al. ArrayExpress update--trends in database growth and links to data analysis tools. Nucleic Acids Res. 41, D987–90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrett T et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 41, D991–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sudlow C et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLOS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.National Institutes of Health (NIH) — All of Us. https://allofus.nih.gov/.
- 49.Savage CJ & Vickers AJ Empirical Study of Data Sharing by Authors Publishing in PLoS Journals. PLoS One 4, e7078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood BDK, Müller R & Brown AN Push button replication: Is impact evaluation evidence for international development verifiable? PLoS One 13, e0209416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tenopir C et al. Changes in Data Sharing and Data Reuse Practices and Perceptions among Scientists Worldwide. PLoS One 10, e0134826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell HA, Micheli-Campbell MA & Udyawer V Early Career Researchers Embrace Data Sharing. Trends in Ecology and Evolution vol. 34 95–98 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Learned K et al. Barriers to accessing public cancer genomic data. Sci. data 6, 98 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Institute of Medicine. Sharing Clinical Trial Data: Maximizing Benefits, Minimizing Risk. (The National Academies Press, 2015). doi: 10.17226/18998. [DOI] [PubMed] [Google Scholar]
- 55.Malin BA An Evaluation of the Current State of Genomic Data Privacy Protection Technology and a Roadmap for the Future. J. Am. Med. Informatics Assoc 12, 28–34 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shringarpure SS & Bustamante CD Privacy risks from genomic data-sharing beacons. Am. J. Hum. Genet 97, 631–646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erlich Y, Shor T, Pe’er I & Carmi S Identity inference of genomic data using long-range familial searches. Science (80-. ). 362, 690–694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gürsoy G, Harmanci A, Green ME, Navarro FCP & Gerstein M Sensitive information leakage from functional genomics data: Theoretical quantifications & practical file formats for privacy preservation. bioRxiv 345074 (2018) doi: 10.1101/345074. [DOI] [Google Scholar]
- 59.Kaiser J We will find you: DNA search used to nab Golden State Killer can home in on about 60% of white Americans. Science (80-. ). (2018) doi: 10.1126/science.aav7021. [DOI] [Google Scholar]
- 60.Hill K & Murphy H Your DNA Profile is Private? A Florida Judge Just Said Otherwise - The New York Times. New York Times (2019). [Google Scholar]
- 61.Lloyd-Price J et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Philibert RA et al. Methylation array data can simultaneously identify individuals and convey protected health information: an unrecognized ethical concern. Clin. Epigenetics 6, 28 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edge MD & Coop G Attacks on genetic privacy via uploads to genealogical databases. Elife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schadt EE, Woo S & Hao K Bayesian method to predict individual SNP genotypes from gene expression data. Nat. Genet 44, 603–608 (2012). [DOI] [PubMed] [Google Scholar]
- 65.NOT-OD-19-023: Update to NIH Management of Genomic Summary Results Access. https://grants.nih.gov/grants/guide/notice-files/NOT-OD-19-023.html.
- 66.Methods for De-identification of PHI ∣ HHS.gov. https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html.
- 67.Shokri R, Stronati M, Song C & Shmatikov V Membership Inference Attacks Against Machine Learning Models. in Proceedings - IEEE Symposium on Security and Privacy 3–18 (Institute of Electrical and Electronics Engineers Inc., 2017). doi: 10.1109/SP.2017.41. [DOI] [Google Scholar]
- 68.Abadi M et al. Deep learning with differential privacy. in Proceedings of the ACM Conference on Computer and Communications Security vols 24-28-October-2016 308–318 (Association for Computing Machinery, 2016). [Google Scholar]
- 69.Beaulieu-Jones BK et al. Privacy-Preserving Generative Deep Neural Networks Support Clinical Data Sharing. Circ. Cardiovasc. Qual. Outcomes 12, 159756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mello MM, Lieou V & Goodman SN Clinical Trial Participants’ Views of the Risks and Benefits of Data Sharing. N. Engl. J. Med 378, 2202–2211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Furman JL & Stern S Climbing atop the shoulders of giants: The impact of institutions on cumulative research. Am. Econ. Rev 101, 1933–1963 (2011). [Google Scholar]
- 72.Oxenham S Legal maze threatens to slow data science. Nature vol. 536 16–17 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Himmelstein DS et al. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife 6, e26726 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hagedorn G et al. Creative commons licenses and the non-commercial condition: Implications for the re-use of biodiversity information. ZooKeys vol. 150 127–149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mannheimer S, Pienta A, Kirilova D, Elman C & Wutich A Qualitative Data Sharing: Data Repositories and Academic Libraries as Key Partners in Addressing Challenges. Am. Behav. Sci 63, 643–664 (2019). [PMC free article] [PubMed] [Google Scholar]
- 76.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karczewski KJ et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 531210 (2019) doi: 10.1101/531210. [DOI] [Google Scholar]
- 78.ANA Marketing Maestros: Data is the New Oil. https://ana.blogs.com/maestros/2006/11/data_is_the_new.html.
- 79.Meglena Kuneva - European Consumer Commissioner - Keynote Speech - Roundtable on Online Data Collection, Targeting and Profiling. https://ec.europa.eu/commission/presscorner/detail/en/SPEECH_09_156.
- 80.Lu Qi: Build 2016 - Stories. https://news.microsoft.com/speeches/qi-lu-build-2016/. [Google Scholar]
- 81.Narayanan A & Shmatikov V Robust de-anonymization of large sparse datasets. in Proceedings - IEEE Symposium on Security and Privacy 111–125 (2008). doi: 10.1109/SP.2008.33. [DOI] [Google Scholar]
- 82.Kaye J et al. Dynamic consent: A patient interface for twenty-first century research networks. Eur. J. Hum. Genet 23, 141–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holzmann GJ Points of Truth. IEEE Softw. 32, 18–21 (2015). [Google Scholar]
- 84.Couture JL, Blake RE, McDonald G & Ward CL A funder-imposed data publication requirement seldom inspired data sharing. PLoS One 13, e0199789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mervis J Data check: U.S. government share of basic research funding falls below 50%. Science (80-. ). (2017) doi: 10.1126/science.aal0890. [DOI] [PubMed] [Google Scholar]
- 86.Piwowar HA, Day RS & Fridsma DB Sharing Detailed Research Data Is Associated with Increased Citation Rate. PLoS One 2, e308 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fraser N, Momeni F, Mayr P & Peters I The effect of bioRxiv preprints on citations and altmetrics. bioRxiv 673665 (2019) doi: 10.1101/673665. [DOI] [Google Scholar]
- 88.Piwowar HA & Vision TJ Data reuse and the open data citation advantage. PeerJ 2013, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Byrd JB & Greene CS Data-Sharing Models. N. Engl. J. Med 376, 2305–2306 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Greene CS, Garmire LX, Gilbert JA, Ritchie MD & Hunter LE Celebrating parasites. Nature Genetics vol. 49 483–484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Milham MP et al. Assessment of the impact of shared brain imaging data on the scientific literature. Nat. Commun 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Joly Y, Dyke SOM, Knoppers BM & Pastinen T Are Data Sharing and Privacy Protection Mutually Exclusive? Cell vol. 167 1150–1154 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Levenstein MC & Lyle JA Data: Sharing Is Caring. Adv. Methods Pract. Psychol. Sci 1, 95–103 (2018). [Google Scholar]
- 94.Federer LM et al. Data sharing in PLOS ONE: An analysis of Data Availability Statements. PLoS One 13, e0194768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nuijten MB et al. Journal Data Sharing Policies and Statistical Reporting Inconsistencies in Psychology. Collabra Psychol. 3, 31 (2017). [Google Scholar]
- 96.Wilkinson MD et al. Comment: The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Terry RF, Littler K & Olliaro PL Sharing health research data – The role of funders in improving the impact [version 2; referees: 3 approved]. F1000Research 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stuart D et al. Whitepaper: Practical challenges for researchers in data sharing. (2018) doi: 10.6084/M9.FIGSHARE.5975011.V1. [DOI] [Google Scholar]
- 99.Teytelman L No more excuses for non-reproducible methods. Nature vol. 560 411 (2018). [DOI] [PubMed] [Google Scholar]
- 100.Merson L, Gaye O & Guerin PJ Avoiding Data Dumpsters — Toward Equitable and Useful Data Sharing. N. Engl. J. Med 374, 2414–2415 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Berghmans S et al. Open data: The researcher perspective. (2017). [Google Scholar]
- 102.Popkin G Data sharing and how it can benefit your scientific career. Nature vol. 569 445–447 (2019). [DOI] [PubMed] [Google Scholar]
- 103.DMPTool. https://dmptool.org/. [Google Scholar]
- 104.DMPonline. https://dmponline.dcc.ac.uk/. [Google Scholar]
- 105.Kiley R, Peatfield T, Hansen J & Reddington F Data Sharing from Clinical Trials — A Research Funder’s Perspective. N. Engl. J. Med 377, 1990–1992 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Piwowar H Altmetrics: Value all research products. Nature vol. 493 159 (2013). [DOI] [PubMed] [Google Scholar]
- 107.Ball CA et al. Submission of Microarray Data to Public Repositories. PLoS Biol. 2, e317 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hill WD et al. Genome-wide analysis identifies molecular systems and 149 genetic loci associated with income. Nat. Commun 10, 5741 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.The relationship of cognitive function and negative emotions with morbidity and mortality: an aetiological investigation ∣ UK Biobank. https://www.ukbiobank.ac.uk/2015/06/dr-catherine-gale-university-of-edinburgh/. [Google Scholar]
- 110.Fiume M et al. Federated discovery and sharing of genomic data using Beacons. Nature Biotechnology vol. 37 220–224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Global Alliance for Genomics and Health. A federated ecosystem for sharing genomic, clinical data. Science vol. 352 1278–1280 (2016). [DOI] [PubMed] [Google Scholar]
- 112.Siva N 1000 Genomes project. Nature biotechnology vol. 26 256 (2008). [DOI] [PubMed] [Google Scholar]
- 113.Auton A et al. A global reference for human genetic variation. Nature vol. 526 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ball MP et al. Harvard Personal Genome Project: lessons from participatory public research. Genome Med. 6, 10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Becnel LB et al. An open access pilot freely sharing cancer genomic data from participants in Texas. Sci. Data 3, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hindorff LA et al. Prioritizing diversity in human genomics research. Nature Reviews Genetics vol. 19 175–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sirugo G, Williams SM & Tishkoff SA The Missing Diversity in Human Genetic Studies. Cell vol. 177 26–31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]