Abstract
Endometriosis (EM) is a multifactorial and debilitating chronic benign gynecological disease, but the pathogenesis of the disease is not completely understood. Dysregulated expression of microRNAs (miRNA/miR) is associated with the etiology of EM due to their role in regulating endometrial stromal cell proliferation and invasion. The present study aimed to identify the functions and mechanisms underlying miR-143-3p in EM. To explore the role of miR-143-3p in EM, functional miRNAs were analyzed via bioinformatics analysis. miR-143-3p expression levels in endometriotic stromal cells (ESCs) and normal endometrial stromal cells (NESCs) were measured via reverse transcription-quantitative PCR. The role of miR-143-3p in regulating ESC proliferation and invasion was assessed by performing Cell Counting Kit-8 and Transwell assays, respectively. miR-143-3p expression was significantly upregulated in ESCs compared with NESCs. Functionally, miR-143-3p overexpression inhibited ESC proliferation and invasion, whereas miR-143-3p knockdown promoted ESC proliferation and invasion. Moreover, miR-143-3p inhibited autophagy activation in ESCs, as indicated by decreased green puncta, which represented autophagic vacuoles, decreased microtubule associated protein 1 light chain 3α expression and increased p62 expression in the miR-143-4p mimic group compared with the control group. Moreover, compared with the control group, miR-143-3p overexpression significantly decreased the expression levels of autophagy-related 2B (ATG2B), a newly identified target gene of miR-143-3p, in ESCs. ATG2B overexpression reversed miR-143-3p overexpression-mediated inhibition of ESC proliferation and invasion. Collectively, the results of the present study suggested that miR-143-3p inhibited EM progression, thus providing a novel target for the development of therapeutic agents against EM.
Keywords: endometriosis, endometriotic stromal cells, microRNA-143-3p, autophagy, ATG2B
Introduction
Endometriosis (EM) is a common gynecological disease characterized by the growth of endometrial tissue outside the uterine cavity. EM affected ~10% of women of reproductive age and 5–50% of infertile women worldwide in 1997 (1). As a common gynecological disease, EM results in a number of clinical symptoms, including chronic pelvic pain, dysmenorrhea, dyspareunia, menorrhagia and mental suffering, which affect the quality of life of patients (2,3). At present, the primary treatment strategy for EM involves relieving pain and other symptoms, but does not involve curing EM (4). Therefore, identifying the molecular mechanism underlying EM is important for the development of novel effective therapeutic strategies.
MicroRNAs (miRNA/miR) are non-coding short RNAs that modulate diverse life processes via regulating target genes at the post-transcriptional level (5). Previous studies identified abnormal expression of miRNAs in the eutopic endometrium, which indicated that miRNAs serve a key role in modulating the progression of EM (6–8). miR-143-3p is involved in cell proliferation, apoptosis, adhesion, invasion and other cellular processes (9,10). Previous studies have reported that miR-143-3p was markedly dysregulated in EM (11–13).
Autophagy is a complex process, which is crucial for cellular self-regulation, and dysfunction of autophagy is associated with multiple human diseases, including cardiovascular diseases, neurodegeneration, metabolic diseases, infectious diseases and cancer (14–16). Several studies have demonstrated abnormal activation of autophagy in ovarian EM (17,18). Therefore, the present study aimed to identify the functions and mechanisms underlying miR-143-3p in EM.
Materials and methods
Clinical samples, and cell isolation and culture
The present study was approved by the Protection of Human Subjects Committee of Shanghai Shuguang Hospital. All patients provided written informed consent. Ectopic (n=10), eutopic (n=10) and normal endometrial (n=10) tissues from patients with or without EM (mean age, 43.2 years; age range, 28–51 years) who had undergone a laparoscopy and uterine curettage were obtained from the Shanghai Shuguang Hospital Affiliated with Shanghai University of Traditional Chinese Medicine between November 2017 and March 2019. Endometriotic stromal cells (ESCs) were isolated from endometriotic tissues as previously described (19). Normal endometrial stromal cells (NESCs) were isolated from eutopic endometrial tissues obtained from four individuals without EM (age range, 30–53 years) who underwent a hysterectomy procedure. Briefly, the endometrium was minced and digested with collagenase type I (Gibco; Thermo Fisher Scientific, Inc.) for 45 min at room temperature. After filtrating through a stainless wire mesh (200 µm) and gentle centrifugation at 250 × g for 25 min at room temperature, ESCs or NESCs were isolated via passing over a stainless wire mesh (400 µm). Subsequently, resuspended cells were layered over Ficoll (Beijing Solarbio Science & Technology Co., Ltd.), centrifuged at 2,000 × g for 25 min at room temperature and cells in the middle layer were collected. ESCs or NESCs were cultured in DMEM/F-12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; GE Healthcare Life Sciences) with 5% CO2 at 37°C.
EM mouse model, and isolation of ESCs and NESCs
The EM mouse model was established using C57BL/6 female mice (age, 8 weeks; weight, 25–30 g; n=10; n=5 per group; Shanghai Model Organisms Center, Inc.). Mice were divided into two groups (n=5 per group): i) EM and ii) Control. All experimental procedures were approved by the Ethics Committee for Animal Experimentation of Shanghai Shuguang Hospital. Mice were housed in a barrier unit in a sterile environment at 22°C with 40–80% humidity, 12-h light/dark cycles, and ad libitum access to food and water. Mice were anesthetized with an intraperitoneal injection of chloral hydrate (350 mg/kg). Animals did not exhibit signs of peritonitis following the administration of chloral hydrate. Endometrial pieces (1 mm3) isolated from ovarian endometriotic samples from patients with EM were suspended in saline and 400 µl suspension was injected into the peritoneal cavity of the mice. At 3 weeks after model establishment, mice were euthanized by cervical dislocation. Sections of endometrial tissue were isolated from control mice and EM model mice. Tissue sections were washed twice with PBS and cultured in DMEM/F12 medium supplemented with 20% FBS at 37°C with 5% CO2. The culture medium was changed with the appearance of a large number of desquamated endothelial cells (every 3 days). Non-adherent cells were removed carefully. At 80–90% confluence, the culture medium was removed, cells were washed twice with PBS and then treated with 0.25% trypsin. Subsequently, the cell concentration was adjusted to 1.0×106/ml using DMEM medium supplemented with 10% FBS. Cells were cultured in DMEM/F12 supplemented with 10% FBS at 37°C with 5% CO2.
Transfection
miR-143-3p mimic (5′-UGAGAUGAAGCACUGUAGCUC-3′), 2′-O-methyl-modified anti-miR-143-3p (5′-GAGCUACAGUGCUUCAUCUCA-3′), miR-143-3p mimic negative control (NC; 5′-UCACAACCUCCUAGAAAGAGUAGA-3′) and anti-miR-143-3p NC (5′-UACUCUUUCUAGGAGGUUGUGAUU-3′) were obtained from Shanghai GenePharma Co., Ltd. Cells (1×106) were transfected with 20 ng/ml miR-143-3p mimic, anti-miR-143-3p, miR-143-3p mimic NC or anti-miR-143-3p NC using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h at 37°C. At 48 h post-transfection, cells were collected for subsequent experiments.
Total RNA was extracted from 293T cells using TRIzol® reagent (Takara Bio, Inc.). Total RNA was reverse transcribed into cDNA using the PrimeScript™ RT reagent kit (Takara Bio, Inc.). The full-length cDNA of Autophagy-related 2B (ATG2B) was cloned into the pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.). The sequences of the primers used to amplify ATG2B were as follows: forward, 5′-GGAGCCACTCTCCAGCATAG-3′ and reverse, 5′-GTGCACAGCTCCAAAGATGA-3′. The following thermocycling conditions were used: Incubation at 50°C for 2 min; 95°C for 2 min; followed by 40 cycles of 95°C for 15 sec and 60°C for 32 sec. NESCs were seeded into multiple-well plates at ~80% confluence. Cells were transfected with recombinant plasmids (1.5 µg per well) using Lipofectamine® 2000 (Invitrogen) at room temperature for 6 h according to the manufacturer's protocol. At 48 h post-transfection, subsequent experiments were performed. pcDNA3.1 was used as a negative control.
Luciferase reporter assay
To confirm the potential target genes of miR-143-3p in ESCs, the present study searched for candidate genes using TargetScan (version 7.1; www.targetscan.org/vert_71) and miRBase22 (www.mirbase.org) databases. A 3′-untranslated region (UTR) luciferase reporter vector of ATG2B containing the predicted binding sites of miR-143-3p was produced by cloning the 3′-UTR of the corresponding mRNA into the pGL3-promoter vector (Promega Corporation). Subsequently, 293T cells (1×104) were co-transfected with 1 ng pRL-TK (Promega Corporation), 100 ng pGL3-ATG2B-3′-UTR-wild-type (WT) or pGL3-ATG2B-3′-UTR-mutant (mut) and 20 nM miR-143-3p mimic or miR-143-3p mimic NC using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.). At 72 h post-transfection, luciferase activities were measured using a Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Immunofluorescence
Autophagy vacuoles were assessed using an Autophagy Detection kit (cat. no. ab139484; Abcam) according to the manufacturer's protocol. A coverslip was placed into each well of a 24-well plate. ESCs were seeded (2×104) into the 24-well plate and incubated at 37°C for 1 day. Following washing three times with PBS, cells were fixed with 4% cold paraformaldehyde for 15 min and then washed three times with PBS. After blocking with 1% BSA (Sigma-Aldrich; Merck KGaA) for 30 min at room temperature, the fluorescent dyes for nuclei staining and autophagy detection were added and incubated for 30 min at room temperature. After washing three times using PBS, the slides were observed using a FV1000s-SIM/IX81 confocal laser scanning microscope (Olympus Corporation). The ratio of green to blue fluorescence was calculated to assess the degree of autophagy.
RNA isolation and reverse transcription quantitative PCR (qPCR)
Total RNA was extracted from ESCs and NESCs using TRIzol® reagent (Takara Bio, Inc.). Total RNA was reverse transcribed into cDNA using the PrimeScript™ RT reagent kit (Takara Bio, Inc.) at 37°C. Subsequently, qPCR was performed using SYBR-Green PCR Master Mix (Takara Bio, Inc.) on an ABI Step One Plus™ real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for qPCR: 95°C for 10 min; followed by 35 cycles of 95°C for 10 sec, 58°C for 15 sec and 72°C for 20 sec; and final extension at 72°C for 20 min. The following primers were used for qPCR: miR-143-3p forward, 5′-CTGGCGTTGAGATGAAGCAC-3′ and reverse, 5′-CAGAGCAGGGTCCGAGGTA-3′; miR-125b-5p forward, 5′-TCCCTGAGACCCTAACTTGTGA-3′ and reverse, 5′-AGTCTCAGGGTCCGAGGTATTC-3′; miR-150-5p forward, 5′-TCGGCGTCTCCCAACCCTTGTAC-3′ and reverse, 5′-GTCGTATCCAGTGCAGGGTCCGAGGT-3′; ATG2B forward, 5′-TCCTTCAGGAAGAACAAAGCA-3′ and reverse, 5′-AAGCCTTACACGTGTGTCCA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and GAPDH forward, 5′-ATTCCACCCATGGCAAATTC-3′ and reverse, 5′-TGGGATTTCCATTGATGACAAG-3′. miRNA and mRNA expression levels were quantified using the 2−ΔΔCq method (20) and normalized to the internal reference genes U6 and GAPDH, respectively.
Western blotting
Total protein was extracted from ESCs using RIPA buffer (Cell Signaling Technology, Inc.). Total protein was quantified using a BCA kit (Takara Bio, Inc.). Proteins (20 µg) were separated via 12% SDS-PAGE and transferred to PVDF membranes. After blocking with 5% BSA (in TBS/0.05% Tween-20 buffer) for 2 h at room temperature, the membranes were incubated at 4°C overnight with primary antibodies (all purchased from Abcam) targeted against the following: ATG2B (1:5,000; cat. no. ab226832), p62 (1:1,000; cat. no. ab109012) and microtubule associated protein 1 light chain 3α (LC3)-I/II (1:2,000; cat. no. ab128025) and actin (1:5,000; cat. no. ab8226). Following primary incubation, the membranes were incubated with a Goat Anti-Mouse IgG H&L (HRP) secondary antibody (1:2,000; cat. no. ab205719; Abcam) for 1 h at room temperature. Protein bands were visualized using an ECL kit (Pierce; Thermo Fisher Scientific, Inc.). Protein expression levels were semi-quantified using the Odyssey Infrared Imaging System (version 3.0; LI-COR Biosciences) with actin as the loading control.
Transwell assays
A Transwell assay was performed to assess cell invasion. The Transwell insert (pore size, 8 µm; Corning Life Sciences) was precoated with Matrigel for 4 h at 37°C. ESCs were seeded (5×104) into the upper chamber in serum-free medium and DMEM/F-12 medium supplemented with 10% FBS was plated into the lower chamber. Cells were incubated at 37°C with 5% CO2 for 1 day. Subsequently, invading cells were fixed with 4% paraformaldehyde for 40 min at room temperature and stained with 1% crystal violet staining solution for 15 min at room temperature. Stained cells were quantified by counting the number of cells in five randomly selected fields of view using a light microscope.
Cell proliferation assay
Cell proliferation was assessed by conducting a Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocol. Briefly, cells were seeded (5×103 cells/well) into 96-well plates. Subsequently, at 0, 24, 48 and 72 h, 10 µl CCK-8 reagent was added to each well and incubated for 4 h. The absorbance was measured at a wavelength of 450 nm using a VersaMax microplate reader (Molecular Devices, LLC).
Statistical analysis
Data are presented as the mean ± SD. All experiments were repeated at least three times. Statistical analyses were performed using SPSS software (version 15; SPSS, Inc.). Comparisons between two groups were analyzed using the Mann-Whitney U test (Fig. 1) or unpaired Student's t-test (Figs. 2, 3, and 4B, C and F). Comparisons among multiple groups were analyzed using one-way ANOVA followed by Scheffé's post hoc test (Fig. 4D and E). P<0.05 was considered to indicate a statistically significant difference.
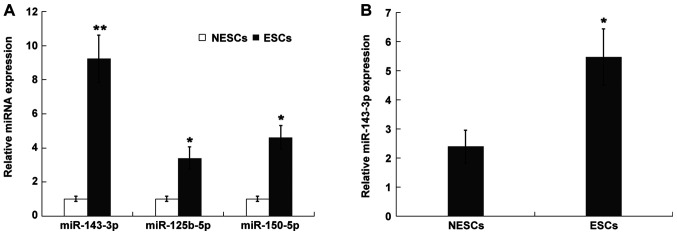
Figure 1.
miR-143-3p is significantly upregulated in ESCs. (A) miR-143-3p, miR-125b-5p and miR-449b-5p expression levels in ESCs and NESCs. (B) miR-143-3p expression levels in ESCs derived from endometriosis model mice and NESCs derived from healthy control mice. *P<0.05 and **P<0.01 vs. NESCs. miR, microRNA; ESC, endometriotic stromal cell; NESC, normal endometrial stromal cell.
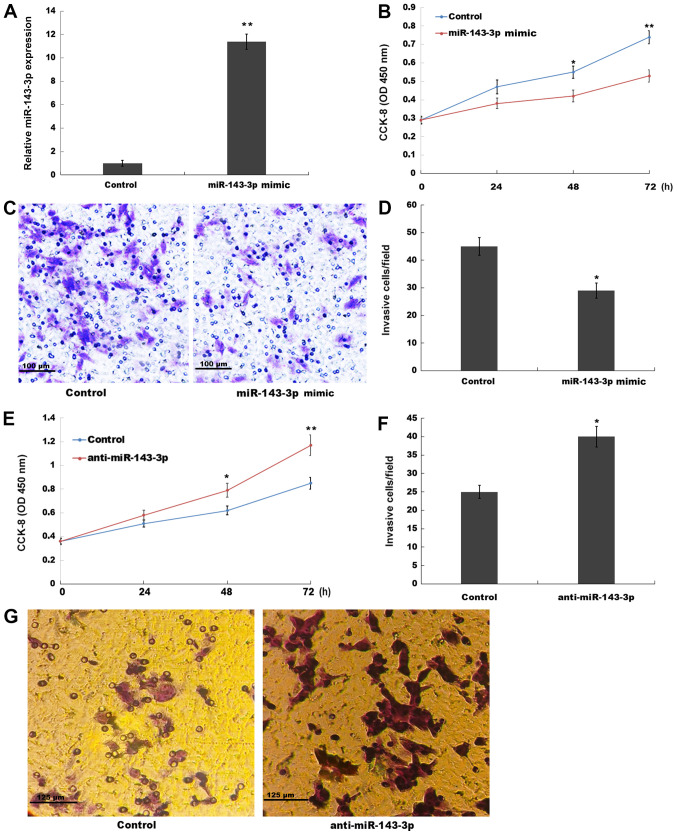
Figure 2.
miR-143-3p inhibits ESC proliferation and invasion. (A) Transfection efficiency of miR-143-3p mimic. (B) Cell proliferation in miR-143-3p mimic-transfected ESCs was assessed by performing the CCK-8 assay. Cell invasion was (C) assessed by performing a Transwell invasion assay and (D) quantified. (E) Cell proliferation in anti-miR-143-3p-transfected ESCs was assessed by performing the CCK-8 assay. (F) Cell invasion was assessed by performing a Transwell invasion assay. (G) Representative images of the Transwell invasion assay. *P<0.05 and **P<0.01 vs. control. miR, microRNA; ESC, endometriotic stromal cell; CCK-8, Cell Counting Kit-8; OD, optical density.
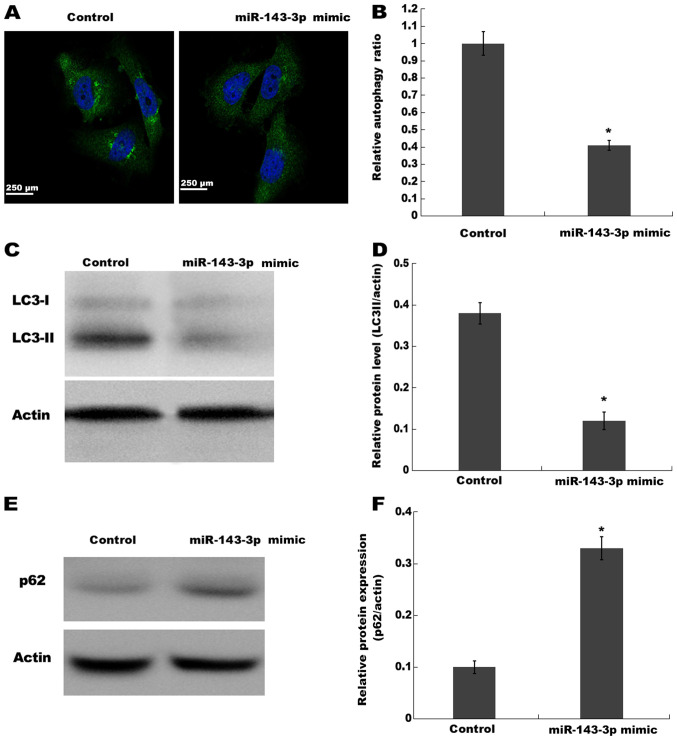
Figure 3.
miR-143-3p suppresses autophagy activation in endometriotic stromal cells. Autophagic vacuoles were identified via (A) immunofluorescence staining and (B) the autophagy ratio was calculated (ratio of green fluorescence to blue fluorescence). LC3-II protein expression levels were determined via (C) western blotting and (D) semi-quantified. p62 protein expression levels were determined via (E) western blotting and (F) semi-quantified. *P<0.05 vs. control. miR, microRNA; LC3, microtubule associated protein 1 light chain 3α.
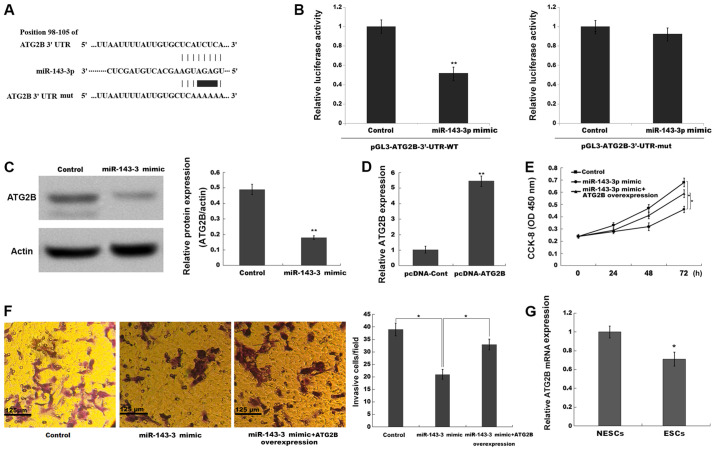
Figure 4.
miR-143-3p inhibits ESC proliferation and invasion by repressing ATG2B. (A) The binding sites between miR-143-3p and the 3′-UTR of ATG2B. (B) The luciferase reporter assay was performed in ESCs co-transfected with miR-143-3p mimic or negative control, pGL3-ATG2B-3′-UTR-WT or pGL3-ATG2B-3′-UTR-mut and pRL-TK. **P<0.001 vs. control. (C) ATG2B protein expression levels were determined via western blotting and semi-quantified. (D) Transfection efficiency of pcDNA-ATG2B. Compared with the control group, miR-143-3p overexpression repressed ESC (E) proliferation and (F) invasion, whereas ATG2B overexpression reversed miR-143-3p-mimic mediated effects. (G) ATG2B mRNA expression levels in ESCs and NESCs isolated from endometriosis model mice. *P<0.05 and **P<0.01 vs. control or NESCs. miR, microRNA; ESC, endometriotic stromal cell; ATG2B, autophagy-related 2B; UTR, untranslated region; WT, wild-type; mut, mutant; CCK-8, Cell Counting Kit-8; OD, optical density; NESC, normal endometrial stromal cell; cont, control.
Results
miR-143-3p is markedly upregulated in ESCs
A number of studies have demonstrated that miRNAs participate in the development and progression of EM via multiple mechanisms (21,22). Abdel-Rasheed et al (23) reported that 32 miRNAs were significantly dysregulated in EM and Cosar et al (13) demonstrated that there are 10 miRNAs that may serve as diagnostic markers of EM. The present study re-analyzed the expression level of three upregulated miRNAs (miR-125b-5p, miR-150-5p and miR-143-3p) that appeared in both datasets (Fig. S1A). The result was verified in ESCs and NESCs. The expression levels of miR-125b-5p, miR150-5p and miR-143-3p were significantly increased in ESCs compared with NESCs (Fig. 1A). In the present study, as miR-143-3p was the most upregulated miRNA among the three miRNAs in ESCs, miR-143-3p was further investigated. Previous studies have also demonstrated that miR-143-3p is upregulated in ectopic endometrial tissues compared with eutopic endometrial tissues (7,24). To further verify the in vitro results, the present study established an EM mouse model and assessed miR-143-3p expression levels in isolated ESCs. miR-143-3p expression levels were significantly increased in ESCs compared with NESCs (Fig. 1B).
miR-143-3p inhibits ESC proliferation and invasion
The role of miR-143-3p in ESC proliferation and invasion was investigated in the present study. miR-143-3p was overexpressed in ESCs and subsequently, cell proliferation and invasion were assessed by performing CCK-8 and Transwell invasion assays, respectively. miR-143-3p mimic significantly increased miR-143-3p expression levels in ESCs compared with the control group (Fig. 2A). The CCK-8 assay results indicated that miR-143-3p overexpression also significantly suppressed ESC proliferation at the 48 and 72 h time points compared with the control group (Fig. 2B). The present study also assessed the role of miR-143-3p overexpression in ESC invasion. The Transwell invasion assay results indicated that miR-143-3p overexpression significantly inhibited ESC invasion compared with the control group (Fig. 2C and D). By contrast, compared with the control group, miR-143-3p knockdown significantly enhanced ESC proliferation at 48 and 72 h, and significantly increased ESC invasion (Figs. S1B and 2E-G). The aforementioned results suggested that miR-143-3p overexpression inhibited EM progression.
miR-143-3p suppresses autophagy activation in ESCs
Previous studies have observed autophagy activation in ectopic endometrium of patients with ovarian endometriosis (17,25,26). Based on the finding that miR-143-3p displays the potential to regulate autophagy in other diseases (27,28), the present study investigated whether miR-143-3p inhibited ESC proliferation and invasion via inactivating autophagy. The activation of autophagy was analyzed using an autophagy detection kit and western blotting. Compared with the control group, miR-143-3p overexpression significantly decreased the autophagy ratio, as evidenced by a decreased number of green puncta, which represented autophagic vacuoles (Fig. 3A and B). miR-143-3p overexpression also significantly decreased LC3-II expression levels (a reliable indicator of autophagy) in ESCs compared with the control group, indicating inhibition of autophagy (Fig. 3C and D). Furthermore, miR-143-3p overexpression significantly increased p62 (an autophagy substrate) protein expression levels in ESCs compared with the control group (Fig. 3E and F). The results indicated that miR-143-3p overexpression inhibited autophagy activation in ESCs.
miR-143-3p suppresses ESC proliferation and invasion by repressing ATG2B
To confirm the potential target genes of miR-143-3p in ESCs, the present study searched for candidate genes using TargetScan (version 7.1; www.targetscan.org/vert_71) and miRBase22 (www.mirbase.org) databases. Bioinformatics analysis using TargetScan and miRBase identified 487 potential target genes. Among the identified target genes, ATG2B was the only gene associated with the autophagy signaling pathway (29). The bioinformatics analysis indicated that miR-143-3p directly targeted the 3′-UTR of the ATG2B gene, an essential regulator of autophagy activation (30) (Fig. 4A). To verify whether miR-143-3p directly bound to the 3′-UTR of ATG2B and repressed its expression, the present study constructed a luciferase reporter vector containing the 3′-UTR of ATG2B. The results indicated that miR-143-3p mimic significantly inhibited the luciferase activity of pGL3-ATG2B-3′-UTR-WT compared with the control group, whereas miR-143-4p mimic displayed no significant effect on pGL3-ATG2B-3′-UTR-mut compared with the control group (Fig. 4B). Furthermore, miR-143-3p overexpression significantly decreased the protein expression levels of ATG2B in ESCs compared with the control group (Fig. 4C). In addition, compared with the control group, miR-143-3p overexpression significantly inhibited ESC proliferation and invasion, whereas ATG2B overexpression significantly alleviated miR-143-3p mimic-mediated effects (Fig. 4D-F). The present study also assessed the expression levels of ATG2B in ESCs derived from the EM mouse model. The results indicated that ATG2B expression levels were significantly decreased in ESCs compared with NESCs (Fig. 4G). Collectively, the aforementioned results suggested that miR-143-3p overexpression inhibited EM progression by repressing ATG2B expression, thus inactivating autophagy.
Discussion
In the present study, the effect of miRNAs on regulating EM progression was investigated. miR-143-3p expression was significantly upregulated in ESCs compared with NESCs. miR-143-3p overexpression markedly decreased ESC cell proliferation and invasion compared with the control group. In addition, compared with the control group, miR-143-3p overexpression significantly decreased LC3-II expression levels and increased p62 expression levels, indicating that miR-143-3p may serve as an inhibitor of autophagy activation in ESCs. Furthermore, the present study verified that miR-143-3p directly targeted the 3′-UTR of ATG2B, and miR-143-3p overexpression significantly decreased the protein expression levels of ATG2B compared with the control group. ATG2B overexpression partially reversed miR-143-3p mimic-mediated effects on ESC proliferation and invasion.
As a gynecological disease, EM frequently results in infertility and chronic pelvic pain (31). Immune disorders affect ectopic endometrial lesions. Following dysfunction of the immune system, the number of immune cells increases, and various growth factors, cytokines, non-specific immunoglobulins and proinflammatory mediators are present in the peritoneum (32–34). A theory of EM is the local hypoxia microenvironment, whereby the first indicator of EM is the topical shifted hypoxic microenvironment, when the shed endometrial fragments retrograde to the peritoneal cavity (35–37).
Autophagy is a highly conserved cellular process, which is activated by various factors, such as hypoxia (38). For the past few years, research has focused on autophagy progression in tumorigenesis (39,40). EM displays certain biological characteristics of tumor diseases, such as hyperproliferation and metastasis (41) Increasing evidence has demonstrated that autophagy is dysregulated in the uterine horns and eutopic endometria of EM model mice (42), and is correlated with endometrial regulation and the pathophysiology of EM (43,44). However, the role of autophagy in EM is controversial, thus whether autophagy in EM is beneficial or detrimental remains to be elucidated. Several studies have demonstrated that the expression levels of autophagy-related genes (for example, Beclin-1 and LC3-II) are markedly decreased and autophagy activation is downregulated in endometrial stromal cells of patients with EM compared with healthy controls (45–47). Functionally, autophagy inhibition contributes to endometrial cell invasion, whereas autophagy activation represses ESC proliferation, colony formation and invasion (48). By contrast, other studies have reported that the expression levels of autophagy-associated genes [for example, Beclin-1, autophagy-related (ATG)14, ATG7 and LC3-II] are increased and autophagy activation is upregulated in ESCs in EM (17). Liu et al (44) further demonstrated that HIF-1α facilitates endometrial stromal cell invasion by activating autophagy, whereas autophagy inhibition alleviates hypoxia-induced cell invasion. The present study demonstrated that miR-143-3p overexpression inhibited ESC proliferation and invasion by regulating ATG2B in EM, whereas ATG2B overexpression partially reversed miR-143-3p overexpression-mediated effects, indicating that autophagy may be beneficial in EM. Although the effect of miR-143-3p on inhibiting autophagy has been verified in Crohn's disease (27), the present study aimed to investigate the association of miR-143-3p with ATG2B and autophagy in EM. The present study further clarified the function of miR-143-3p in EM. However, a key limitation of the present study was that the role of miR-143-3p in NESC autophagy was not investigated.
A previous study demonstrated that miRNAs are crucial modulators in the occurrence and development of various diseases (49–52). Several studies have implicated that the aberrant expression of miRNAs is a potential regulator of EM pathogenesis (11,23). It has been verified that the expression levels of HIF-1α and VEGF were elevated in ectopic endometrial tissues compared with eutopic endometrial tissues, which was induced by hypoxia stress. miR-17-5p/20a is a regulator of HIF-1α and VEGF via directly targeting the 3′-UTR, and further regulates downstream hypoxic stress-associated proteins (53–55). The present study investigated the function of miR-143-3p in EM and demonstrated that miR-143-3p overexpression notably inhibited ESC proliferation and invasion in EM compared with the control group. In hepatocellular carcinoma, upregulated miR-143-3p promotes cancer cell migration and invasion by repressing fibronectin type III domain containing 3B (56). Therefore, the results of the present study and the aforementioned previous studies suggested that miR-143-3p may serve various regulatory roles in different biological processes or diseases.
In summary, miR-143-3p was significantly upregulated in ESCs compared with NESCs. miR-143-3p regulated the phenotype of EM, suppressing autophagy activation in ESCs, and inhibiting ESC proliferation and invasion by directly targeting ATG2B. The results of the present study may further the current understanding of the role of miR-143-3p in the pathogenesis of EM.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- miRNAs
microRNAs
- EM
endometriosis
- ESCs
endometriotic stromal cells
- NESCs
normal endometrial stromal cells
- ATG2B
autophagy-related 2B
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant no. 81704108).
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81704108).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HY and LQ made substantial contributions to the conception and design of the present study. TH, PH, CQ and LQ collected, analyzed and interpreted the data. PH, CQ and LQ drafted the work and made critical modifications to the manuscript. All authors agreed to be accountable for the work in ensuring that questions related to the integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Protection of Human Subjects Committee of Shanghai Shuguang Hospital and the Ethics Committee for Animal Experimentation of Shanghai Shuguang Hospital (approval no. 2018-618-47-01). All patients or their legal guardians provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Eskenazi B, Warner ML. Epidemiology of endometriosis. Obstet Gynecol Clin North Am. 1997;24:235–258. doi: 10.1016/S0889-8545(05)70302-8. [DOI] [PubMed] [Google Scholar]
- 2.Simoens S, Dunselman G, Dirksen C, Hummelshoj L, Bokor A, Brandes I, Brodszky V, Canis M, Colombo GL, DeLeire T, et al. The burden of endometriosis: Costs and quality of life of women with endometriosis and treated in referral centres. Hum Reprod. 2012;27:1292–1299. doi: 10.1093/humrep/des073. [DOI] [PubMed] [Google Scholar]
- 3.Shafrir AL, Farland LV, Shah DK, Harris HR, Kvaskoff M, Zondervan K, Missmer SA. Risk for and consequences of endometriosis: A critical epidemiologic review. Best Pract Res Clin Obstet Gynaecol. 2018;51:1–15. doi: 10.1016/j.bpobgyn.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Friend DR. Drug delivery for the treatment of endometriosis and uterine fibroids. Drug Deliv Transl Res. 2017;7:829–839. doi: 10.1007/s13346-017-0423-2. [DOI] [PubMed] [Google Scholar]
- 5.Vishnoi A, Rani S. miRNA biogenesis and regulation of diseases: An overview. Methods Mol Biol. 2017;1509:1–10. doi: 10.1007/978-1-4939-6524-3_1. [DOI] [PubMed] [Google Scholar]
- 6.Arora S, Rana R, Chhabra A, Jaiswal A, Rani V. miRNA-transcription factor interactions: A combinatorial regulation of gene expression. Mol Genet Genomics. 2013;288:77–87. doi: 10.1007/s00438-013-0734-z. [DOI] [PubMed] [Google Scholar]
- 7.Filigheddu N, Gregnanin I, Porporato PE, Surico D, Perego B, Galli L, Patrignani C, Graziani A, Surico N. Differential expression of microRNAs between eutopic and ectopic endometrium in ovarian endometriosis. J Biomed Biotechnol. 2010;2010:369549. doi: 10.1155/2010/369549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao L, Gu C, Ye M, Zhang Z, Li L, Fan W, Meng Y. Integration analysis of microRNA and mRNA paired expression profiling identifies deregulated microRNA-transcription factor-gene regulatory networks in ovarian endometriosis. Reprod Biol Endocrinol. 2018;16:4. doi: 10.1186/s12958-017-0319-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H, Shen H, Xu J, Zhao S, Yao S, Jiang N. MiR-143-3p suppresses the progression of ovarian cancer. Am J Transl Res. 2018;10:866–874. [PMC free article] [PubMed] [Google Scholar]
- 10.Deng L, Blanco FJ, Stevens H, Lu R, Caudrillier A, McBride M, McClure JD, Grant J, Thomas M, Frid M, et al. MicroRNA-143 Activation regulates smooth muscle and endothelial cell crosstalk in pulmonary arterial hypertension. Circ Res. 2015;117:870–883. doi: 10.1161/CIRCRESAHA.115.306806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2010;16:142–165. doi: 10.1093/humupd/dmp034. [DOI] [PubMed] [Google Scholar]
- 12.Zheng B, Xue X, Zhao Y, Chen J, Xu CY, Duan P. The differential expression of microRNA-143,145 in endometriosis. Iran J Reprod Med. 2014;12:555–560. [PMC free article] [PubMed] [Google Scholar]
- 13.Cosar E, Mamillapalli R, Ersoy GS, Cho S, Seifer B, Taylor HS. Serum microRNAs as diagnostic markers of endometriosis: A comprehensive array-based analysis. Fertil Steril. 2016;106:402–409. doi: 10.1016/j.fertnstert.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Bravo-San Pedro JM, Kroemer G, Galluzzi L. Autophagy and mitophagy in cardiovascular disease. Circ Res. 2017;120:1812–1824. doi: 10.1161/CIRCRESAHA.117.311082. [DOI] [PubMed] [Google Scholar]
- 15.Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, Füllgrabe J, Jackson A, Jimenez Sanchez M, Karabiyik C, et al. Autophagy and neurodegeneration: Pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93:1015–1034. doi: 10.1016/j.neuron.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 16.White E, Mehnert JM, Chan CS. Autophagy, metabolism, and cancer. Clin Cancer Res. 2015;21:5037–5046. doi: 10.1158/1078-0432.CCR-15-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allavena G, Carrarelli P, Del Bello B, Luisi S, Petraglia F, Maellaro E. Autophagy is upregulated in ovarian endometriosis: A possible interplay with p53 and heme oxygenase-1. Fertil Steril. 2015;103:1244–51.e1. doi: 10.1016/j.fertnstert.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Neshkov NS. Use of ultrasound in therapy of neuroreceptor forms of impotence. Vopr Kurortol Fizioter Lech Fiz Kult. 1970;35:270. (In Russian) [PubMed] [Google Scholar]
- 19.Shi YL, Luo XZ, Zhu XY, Hua KQ, Zhu Y, Li DJ. Effects of combined 17beta-estradiol with TCDD on secretion of chemokine IL-8 and expression of its receptor CXCR1 in endometriotic focus-associated cells in co-culture. Hum Reprod. 2006;21:870–879. doi: 10.1093/humrep/dei414. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Mari-Alexandre J, Sanchez-Izquierdo D, Gilabert-Estelles J, Barcelo-Molina M, Braza-Boils A, Sandoval J. miRNAs regulation and its role as biomarkers in endometriosis. Int J Mol Sci. 2016;17:93. doi: 10.3390/ijms17010093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto M, Nasu K, Abe W, Aoyagi Y, Kawano Y, Kai K, Moriyama M, Narahara H. Enhanced miR-210 expression promotes the pathogenesis of endometriosis through activation of signal transducer and activator of transcription 3. Hum Reprod. 2015;30:632–641. doi: 10.1093/humrep/deu332. [DOI] [PubMed] [Google Scholar]
- 23.Abdel-Rasheed M., Nour Eldeen G., Mahmoud M., El Hefnawi M., Abu-Shahba N., Reda M., Elsetohy K., Nabil M., Elnoury, et al. MicroRNA expression analysis in endometriotic serum treated mesenchymal stem cells. EXCLI journal. 2017;16:852–867. doi: 10.17179/excli2017-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, Print CG, Hull LM. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23:265–275. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He R, Liu X, Zhang J, Wang Z, Wang W, Fu L, Fan Y, Sun S, Cao Y, Zhan L, et al. NLRC5 inhibits inflammation of secretory phase ectopic endometrial stromal cells by up-regulating autophagy in ovarian endometriosis. Front Pharmacol. 2020;11:1281. doi: 10.3389/fphar.2020.01281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding Y, Zhu Q, He Y, Lu Y, Wang Y, Qi J, Wu H, Xu R, Li J, Li X, et al. Induction of autophagy by Beclin-1 in granulosa cells contributes to follicular progesterone elevation in ovarian endometriosis. Transl Res. 2021;227:15–29. doi: 10.1016/j.trsl.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Ma W, Ding F, Wang X, Huang Q, Zhang L, Bi C, Hua B, Yuan Y, Han Z, Jin M, et al. By Targeting Atg7 MicroRNA-143 mediates oxidative stress-induced autophagy of c-Kit+ mouse cardiac progenitor cells. EBioMedicine. 2018;32:182–191. doi: 10.1016/j.ebiom.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin XT, Zheng XB, Fan DJ, Yao QQ, Hu JC, Lian L, Wu XJ, Lan P, He XS. MicroRNA-143 targets ATG2B to inhibit autophagy and increase inflammatory responses in Crohn's disease. Inflamm Bowel Dis. 2018;24:781–791. doi: 10.1093/ibd/izx075. [DOI] [PubMed] [Google Scholar]
- 29.Wei J, Ma Z, Li Y, Zhao B, Wang D, Jin Y, Jin Y. miR-143 inhibits cell proliferation by targeting autophagy-related 2B in non-small cell lung cancer H1299 cells. Mol Med Rep. 2015;11:571–576. doi: 10.3892/mmr.2014.2675. [DOI] [PubMed] [Google Scholar]
- 30.Gao S, Wang K, Wang X. miR-375 targeting autophagy-related 2B (ATG2B) suppresses autophagy and tumorigenesis in cisplatin-resistant osteosarcoma cells. Neoplasma. 2020;67:724–734. doi: 10.4149/neo_2020_190423N366. [DOI] [PubMed] [Google Scholar]
- 31.Tanbo T, Fedorcsak P. Endometriosis-associated infertility: Aspects of pathophysiological mechanisms and treatment options. Acta Obstet Gynecol Scand. 2017;96:659–667. doi: 10.1111/aogs.13082. [DOI] [PubMed] [Google Scholar]
- 32.Ahn SH, Monsanto SP, Miller C, Singh SS, Thomas R, Tayade C. Pathophysiology and immune dysfunction in endometriosis. BioMed Res Int. 2015;2015:795976. doi: 10.1155/2015/795976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Králíčková M, Vetvicka V. Immunological aspects of endometriosis: A review. Ann Transl Med. 2015;3:153. doi: 10.3978/j.issn.2305-5839.2015.06.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olovsson M. Immunological aspects of endometriosis: An update. Am J Reprod Immunol. 2011;66(Suppl 1):101–104. doi: 10.1111/j.1600-0897.2011.01045.x. [DOI] [PubMed] [Google Scholar]
- 35.Filippi I, Carrarelli P, Luisi S, Batteux F, Chapron C, Naldini A, Petraglia F. Different expression of hypoxic and angiogenic factors in human endometriotic lesions. Reprod Sci. 2016;23:492–497. doi: 10.1177/1933719115607978. [DOI] [PubMed] [Google Scholar]
- 36.Zhan L, Wang W, Zhang Y, Song E, Fan Y, Wei B. Hypoxia-inducible factor-1alpha: A promising therapeutic target in endometriosis. Biochimie. 2016;123:130–137. doi: 10.1016/j.biochi.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Tsuzuki T, Okada H, Cho H, Tsuji S, Nishigaki A, Yasuda K, Kanzaki H. Hypoxic stress simultaneously stimulates vascular endothelial growth factor via hypoxia-inducible factor-1α and inhibits stromal cell-derived factor-1 in human endometrial stromal cells. Hum Reprod. 2012;27:523–530. doi: 10.1093/humrep/der405. [DOI] [PubMed] [Google Scholar]
- 38.Daskalaki I, Gkikas I, Tavernarakis N. Hypoxia and selective autophagy in cancer development and therapy. Front Cell Dev Biol. 2018;6:104. doi: 10.3389/fcell.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W, Meng Y, Zong C, Zhang S, Wei L. Autophagy and tumorigenesis. Adv Exp Med Biol. 2020;1207:275–299. doi: 10.1007/978-981-15-4272-5_20. [DOI] [PubMed] [Google Scholar]
- 40.White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leyendecker G, Kunz G, Noe M, Herbertz M, Mall G. Endometriosis: A dysfunction and disease of the archimetra. Hum Reprod Update. 1998;4:752–762. doi: 10.1093/humupd/4.5.752. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz A, Rockfield S, Taran N, Haller E, Engelman RW, Flores I, Panina-Bordignon P, Nanjundan M. Effect of hydroxychloroquine and characterization of autophagy in a mouse model of endometriosis. Cell Death Dis. 2016;7:e2059. doi: 10.1038/cddis.2015.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhan L, Li J, Wei B. Autophagy in endometriosis: Friend or foe? Biochem Biophys Res Commun. 2018;495:60–63. doi: 10.1016/j.bbrc.2017.10.145. [DOI] [PubMed] [Google Scholar]
- 44.Liu H, Zhang Z, Xiong W, Zhang L, Xiong Y, Li N, He H, Du Y, Liu Y. Hypoxia-inducible factor-1alpha promotes endometrial stromal cells migration and invasion by upregulating autophagy in endometriosis. Reproduction. 2017;153:809–820. doi: 10.1530/REP-16-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pei T, Huang X, Long Y, Duan C, Liu T, Li Y, Huang W. Increased expression of YAP is associated with decreased cell autophagy in the eutopic endometrial stromal cells of endometriosis. Mol Cell Endocrinol. 2019;491:110432. doi: 10.1016/j.mce.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 46.Sui X, Li Y, Sun Y, Li C, Li X, Zhang G. Expression and significance of autophagy genes LC3, Beclin1 and MMP-2 in endometriosis. Exp Ther Med. 2018;16:1958–1962. doi: 10.3892/etm.2018.6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mei J, Zhou WJ, Zhu XY, Lu H, Wu K, Yang HL, Fu Q, Wei CY, Chang KK, Jin LP, et al. Suppression of autophagy and HCK signaling promotes PTGS2high FCGR3- NK cell differentiation triggered by ectopic endometrial stromal cells. Autophagy. 2018;14:1376–1397. doi: 10.1080/15548627.2018.1476809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo X, Cheng W, Wang S, Chen Z, Tan J. Autophagy suppresses invasiveness of endometrial cells through reduction of Fascin-1. BioMed Res Int. 2018;2018:8615435. doi: 10.1155/2018/8615435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 50.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 51.Stolzenburg LR, Harris A. The role of microRNAs in chronic respiratory disease: Recent insights. Biol Chem. 2018;399:219–234. doi: 10.1515/hsz-2017-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soroosh A, Koutsioumpa M, Pothoulakis C, Iliopoulos D. Functional role and therapeutic targeting of microRNAs in inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol. 2018;314:G256–G262. doi: 10.1152/ajpgi.00268.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu MH, Chen KF, Lin SC, Lgu CW, Tsai SJ. Aberrant expression of leptin in human endometriotic stromal cells is induced by elevated levels of hypoxia inducible factor-1alpha. Am J Pathol. 2007;170:590–598. doi: 10.2353/ajpath.2007.060477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsiao KY, Lin SC, Wu MH, Tsai SJ. Pathological functions of hypoxia in endometriosis. Front Biosci (Elite Ed) 2015;7:309–321. doi: 10.2741/E736. [DOI] [PubMed] [Google Scholar]
- 55.Donnez J, Smoes P, Gillerot S, Casanas-Roux F, Nisolle M. Vascular endothelial growth factor (VEGF) in endometriosis. Hum Reprod. 1998;13:1686–1690. doi: 10.1093/humrep/13.6.1686. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–499. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.