Abstract
Circular RNAs (circRNAs) are a class of non-coding RNAs formed by covalently closed loops through back-splicing and exon-skipping. circRNAs have been confirmed to play a vital role in various biological functions, acting as microRNA sponges and reservoirs, as well as combining with RNA-binding proteins during the progression of multiple cancer types. Therefore, the present review evaluated recent research articles in PubMed that were published between November 2017 and September 2020. Key word search strings included: ‘Circular RNA (circRNA) AND bladder cancer (BC)’, ‘circular RNA (circRNA) AND prostate cancer (PCa)’ and ‘circular RNA (circRNA) AND renal cell cancer (RCC)’. In total, >58 circRNAs were found to be implicated in urological cancers, with several of the circRNAs targeting common carcinogenic pathways, such as the AKT, TGF-β, MAPK, VEGF and even metabolic pathways. circRNAs are important modulators of BC, PCa and RCC. circRNAs are functionally implicated in the pathogenesis of these cancer types, and have been found to act as biomarkers for the diagnosis and prognosis of urological cancer. However, to the best of our knowledge, the functions of circRNAs in tumors of the urinary system remain largely unknown and require further research.
Keywords: circular RNA, bladder cancer, prostate cancer, renal cell cancer
1. Introduction
Cancer is one of the leading cause of mortality worldwide; however, with incidences of cardiovascular disease decreasing in several countries, cancer is most likely to soon become the leading cause (1). According to estimates, there were 17.0 million new cases and 9.5 million cancer-related deaths worldwide in 2018, with bladder cancer (BC), prostate cancer (PCa) and renal cell cancer (RCC) ranking 12th, 3rd and 16th, respectively, in terms of global incidence (2). Surgical resection is the most common treatment method for these diseases, but its effect remains unsatisfactory. For example, the recurrence rates of BC and RCC are 50 and 40%, respectively (3,4). Furthermore, although 80% of newly diagnosed PCa cases are sensitive to androgen deprivation therapy, >50% of patients with PCa develop recurrence, infiltration or metastasis, or progress to hormone refractory PCa (5). Therefore, determining a new preventive and therapeutic target for these malignant tumors is an urgent requirement.
Circular RNAs (circRNAs), a class of non-coding RNAs ubiquitous in the cytoplasm of various eukaryotic cells, commonly originate from back-splicing events of exons or introns (6). ‘Exon skipping’ and ‘direct back-splicing’ are the two mechanisms that lead to the formation of exonic or exon-intron circRNAs, in which the 3′ and 5′ ends of circRNAs are covalently joined together to form single-stranded continuous loop structures, and can be regulated by certain splicing factors (Fig. 1). The outstanding characteristics of circRNAs include their marked stability, high abundance, evolutionary conservation and tissue-specific expression (7). In addition, circRNAs are different from microRNAs (miRNAs/miR) and long non-coding RNAs (lncRNAs), as they lack a 5′cap and a 3′polyadenylated tail, and they have the ability to encode regulatory peptides (8). To date, ~15,000 circRNAs have been identified in both humans and mice, representing 15 and 40% of the total circRNAs in humans and mice, respectively (9).
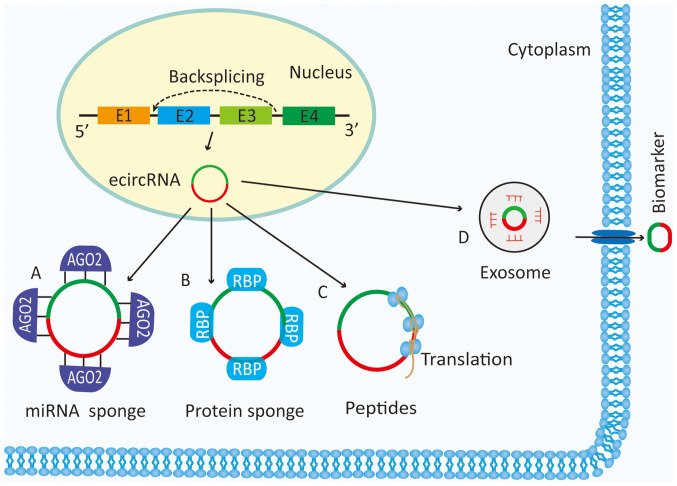
Figure 1.
Functions of circRNAs. (A) circRNAs can act as miRNA sponges and subsequently regulate the expression of relevant target genes. (B) circRNAs can bind to several proteins and mediate their actions. (C) circRNAs can be translated into peptides or proteins. (D) circRNAs exist in the serum and other bodily fluids, and can function as molecular biomarkers for the diagnosis and treatment of cancer. ecircRNA, exonic circRNA; circRNA, circular RNA; AGO2, Argonaute-2; RBP, RNA-binding protein.
One of the main functions of circRNAs is to sponge miRNAs that regulate the function of target genes, with common features such as being derived from one or more exons of known protein-coding genes and being formed by a back-splice event. In addition, these circRNAs are predominantly localized in the cytoplasm, occupying the same space as miRNAs (10). circRNAs also act as a reservoir for miRNAs, which means that they can increase the availability of miRNAs for binding to and inhibiting their target mRNAs (Fig. 1) (11). Another study confirmed that circRNAs play crucial roles in tumor growth, metastasis, epithelial-mesenchymal transition (EMT) and treatment resistance (12). However, the function of circRNAs in tumors of the urinary system remains unclear. The aim of the present review was to illustrate the roles of circRNAs in BC, PCa and RCC.
2. circRNAs in BC
circRNAs act as oncogenes in BC
circRNA-miRNA-mRNA interaction networks, as a major function of circRNAs, have been associated with cell signaling transduction in BC. For instance, the expression levels of TGF-β2, Smad3 and phosphorylated (p)-Smad3 could be increased by circ_0005777 (circRIP2), which reverses the miR-1305-induced suppression of BC progression (Fig. 2) (13). AKT signaling is another regulator of cancer metastasis. The expression of p-AKT/PI3K was increased by circKIF4A (circ_0007255), which promoted BC growth and metastasis in vitro and in vivo. circKIF4A could act as a sponge for miR-375 and miR-1231 to enhance the level of Notch2, which has been found to play an oncogenic role in BC (14). Likewise, Notch1, NICD-1 and HES1, all downstream genes of the Notch signaling pathway, have been shown to be highly expressed in BCa and repressed by circ_0008532 and MTGR1. In addition, circ_0008532 was shown to increase the progression of BC by regulating MTGR1 expression, an effect that could be reversed by miR-155-5p/miR-330-5p. circ_0008532 is derived from the MTGR1 gene (15). circ_0068871 produced at the fibroblast growth factor receptor 3 gene (FGFR3) was highly expressed in BC, activated p-STAT3 and facilitated tumor development; however, its effects could be reversed by miR-181a-5p (16). The incidence and metastasis of BCa is 4-fold higher in men compared with that in women, indicating that sex steroid pathways play a vital role in BC progression (2). Indeed, estrogen receptor α (ERα) exhibits low expression in BC, decreases the expression of epidermal growth factor receptor, and is mediated by circ_0023642 and miR-490-5p. Moreover, circ_0023642 was found to promote the metastasis of BC, but its effect could be inhibited by ERα (17).
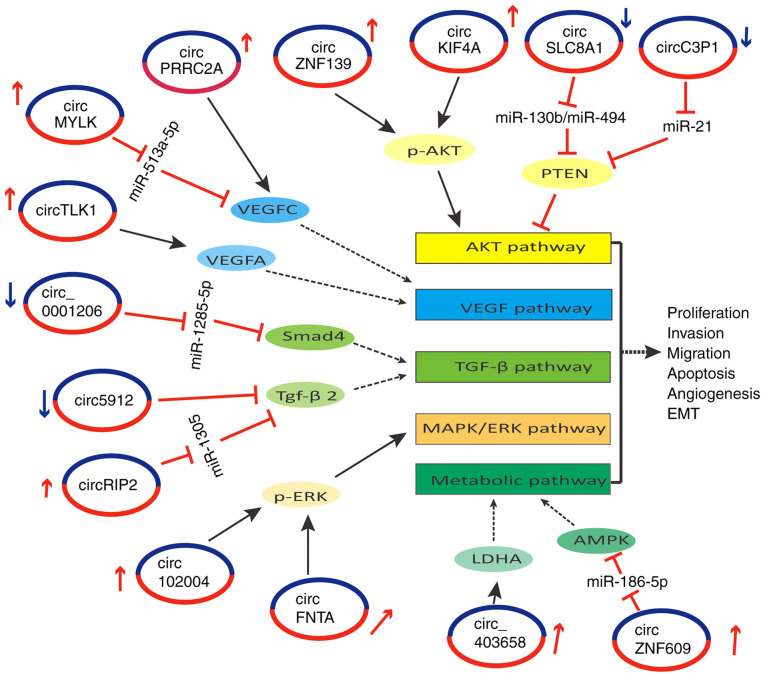
Figure 2.
Representative diagram of circRNAs that interact with the circRNA-miRNA-mRNA regulatory network/RBPs and regulate oncogene or tumor suppressor signaling pathways. The diagram mainly shows that circZNF139 and circKIF4A activate the AKT pathway through p-AKT in BC, and that circFNTA, circ_403658 and circRIP2 activate the MAPK, metabolic and TGF-β signaling pathways through p-ERK, LDHA and miR-1305/TGF-β2, respectively, in BC. Furthermore, circ102004 and circZNF609 activate the MAPK and metabolic pathways through p-EKT and miR-186-5p/AMPK, respectively, in PCa. Moreover, circMYLK, circTLK1 and circPRRC2A activate the VEGF pathway through miR-513a-5p/VEGFC, VEGFA and VEGFC, respectively, in RCC. By contrast, circSLC8A1 and circ5912 deactivate the AKT and TGF-β pathways through miR-130b/miR-494/PTEN and TGF-β2, respectively, in BC. In addition, circ_0001206 deactivates the TGF-β pathway through miR-1285-5p/Smad4 in PCa, and circC3P1 deactivates the AKT pathway through miR-21/PTEN in RCC, which suggests that the circRNA-miRNA-mRNA interaction networks and RBPs that sponge circRNA serve a critical role in BC, PCa and RCC progression through signaling pathways. circRNA, circular RNA; EMT, epithelial-mesenchymal transition; RBP, RNA-binding protein; PCa, prostate cancer; BC, bladder cancer; RCC, renal cell cancer; miR, microRNA.
Unlike healthy cells, tumor cells do not undergo apoptosis when mitosis arrests, thus the dysfunction of mitosis and apoptosis participates in tumor progression (18). For instance, kinesin family member 2C (KIF2C), a mitotic centromere-associated kinesin, is highly expressed in BC, is upregulated by circRGNEF (circ_0072995) and is downregulated by miR-548; it also enhances tumor progression following circRGNEF upregulation or miR-548 downregulation, and circ_0072995 is derived from the RGNEF gene (Table I) (19). Another study demonstrated that insulin-like growth factor binding protein 2 (IGFBP2), a key anti-apoptotic regulator, is upregulated by circVANGL1 or inhibited by miR-1184 in BC. In addition, circVANGL1 facilitates tumor progression by upregulating IGFBP2, while its effect could be reversed by miR-1184; circVANGL1 is derived from the VANGL1 gene (20). Notably, BC stem cells (BCSCs), which have self-renewal and differentiation capacities, may contribute to the tumor initiation, metastasis, recurrence and drug resistance of BC (21). For example, circ_103809, which is derived from the zinc finger RNA-binding protein gene, is highly expressed in BCSCs and has been shown to increase the cell oncosphere formation and aggressiveness of BC, and decrease the expression of miR-511 (22).
Table I.
circRNAs act as oncogenes in bladder, prostate and renal cancer: Summary of circRNAs, including their ID number, host genes, mechanisms, functions and clinical applications.
| A, Bladder cancer | |||||
|---|---|---|---|---|---|
| circRNAs | Host gene | Biological function | Target | Clinical applicant | (Refs.) |
| circ_403658 | ZNF292 | Promote tumor growth, increase lactate production, LDH activity, ATP production and glucose uptake of cell | LDHA, VEGFR and VEGF | Target | (10) |
| circPRMT5 (circ_101320) | PRMT5 | Enhance tumor metastasis, increase expression of Vimentin, SNAIL1 | miR-30c | DFS, target | (65) |
| cTFRC (circ_0001445) | TFRC | Promote cell proliferation and invasion | E-cadherin, miR-107 | OS, target | (66) |
| circRIP2 (circ_0005777) | RIP2 | Promote tumor growth, metastasis and increase expression of TGF-β2, N-cadherin, Vimentin, smad3 and p-smad3 | miR-1305, TGF-β2 and smad3 | (13) | |
| circRGNEF (circ_0072995) | RGNEF | Enhance tumor and metastasis | miR-548 and KIF2C | (19) | |
| circVANGL1 | VANGL1 | Promote tumor growth | miR-1184 and IGFBP2 | (20) | |
| circFNTA (circ_0084171) | FNTA | Promote tumor metastasis and decrease cisplatin chemosensitivity, increases the expression of p-ERK1/2 and p-MEK1/2 | miR-370-3p and KRAS | (70) | |
| circ_0023642 | UVARG | Promote tumor growth and metastasis, increase the expression of EGFR | miR-490-5p and ERα | (17) | |
| circ_103809 | ZFR | Increase BCSC oncosphere formation, migration and invasion | miR-511 | (22) | |
| circELP3 | ELP3 | Promote tumor growth and cisplatin resistance | hypoxia | (71) | |
| circ_0068871 | FGFR3 | Enhance tumor growth and the expression of p-STAT3 FGFR3 | miR-181a-5p and | (16) | |
| circZNF139 (circ_0001727) | ZNF139 | Increase cell proliferation, migration and invasion. Promote the expression level of p-AKT and PI3K | p-AKT and PI3K | (23) | |
| circKIF4A (circ_0007255) | KIF4A | Promote tumor growth and metastasis, enhance expression of p-AKT and PI3K | miR-375/1231 and NOTCH2 | (14) | |
| circ_0008532 | MTGR1 | Promote tumor growth and metastasis, reduce the expression of Notch1, NICD-1 and HES1 | miR-155-5p/330-5p | (15) | |
| circ_0003221 (circPTK2) | PTK2 | Promote cell proliferation and migration | Biomarker | (61) | |
| B, Prostate cancer | |||||
| circRNAs | Host gene | Biological function | Target | Clinical applicant | (Refs.) |
| circHIPK3 (circ_0000284) | HIPK3 | Increase cell proliferation and invasion, and inhibit apoptosis | miRNA-338-3p and ADAM17 | (5,42) | |
| circ0005276 | XIAP | Increase tumor growth and increase expression of N-cadherin | XIAP and FUS | (41) | |
| circ102004 | USP22 | Increase cell proliferation, invasion and migration, improve expression of p-ERK, p-AKT, p-JNK, JNK, β-catenin, BCL2 and MMP2 | (43) | ||
| circABCC4 (circ_0030586)) | ABCC4 | Increase cell proliferation, invasion and migration | miR-1182 and FOXP4 | (39) | |
| circFOXO3 (circ_0006404) | FOXO3 | Increase cell proliferation and inhibit apoptosis | miR-29a-3p and SLC25A15 | (63) | |
| circHIPK3 (circ_0000284) | HIPK3 | Increase cell proliferation and invasion, and inhibit apoptosis | miRNA-338-3p and | (5,42) | |
| circFMN2 (circ_0005100) | FMN2 | Increase tumor growth and reduce the expression of E-cadherin | miR-1238 and LHX2 | (64) | |
| circZNF609 | ZNF609 | Increase cell proliferation, invasion and migration, improve ratio of p/t-AMPK and the level of YAP1 | miR-186-5p | (38) | |
| circ_0044516 | COL1A1 | Increase cell proliferation, invasion and migration | miR-29a-3p | Diagnosis | (62) |
| C, Renal cancer | |||||
| circRNAs | Host gene | Biological function | Target | Clinical applicant | (Refs.) |
| circPCNXL2 (circ_406752) | PCNXL2 | Enhance tumor growth | miR-153 and ZEB2 | Target and diagnosis | (49) |
| circPRRC2A (circ_406752) | PRRC2A | Enhance tumor growth, increase expression of N-cadherin, Snail and vimentin, VEGFC | miR-514a-5p/6776-5p and TRPM3 | OS | (50) |
| circMYLK (circ_0141940) | MYLK | Enhance tumor growth and metastasis, increase expression of VEGFC | miR-513a-5p and VEGFC | (51) | |
| circTLK1 (circ_0004442) | TLK1 | Improve tumor growth and metastasis, increase expression of N-cadherin, vimentin and VEGFA | miR-136-5p and CBX4 | (52) | |
| circ_001895 | CTBP1 | Increase tumor growth, improve expression of N-cadherin | miR-296-5p and SOX12 | OS | (53) |
| circ-EGLN3 (circ_0031594) | EGLN3 | Increase cell proliferation, invasion and inhibit apoptosis | miR-1299 and IRF7 | OS, biomarker/target | (55,60) |
| circ_0039569 | CCL2 | Promote cell proliferation, migration and invasion | miR-34a-5p and CCL22 | Biomarker | (72) |
| circ_0085576 | ASAP1 | Enhance tumor growth and metastasis | miR-498 and YAP1 | OS, DFS | (67) |
circRNA, circular RNA; BCSC, bladder cancer stem cell; OS, overall survival; DFS, disease-free survival; miR, microRNA.
circRNAs can bind to numerous RNA-binding proteins (RBPs), acting as protein sponges or decoys to regulate protein functions. Hypoxia-inducible factor-1α enhances the expression of circ_403658, and promotes BC growth in vitro and in vivo. In addition, circ_403658 improves the expression of L-lactate dehydrogenase A chain (LDHA), vascular endothelial growth factor and VEGF receptor in BC, and increases lactate production, LDH activity, ATP production and glucose uptake in tumor cells. LDHA serves as a key checkpoint of glycolysis (Fig. 2) (10). In addition, as a member of the zinc finger protein (ZNF) family, circZNF139 (circ_0001727) is derived from the ZNF139 gene, which is highly expressed in BC, and promotes tumor cell development by improving the level of p-AKT and PI3K (23).
circRNAs act as tumor suppressors in BC
The abnormal activation of transcription factors promotes the proliferation and differentiation of tumor cells (24). For instance, ΔNP63, a transcription factor of the p53 family, is upregulated by circ family with sequence similarity 114 member A2 (FAM114A2), thus suppressing the miR-762-induced growth of urothelial carcinoma of the bladder. circFAM114A2 (circ_0001546) is derived from the FAM114A2 gene (25). circ activin A receptor type 2A attenuates tumor growth and aggressiveness in vitro and in vivo by improving the expression of eye absent 4 (EYA4), and reduces miR-626-induced BC progression. EYA4 is a transcription factor (26). A different study illustrated that Krüppel-like factor (KLF)9 and KLF10, which are zinc finger transcription factors, repress the progress of miR-636- and miR-570-3p-induced BC through the upregulation of circ protein tyrosine phosphatase receptor type A and circFUT8 (circ_0003028), respectively (27,28). c-Myc is a famous oncogene and transcription factor, that has been reported to participate in the progression of bladder cancer. For example, c-Myc promotes the growth and mobility of BCSCs, an effect that could be improved by miR-147 or repressed by circ_0068307; circ_0068307 is derived from the ALG3 α-1,3-mannosyltransferase gene (29). In addition, c-Myc sabotages the G0/G1 phase of circ chromodomain Y-like (CDYL)-induced cell cycle arrest, consequently facilitating the progression of BC; circCDYL is generated from the CDYL gene (Table II) (7).
Table II.
circRNAs act as tumor suppressors in bladder, prostate and renal cancer: Summary of circRNA names, including their ID number, host genes, mechanisms, functions and clinical applications.
| A, Bladder cancer | |||||
|---|---|---|---|---|---|
| circRNAs | Host gene | Biological function | Target | Clinical applicant | (Refs.) |
| circFAM114A2 (circ_0001546) | FAM114A2 | Reduce tumor growth | miR-762 and ∆NP63 | Biomarker and target | (25) |
| circACVR2A | ACVR2A | Reduce tumor growth and metastasis | miR-626 and EYA4 | OS and target | (26) |
| circPTPRA (circ_102984/0006117) | PTPRA | Reduce tumor growth | KLF9 and miR-636 | Biomarker and target | (27) |
| circFUT8 (circ_0003028) | FUT8 | Suppress tumor metastasis and inhibit expression of Slug | miR-570-3p and KLF10 | (28) | |
| circ_0071662 | TPPP | Reduce cell proliferation and invasion, and deactivate AKT pathway | miR-146-3p, HPGD and NF2 | Biomarker | (30) |
| circSLC8A1 (circ_0000994) | SLC8A1 | Reduce tumor growth and metastasis, inhibit expression of p-Akt and MMP-9 | miR-130b/494 and PTEN | Biomarker and target | (31) |
| circ5912 (circ_0005912) | FIP1L1 | Increase tumor growth and metastasis, reverses TGF-β2-induced EMT | (37) | ||
| circCdr1as (CiRS-7) | LINC00632 | Reduce tumor growth and increase cisplatin chemosensitivity | miR-1270 and APAF1 | Biomarker | (74) |
| circ_0006260 | SLC41A2 | Reduce tumor growth | miR-653 and March1 | (32) | |
| circPICALM (circ_0023919) | PICALM | Reduce tumor growth and metastasis, decrease pFAK/FAK ratio and decrease expression of β-catenin, Vimentin, ZEB1, Slug and N-cadherin | miR-1265 and STEAP4 | (36) | |
| circNR3C1(circ_0001543) | NR3C1 | Reduce tumor growth | miR-27a-3p and cyclin D1 | (33) | |
| circ-ZKSCAN1 (circ_0001727) | ZKSCAN1 | Reduce tumor growth and metastasis | miR-1178-3p and p21 | OS, DFS | (34) |
| circ-Foxo3 (circ_0006404) | Foxo3 | Promote cell apoptosis and inhibit proliferation, enhance expression of cleaved-caspase 3 and ratio of Bax/Bcl2 | miR-191-5p | (73) | |
| circ_0068307 | ALG3 | Reduce tumor growth | miR-147 and c-Myc | (29) | |
| circCDYL | CDYL | Decrease cell proliferation and migration, reduce expression of c-Myc | (7) | ||
| circ_0077837/0004826 | EPB41L2/UTRN | Reduce cell proliferation, migration and invasion | OS, RFS | (68) | |
| circHIPK3 (circ_0000284) | HIPK3 | Inhibit cell viability and enhance cell apoptosis induced by gemcitabine | Target, DFS | (75) | |
| B, Prostate cancer | |||||
| circRNAs | Host gene | Biological function | Target | Clinical applicant | (Refs.) |
| circRNA17 (circ_0001427) | PDLIM5 | Inhibit tumor growth, metastasis and enzalutamide-resistant | miRNA-181c-5p and ARv7 | (11) | |
| circAMOTL1L (circ_000350) | AMOTL1L | Inhibit tumor growth, increase expression of Pcdha8 and E-cadherin | miR-193a-5p and Pcdha8 | (46) | |
| circ_0001206 | CRKL | Inhibit tumor growth | miR-1285-5p and Smad4 | (45) | |
| circ-ITCH | ITCH | Inhibit tumor growth | miR-17-5p and HOXB13 | (47) | |
| circFoxo3 | Foxo3 | Inhibit tumor growth, metastasis and chemoresistance to docetaxel | (76) | ||
| circ_0004870 | RBM39 | Biomarker | (69) | ||
| C, Renal cancer | |||||
| circ-AKT3 (circ_0017252) | AKT3 | Inhibit tumor growth and metastasis | miR-296-3p and E-cadherin | (58) | |
| circC3P1 | C3P1 | Inhibit cell proliferation, migration, invasion and ratio of p/t-PI3K, p/t-AKT, p/t-p65, and p/t-IκBα | miR-21 and PTEN | (56) | |
| cRAPGEF5 (circ_0001681) | RAPGEF5 | Inhibit tumor growth and metastasis | miR-27a-3p and TXNIP | OS, RFS | (59) |
| circUBAP2 (circ_0001846) | UBAP2 | Reduce cell proliferation, migration and invasion | miR-148a-3p and FOXK2 | (57) | |
circRNA, circular RNA; BCSC, bladder cancer stem cell; OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival; miR, microRNA.
Abnormal cell signal transduction is closely associated with the occurrence of BC. For example, circ_0071662 abolishes the activation of AKT by increasing the expression of 15-hydroxyprostaglandin dehydrogenase, and inhibits BC development; however, its effect could be reversed by miR-146-3p (30). Furthermore, the expression of PETN is upregulated by circ solute carrier family 8 member A1 (SLC8A1) or downregulated by miR-130b/miR-494 in BC. circSLC8A1 blocks BC progression by sponging miR-130b/miR-494, and impairs the expression of p-AKT; circSLC8A1 (circ_0000994) is derived from the SLC8A1 gene (Fig. 2) (31). Another study showed that circ_0006260 inhibits tumor progression by upregulating membrane-associated ring finger 1 (MARCH1) in BC, an effect that could be reversed by miR-653. MARCH1 belongs to the E3 ligase family (32). In addition, cyclin D1 and p21, which control the cell cycle, are upregulated by miR-27a-3p and miR-1178-3p, resulting in the facilitation of tumor progression. By contrast, circNR3C1 and circZKSCAN1 act as a sponge for miR-27a-3p and miR-1178-3p to repress the cell development induced by miR-27a-3p and miR-1178-3p, respectively, in BC (33,34).
EMT plays a crucial role in the metastasis of BC (35). Indeed, circ phosphatidylinositol binding clathrin assembly protein (PICALM) inhibits cell metastasis and reduces the expression of β-catenin, vimentin, zinc finger E-box binding homeobox 1 (ZEB1), Slug and N-cadherin, which were found to be associated with EMT in BC; however, those effects could be reversed by miR-1265. circPICALM is derived from the PICALM gene (36). Notably, circRNA acts as a double agent in BC. For instance, circ5912 sabotages tumor growth in vitro and in vivo, and reduces the TGF-β2-induced EMT process. In addition, circ5912 attenuates early stage cancer progression, but promotes cancer development following the occurrence of distant metastasis (Fig. 2) (37).
3. circRNAs in PCa
circRNAs act as an oncogene in PCa
The circRNA-miRNA-mRNA axis plays a vital role in PCa progression. A disintegrin and metalloprotease 17, as a downstream of the Notch signaling pathway, was found to be highly expressed in PCa and to increase cell proliferation and invasion. Notably, its effect could be promoted by circ homeodomain interacting protein kinase 3 (HIPK3) or impaired by miR-338-3p. circHIPK3 is derived from the HIPK3 gene (5). Another study demonstrated that circZNF609 enhances PCa cell growth and metastasis, and increases the ratio of p/total-AMP-activated protein kinase (AMPK) by upregulating miR-186-5p, which promoted tumor progression through the activation of the AMPK signaling pathway (38). Furthermore, forkhead box p4 (FOXP4), a member of the FOXO family, was found to promote PCa progression in vitro, an effect that could be reversed by miR-1182 or enhanced by circ ATP-binding cassette subfamily C member 4 (ABCC4). circABCC4 (circ_0030586) is derived from the ABCC4 gene (Table I) (39).
circRNAs also interact with RBPs in PCa. Fused in sarcoma (FUS), a nuclear DNA/RNA-binding protein that regulates different steps of gene expression, acts as an oncogene in multiple cancer types (40), is overexpressed in PCa, and by binding to circ0005276, upregulates X-linked inhibitor of apoptosis (XIAP), the host gene of circ0005276. Furthermore, XIAP and circ0005276 enhance tumor growth and improve the expression of N-cadherin in PCa (41). circHIPK3 facilitates the G2/M transition of PCa cells by modifying cell division cyclin 25, which causes G2/M cell cycle arrest (42). In addition, circ102004 promotes PCa tumor growth in vitro and in vivo by increasing the levels of p-ERK, p-AKT, p-JNK, JNK and β-catenin, which have been found to be positively correlated with tumor aggressiveness; circ102004 is derived from oncogene ubiquitin-specific peptidase 22 (Fig. 2) (43).
circRNAs act as a tumor suppressor in PCa
Previous evidence has confirmed that Smad4, as a central mediator of the TGF-β signaling pathway, represses androgen receptor (AR) transactivation and exhibits low expression in PCa (44). circ_0001206 is derived from the CRKL gene, and was found to reduce PCa development by regulating Smad4; however, this effect could be reversed by miR-1285-5p (Fig. 2) (45). circRNA also acts as a reservoir for miRNA in PCa. For example, circRNA17 (circ_0001427) enhances the function of miR-181c-5p to suppress AR-splicing variant 7 (ARv7) expression and improves the ability of miR-181c-5p to inhibit tumor progression. ARv7 has been found to be positively correlated with enzalutamide resistance in PCa (11).
circRNA can both sponge miRNA and interact with RBPs in PCa. For instance, P53 and RNA-binding protein 25 (RBM25; a transcriptional target of p53) increase the expression of circ_000350 [circ angiomotin-like 1 (circAMOTL1L)] in PCa. circAMOTL1L and RBM25 impair cell mobility and diminish the level of vimentin and β-catenin in PCa. circAMOTL1L is derived from the AMOTL1 gene. In addition, circAMOTL1L inhibits tumor growth and enhances the expression of protocadherin α8 (Pcdha8) in PCa; however, these effects could be reversed by upregulating miR-193a-5p or depleting p53. Pcdha8 is a tumor suppressor and a member of a subset of the cadherin superfamily (Table II) (46). In addition, circ itchy E3 ubiquitin protein ligase-overexpression reduces PCa growth by sponging miR-17-5p to rescue the degradation of homeobox protein Hox-B13 (HOXB13) in vitro and in vivo (47). HOXB13 has been verified to act as an oncogene in PCa.
4. circRNAs in RCC
circRNAs act as an oncogene in RCC
The activation of the EMT process and VEGF signaling pathway contributes to tumor initiation (48). The expression of ZEB2 is upregulated by circ pecanex 2 (PCNXL2) or downregulated by miR-153 in RCC. In addition, circPCNXL2 (circ_406752) is derived from the PCNXL2 gene, and enhances tumor growth in vitro and in vivo, while its effect can be reversed by miR-153 (49). The expression of N-cadherin, Snail and vimentin could be increased by circ proline rich coiled-coil 2A (PRRC2A) and circ myosin light chain kinase (MYLK), which abolish the tumor suppression effect of miR-514a-5p/miR-6776-5p and miR-513a-5p, respectively, in RCC. In addition, circPRRC2A and circMYLK have been shown to facilitate tumor progression by upregulating transient receptor potential cation channel subfamily M member 3 (TRPM3) and VEGFC, respectively. TRPM3 has been reported to act as an oncogene in metastatic RCC. VEGFC is a member of the VEGF family and circPRRC2A is derived from the PRRC2A gene (Fig. 2) (50,51). Chromobox 4, as a SUMO E3 ligase, upregulates the level of VEGFA, and represses cell growth and metastasis in RCC following circTLK1-silencing or miR-136-5p-overexpression. Circ tousled-like kinase 2 (TLK1; circ_0004442) is derived from the back-splicing of the TLK1 mRNA (52).
The abnormal activation of transcription factors promotes the proliferation and differentiation of tumor cells (24). For example, SRY-box transcription factor 12 (SOX-12), which was overexpressed in ccRCC, was upregulated by circ_001895 or downregulated by miR-296-5p. Furthermore, the high expression of circ_001895 led to increased tumor growth and metastasis by regulating SOX-12, and improving the expression of N-cadherin, while those effects could be reversed by miR-296-5p (53). The interferon regulatory factors (IRFs) are a family of master transcription factors that regulate pathogen-induced innate and acquired immune responses (54). By contrast, the IRF7 overexpression in RCC was found to facilitate tumor progression in vitro, and to be increased by circ-egl-9 family hypoxia inducible factor 3 (EGLN3) or repressed by miR-1299; circ-EGLN3 (circ_0031594) is derived from the EGLN3 gene (Table I) (55).
circRNAs act as tumor suppressors in RCC
Cell signaling transduction plays a vital role in the occurrence of RCC. circ complement component 3 precursor pseudogene (C3P1) is derived from exons 27–29 of the complement component 3 precursor pseudogene, decreases the ratio of p/total-PI3K, p/t-AKT, p/total-p65 and p/total-IκBα, promotes the expression of Bcl-2, cleaved caspase-3 and cleaved caspase-9, and consequently inhibits tumor progression in kidney cancer. These circC3P1-induced effects could be reversed by the upregulation of miR-21 through targeting PTEN in RCC (Fig. 2) (56). FOXK2 was found to translocate to the nucleus, be dependent on the AKT-mTOR signaling pathway, be expressed at a low level in ccRCC, and impair tumor cell proliferation and metastasis; however, its effect could be enhanced by circ_0001846 or attenuated by miR-148a-3p (57).
Notably, circRNAs can sponge miRNAs to protect RBP degradation in RCC. The overexpression of circ-AKT3 leads to the inhibition of tumor growth and metastasis, and sponges miR-296-3p to avoid the mRNA degradation of E-cadherin. circ-AKT3 (circ_0017252) is derived from the AKT3 gene locus (58). circ Rap guanine nucleotide exchange factor 5 (RAPGEF5; circ_0001681) is derived from the RAPGEF5 gene, and could be upregulated by thioredxin interacting protein (TXNIP) or downregulated by miR-27a-3p in RCC cells. circRAPGEF5 overexpression leads to a repressive effect on the progression and aggressiveness of RCC by upregulating TXNIP; however, these effects could be reversed by miR-27a-3p. In addition, patients with RCC and high circRAPGEF5 expression have been associated with an improved overall survival (OS) and recurrence-free survival (RFS) rate (Table II) (59).
5. circRNAs act as biomarkers for the diagnosis and prognosis of BC, PCa and RCC
In the past few decades, tumor diagnosis and prognosis have largely depended on radiography testing and pathological examinations. Nowadays, circRNA also acts as a biomarker for the diagnosis and prognosis of urological neoplasms. For example, circEGLN3 can distinguish ccRCC from normal tissues with 97% accuracy (60). circRNAs abundantly exist in exosomes of patients with urological cancer, which indicates that they can play a diagnostic role in liquid biopsies. The level of circ protein tyrosine kinase 2 (circ_0003221) was increased in the lymph nodes of a nude mouse model and in vivo, as well as in the blood of patients with BC, and promoted the progression of BC (61). Similarly, the expression of circ_0044516, circFOXO3 and circ formin 2 (circ_0005100) was increased in the blood of patients with PCa, and promoted tumor progression by sponging miR-29a-3p and miR-1238, respectively (Table I) (62–64).
circ protein arginine methyltransferase 5 (PRMT5; circ_101320) and circ_0001445 have been shown to facilitate the progression of BC by sponging miR-30c and miR-107, respectively. Furthermore, patients with BC and a high level of circPRMT5 and circ_0001445 displayed poor disease-free survival (DFS) and OS rate (65,66). The overexpression of circ_0085576 has been shown to increase RCC tumor progression following Yes1-associated transcriptional regulator upregulation and miR-498-silencing. In addition, patients with ccRCC and a high level of circ_0085576 had a poor OS and DFS rate; circ_0085576 is derived from the ArfGAP with SH3 domain ankyrin repeat and PH domain 1 gene (67). By contrast, circ_0004826 and circ_0077837, which are spliced from the utrophin and erythrocyte membrane protein band 4.1-like 2 genes, respectively, are usually expressed at a low level in BC, inhibit tumor cell development and act as a biomarker for OS and RFS in patients with BC (Table II) (68). circRNAs can also act as biomarkers for the effect of drug treatment. circ_0004870 is an exonic circRNA located on chromosome 20, which has been found to be downregulated in enzalutamide-resistant cells and expressed at a low level in an AR-positive cell line (LNCaP clone 1/9), thus acting as a biomarker for the effect of enzalutamide treatment in PCa (69)
6. circRNAs act as therapeutic targets for BC, PCa and RCC
The overexpression of circ_0084171 and circELP3 result in an increase in tumor growth and promotion of cisplatin resistance in BC. In addition, circ_0084171 improves the expression of p-ERK1/2 and p-MEK1/2 in BC, while those effects could be reversed by miR-370-3p (Fig. 2) (70,71). Furthermore, circ_0039569 is derived from the C-C motif chemokine ligand 22 (CCL22) gene, is highly expressed in RCC and promotes tumor development after sponging miR-34a-5p or over-regulating CCL22. Furthermore, the inhibition of circ_0039569 may enhance the drug sensitivity of RCC cells; however, the study suggesting this did not provide any details to prove it (Table II) (72).
By contrast, the overexpression of CDR1 antisense RNA (Cdr1as; circ_0001946) and circ-Foxo3 has been shown to increase cell apoptosis and decrease cisplatin chemoresistance reduced by miR-1270 and miR-191 in BC, consequently suppressing tumor development. circ-Foxo3 is derived from FOXO3 mRNA, and Cdr1as is formed by back-splicing of the cerebellar degeneration-related protein 1 gene (73,74). A different study illustrated that the overexpression of circHIPK3 has been shown to lead to an increased sensitization of BC cells to gemcitabine and act as a biomarker for DFS in patients with BC (75). In addition, the apoptosis and chemosensitivity of docetaxel in androgen-dependent PCa cells could be increased by the overexpression of circFoxo3. Furthermore, the upregulation of circfoxo3 enhanced the chemosensitivity of PCa-bearing mice to docetaxel and prolonged the life span of these mice (Table II) (76).
7. Conclusion
In conclusion, circRNAs, as a family of non-coding RNAs that can form a loop with joined 3′heads and 5′tails, are characterized by abundant, highly stable, evolutionarily conserved and tissue-specific expression. Several studies have shown that circRNAs play a crucial role in tumor growth, metastasis and treatment resistance (12). circRNAs have shown an ability to help determine the pathogenesis, and serve as a biomarker for the diagnosis and prognosis, of urological cancer. Clinical trials using treatment and diagnostic methods involving circRNAs are now being conducted, including trials in hepatocellular carcinoma and myocardial infarction (77,78). However, a number of limitations in the recently published studies of circRNAs were identified. For example, circZNF139 and circ-ZKSCAN1 are identical circRNAs that are known as circ_0001727, but display opposite roles in BC (23,34). In addition, circ-Foxo3 serves as both an oncogene and tumor suppressor in PCa (63,76), and circ5912 serves as a double agent in BC (37). Therefore, the function of circRNAs in tumors of the urinary system remains largely unclear and further research is required.
Acknowledgements
Not applicable.
Funding Statement
The study was supported by the National Natural Science Foundation of China (grant nos. 81760462 and 81860456), and the Science and Technology Project Founded by the Education Department of Jiangxi Province (grant nos. GJJ180789 and GJJ180788).
Funding
The study was supported by the National Natural Science Foundation of China (grant nos. 81760462 and 81860456), and the Science and Technology Project Founded by the Education Department of Jiangxi Province (grant nos. GJJ180789 and GJJ180788).
Availability of data and materials
Not applicable.
Authors' contributions
LY reviewed the publications and drafted the manuscript. LY and JZ designed and drew the diagram. GZ and XZ revised the manuscript and confirmed the authenticity of all the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dagenais GR, Leong DP, Rangarajan S, Lanas F, Lopez-Jaramillo P, Gupta R, Diaz R, Avezum A, Oliveira GBF, Wielgosz A, et al. Variations in common diseases, hospital admissions, and deaths in middle-aged adults in 21 countries from five continents (PURE): A prospective cohort study. Lancet. 2019;395:785–794. doi: 10.1016/S0140-6736(19)32007-0. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Liu F, Zhang H, Xie F, Tao D, Xiao X, Huang C, Wang M, Gu C, Zhang X, Jiang G. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene. 2019;39:1696–1709. doi: 10.1038/s41388-019-1092-z. [DOI] [PubMed] [Google Scholar]
- 4.Massari F, Di Nunno V, Ciccarese C, Graham J, Porta C, Comito F, Cubelli M, Iacovelli R, Heng DYC. Adjuvant therapy in renal cell carcinoma. Cancer Treat Rev. 2017;60:152–157. doi: 10.1016/j.ctrv.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Cai C, Zhi Y, Wang K, Zhang P, Ji Z, Xie C, Sun F. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. Onco Targets Ther. 2019;12:3363–3372. doi: 10.2147/OTT.S196931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi J, Liu H, Cai Z, Dong W, Jiang N, Yang M, Huang J, Lin T. Circ-BPTF promotes bladder cancer progression and recurrence through the miR-31-5p/RAB27A axis. Aging (Albany NY) 2018;10:1964–1976. doi: 10.18632/aging.101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J, Zhang H, Tao D, Xie F, Liu F, Gu C, Wang M, Wang L, Jiang G, Wang Z, Xiao X. CircCDYL inhibits the expression of C-MYC to suppress cell growth and migration in bladder cancer. Artif Cells Nanomed Biotechnol. 2019;47:1349–1356. doi: 10.1080/21691401.2019.1596941. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, Liu H, Xu J, Xiao F, Zhou H, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong R, Ma XK, Chen LL, Yang L. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017;14:1064–1074. doi: 10.1080/15476286.2016.1269999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei Y, Zhang Y, Meng Q, Cui L, Xu C. Hypoxia-induced circular RNA has_circRNA_403658 promotes bladder cancer cell growth through activation of LDHA. Am J Transl Res. 2019;11:6838–6849. [PMC free article] [PubMed] [Google Scholar]
- 11.Wu G, Sun Y, Xiang Z, Wang K, Liu B, Xiao G, Niu Y, Wu D, Chang C. Preclinical study using circular RNA 17 and micro RNA 181c-5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis. 2019;10:37. doi: 10.1038/s41419-018-1048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 13.Su Y, Feng W, Shi J, Chen L, Huang J, Lin T. circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-β2/smad3 pathway. Mol Cancer. 2020;19:23. doi: 10.1186/s12943-019-1129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi YR, Wu Z, Xiong K, Liao QJ, Ye X, Yang P, Zu XB. Circular RNA circKIF4A Sponges miR-375/1231 to promote bladder cancer progression by upregulating NOTCH2 expression. Front Pharmacol. 2020;11:605. doi: 10.3389/fphar.2020.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Yang X, Zhao J, Xiong M, Almaraihah R, Chen Z, Hou T. Circ_0008532 promotes bladder cancer progression by regulation of the miR-155-5p/miR-330-5p/MTGR1 axis. J Exp Clin Cancer Res. 2020;39:94. doi: 10.1186/s13046-020-01592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao W, Huang X, Wang L, Zhang Z, Liu M, Li Y, Luo M, Yao X, Fan J, Geng J. Circular RNA hsa_circ_0068871 regulates FGFR3 expression and activates STAT3 by targeting miR-181a-5p to promote bladder cancer progression. J Exp Clin Cancer Res. 2019;38:169. doi: 10.1186/s13046-019-1136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu L, Zhang M, Qi L, Zu X, Li Y, Liu L, Chen M, Li Y, He W, Hu X, et al. ERα-mediated alterations in circ_0023642 and miR-490-5p signaling suppress bladder cancer invasion. Cell Death Dis. 2019;10:635. doi: 10.1038/s41419-019-1827-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Topham CH, Taylor SS. Mitosis and apoptosis: How is the balance set? Curr Opin Cell Biol. 2013;25:780–785. doi: 10.1016/j.ceb.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Yang C, Li Q, Chen X, Zhang Z, Mou Z, Ye F, Jin S, Jun X, Tang F, Jiang H. Circular RNA circRGNEF promotes bladder cancer progression via miR-548/KIF2C axis regulation. Aging (Albany NY) 2020;12:6865–6879. doi: 10.18632/aging.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D, Qian H, Fang Z, Xu A, Zhao S, Liu B, Li D. Silencing circular RNA VANGL1 inhibits progression of bladder cancer by regulating miR-1184/IGFBP2 axis. Cancer Med. 2019;9:700–710. doi: 10.1002/cam4.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Li C, Fan Z, Liu H, Zhang X, Cai Z, Xu L, Luo J, Huang Y, He L, et al. Single-cell sequencing reveals variants in ARID1A, GPRC5A and MLL2 driving self-renewal of human bladder cancer stem cells. Eur Urol. 2017;71:8–12. doi: 10.1016/j.eururo.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Tao T, Yuan S, Liu J, Shi D, Peng M, Li C, Wu S. Cancer stem cell-specific expression profiles reveal emerging bladder cancer biomarkers and identify circRNA_103809 as an important regulator in bladder cancer. Aging (Albany NY) 2020;12:3354–3370. doi: 10.18632/aging.102816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao J, Qian K, Chen C, Liu X, Yu D, Yan X, Liu T, Li S. ZNF139/circZNF139 promotes cell proliferation, migration and invasion via activation of PI3K/AKT pathway in bladder cancer. Aging (Albany NY) 2020;12:9915–9934. doi: 10.18632/aging.103256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francis R, Guo H, Streutker C, Ahmed M, Yung T, Dirks PB, He HH, Kim TH. Gastrointestinal transcription factors drive lineage-specific developmental programs in organ specification and cancer. Sci Adv. 2019;5:eaax8898. doi: 10.1126/sciadv.aax8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T, Lu Q, Liu J, Xie S, Feng B, Zhu W, Liu M, Liu Y, Zhou X, Sun W, et al. Circular RNA FAM114A2 suppresses progression of bladder cancer via regulating ∆NP63 by sponging miR-762. Cell Death Dis. 2020;11:47. doi: 10.1038/s41419-020-2226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q, Wang Q, Xie R, Su Y, Yang M, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer. 2019;18:95. doi: 10.1186/s12943-019-1025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Q, Huang L, Yan D, Bi J, Yang M, Huang J, Lin T. CircPTPRA acts as a tumor suppressor in bladder cancer by sponging miR-636 and upregulating KLF9. Aging (Albany NY) 2019;11:11314–11328. doi: 10.18632/aging.102530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Q, Yan D, Dong W, Bi J, Huang L, Yang M, Huang J, Qin H, Lin T. circRNA circFUT8 upregulates Krüpple-like Factor 10 to inhibit the metastasis of bladder cancer via sponging miR-570-3p. Mol Ther Oncolytics. 2020;16:172–187. doi: 10.1016/j.omto.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q, Yin Q, Mao Y, Zhang Z, Wu S, Cheng Z, Chen X, Xu H, Jin S, Jiang H, Yang C. Hsa_circ_0068307 mediates bladder cancer stem cell-like properties via miR-147/c-Myc axis regulation. Cancer Cell Int. 2020;20:151. doi: 10.1186/s12935-020-01235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abulizi R, Li B, Zhang CG. Circ_0071662, a novel tumor biomarker, suppresses bladder cancer cell proliferation and invasion by sponging miR-146b-3p. Oncol Res. 2019 Nov 18; doi: 10.3727/096504019X15740729375088. (Epub ahead of print). doi: 10.3727/096504019X15740729375088. [DOI] [PubMed] [Google Scholar]
- 31.Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, Jiang B, Qin H, Guo X, Liu M, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18:111. doi: 10.1186/s12943-019-1040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Y, Feng W, Zhong G, Ya Y, Du Z, Shi J, Chen L, Dong W, Lin T. ciRs-6 upregulates March1 to suppress bladder cancer growth by sponging miR-653. Aging (Albany NY) 2019;11:11202–11223. doi: 10.18632/aging.102525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng F, Wang M, Li Y, Huang C, Tao D, Xie F, Zhang H, Sun J, Zhang C, Gu C, et al. CircNR3C1 inhibits proliferation of bladder cancer cells by sponging miR-27a-3p and downregulating cyclin D1 expression. Cancer Lett. 2019;460:139–151. doi: 10.1016/j.canlet.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 34.Bi J, Liu H, Dong W, Xie W, He Q, Cai Z, Huang J, Lin T. Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer. 2019;18:133. doi: 10.1186/s12943-019-1060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M, Du H, Wang L, Yue Y, Zhang P, Huang Z, Lv W, Ma J, Shao Q, Ma M, et al. Thymoquinone suppresses invasion and metastasis in bladder cancer cells by reversing EMT through the Wnt/β-catenin signaling pathway. Chem Biol Interact. 2020;320:109022. doi: 10.1016/j.cbi.2020.109022. [DOI] [PubMed] [Google Scholar]
- 36.Yan D, Dong W, He Q, Yang M, Huang L, Kong J, Qin H, Lin T, Huang J. Circular RNA circPICALM sponges miR-1265 to inhibit bladder cancer metastasis and influence FAK phosphorylation. EBioMedicine. 2019;48:316–331. doi: 10.1016/j.ebiom.2019.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su Y, Du Z, Zhong G, Ya Y, Bi J, Shi J, Chen L, Dong W, Lin T. circ5912 suppresses cancer progression via inducing MET in bladder cancer. Aging (Albany NY) 2019;11:10826–10838. doi: 10.18632/aging.102464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin C, Zhao W, Zhang Z, Liu W. Silencing circular RNA circZNF609 restrains growth, migration and invasion by up-regulating microRNA-186-5p in prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47:3350–3358. doi: 10.1080/21691401.2019.1648281. [DOI] [PubMed] [Google Scholar]
- 39.Huang C, Deng H, Wang Y, Jiang H, Xu R, Zhu X, Huang Z, Zhao X. Circular RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23:6112–6119. doi: 10.1111/jcmm.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward CL, Boggio KJ, Johnson BN, Boyd JB, Douthwright S, Shaffer SA, Landers JE, Glicksman MA, Bosco DA. A loss of FUS/TLS function leads to impaired cellular proliferation. Cell Death Dis. 2014;5:e1572. doi: 10.1038/cddis.2014.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Y, Yang Y, Zhao X, Fan Y, Zhou L, Rong J, Yu Y. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 2019;10:792. doi: 10.1038/s41419-019-2028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu F, Fan Y, Ou L, Li T, Fan J, Duan L, Yang J, Luo C, Wu X. CircHIPK3 facilitates the G2/M transition in prostate cancer cells by sponging miR-338-3p. Onco Targets Ther. 2020;13:4545–4558. doi: 10.2147/OTT.S242482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Si-Tu J, Cai Y, Feng T, Yang D, Yuan S, Yang X, He S, Li Z, Wang Y, Tang Y, et al. Upregulated circular RNA circ-102004 that promotes cell proliferation in prostate cancer. Int J Biol Macromol. 2019;122:1235–1243. doi: 10.1016/j.ijbiomac.2018.09.076. [DOI] [PubMed] [Google Scholar]
- 44.Kang HY, Huang KE, Chang SY, Ma WL, Lin WJ, Chang C. Differential modulation of androgen receptor-mediated transactivation by Smad3 and tumor suppressor Smad4. J Biol Chem. 2002;277:43749–43756. doi: 10.1074/jbc.M205603200. [DOI] [PubMed] [Google Scholar]
- 45.Song Z, Zhuo Z, Ma Z, Hou C, Chen G, Xu G. Hsa_Circ_0001206 is downregulated and inhibits cell proliferation, migration and invasion in prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47:2449–2464. doi: 10.1080/21691401.2019.1626866. [DOI] [PubMed] [Google Scholar]
- 46.Yang Z, Qu CB, Zhang Y, Zhang WF, Wang DD, Gao CC, Ma L, Chen JS, Liu KL, Zheng B, et al. Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene. 2019;38:2516–2532. doi: 10.1038/s41388-018-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Wang R, Wu Z, Bai P. Circular RNA ITCH suppressed prostate cancer progression by increasing HOXB13 expression via spongy miR-17-5p. Cancer Cell Int. 2019;19:328. doi: 10.1186/s12935-019-0994-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Fantozzi A, Gruber DC, Pisarsky L, Heck C, Kunita A, Yilmaz M, Meyer-Schaller N, Cornille K, Hopfer U, Bentires-Alj M, Christofori G. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res. 2014;74:1566–1575. doi: 10.1158/0008-5472.CAN-13-1641. [DOI] [PubMed] [Google Scholar]
- 49.Zhou B, Zheng P, Li Z, Li H, Wang X, Shi Z, Han Q. CircPCNXL2 sponges miR-153 to promote the proliferation and invasion of renal cancer cells through upregulating ZEB2. Cell Cycle. 2018;17:2644–2654. doi: 10.1080/15384101.2018.1553354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Yang FQ, Sun CM, Huang JH, Zhang HM, Li X, Wang GC, Zhang N, Che JP, Zhang WT, et al. circPRRC2A promotes angiogenesis and metastasis through epithelial-mesenchymal transition and upregulates TRPM3 in renal cell carcinoma. Theranostics. 2020;10:4395–4409. doi: 10.7150/thno.43239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Huang C, Zou Y, Yu J, Gui Y. Circular RNA MYLK promotes tumour growth and metastasis via modulating miR-513a-5p/VEGFC signalling in renal cell carcinoma. J Cell Mol Med. 2020;24:6609–6621. doi: 10.1111/jcmm.15308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Huang C, Zou Y, Ye J, Yu J, Gui Y. CircTLK1 promotes the proliferation and metastasis of renal cell carcinoma by sponging miR-136-5p. Mol Cancer. 2020;19:103. doi: 10.1186/s12943-020-01225-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Chen Z, Xiao K, Chen S, Huang Z, Ye Y, Chen T. Circular RNA hsa_circ_001895 serves as a sponge of microRNA-296-5p to promote clear cell renal cell carcinoma progression by regulating SOX12. Cancer Sci. 2020;111:713–726. doi: 10.1111/cas.14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson CD, Matta B, Barnes BJ. Therapeutic targeting of IRFs: Pathway-dependence or structure-based? Front Immunol. 2018;9:2622. doi: 10.3389/fimmu.2018.02622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin L, Cai J. Circular RNA circ-EGLN3 promotes renal cell carcinoma proliferation and aggressiveness via miR-1299-mediated IRF7 activation. J Cell Biochem. 2020;121:4377–4385. doi: 10.1002/jcb.29620. [DOI] [PubMed] [Google Scholar]
- 56.Chen T, Yu Q, Xin L, Guo L. Circular RNA circC3P1 restrains kidney cancer cell activity by regulating miR-21/PTEN axis and inactivating PI3K/AKT and NF-kB pathways. J Cell Physiol. 2020;235:4001–4010. doi: 10.1002/jcp.29296. [DOI] [PubMed] [Google Scholar]
- 57.Sun J, Yin A, Zhang W, Lv J, Liang Y, Li H, Li Y, Li X. CircUBAP2 inhibits proliferation and metastasis of clear cell renal cell carcinoma via targeting miR-148a-3p/FOXK2 pathway. Cell Transplant. 2020;29:963689720925751. doi: 10.1177/0963689720925751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue D, Wang H, Chen Y, Shen D, Lu J, Wang M, Zebibula A, Xu L, Wu H, Li G, Xia L. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol Cancer. 2019;18:151. doi: 10.1186/s12943-019-1072-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Q, Liu T, Bao Y, Zhao T, Wang J, Wang H, Wang A, Gan X, Wu Z, Wang L. circRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 2020;469:68–77. doi: 10.1016/j.canlet.2019.10.017. [DOI] [PubMed] [Google Scholar]
- 60.Franz A, Ralla B, Weickmann S, Jung M, Rochow H, Stephan C, Erbersdobler A, Kilic E, Fendler A, Jung K. Circular RNAs in clear cell renal cell carcinoma: Their microarray-based identification, analytical validation, and potential use in a clinico-genomic model to improve prognostic accuracy. Cancers. 2019;11:1473. doi: 10.3390/cancers11101473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu ZQ, Yang MG, Liu HJ, Su CQ. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem. 2018;119:3317–3325. doi: 10.1002/jcb.26492. [DOI] [PubMed] [Google Scholar]
- 62.Li T, Sun X, Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. 2020;121:2118–2126. doi: 10.1002/jcb.28239. [DOI] [PubMed] [Google Scholar]
- 63.Kong Z, Wan X, Lu Y, Zhang Y, Huang Y, Xu Y, Liu Y, Zhao P, Xiang X, Li L, Li Y. Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J Cell Mol Med. 2020;24:799–813. doi: 10.1111/jcmm.14791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shan G, Shao B, Liu Q, Zeng Y, Fu C, Chen A, Chen Q. circFMN2 Sponges miR-1238 to promote the expression of LIM-homeobox Gene 2 in prostate cancer cells. Mol Ther Nucleic Acids. 2020;21:133–146. doi: 10.1016/j.omtn.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 66.Su H, Tao T, Yang Z, Kang X, Zhang X, Kang D, Wu S, Li C. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18:27. doi: 10.1186/s12943-019-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu G, Zhou J, Piao Y, Zhao X, Zuo Y, Ji Z. Hsa_circ_0085576 promotes clear cell renal cell carcinoma tumorigenesis and metastasis through the miR-498/YAP1 axis. Aging (Albany NY) 2020;12:11530–11549. doi: 10.18632/aging.103300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen C, Wu Z, Wang Y, Gao S, Da L, Xie L, Qie Y, Tian D, Hu H. Downregulated hsa_circ_0077837 and hsa_circ_0004826, facilitate bladder cancer progression and predict poor prognosis for bladder cancer patients. Cancer Med. 2020;9:3885–3903. doi: 10.1002/cam4.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greene J, Baird AM, Casey O, Brady L, Blackshields G, Lim M, O'Brien O, Gray SG, McDermott R, Finn SP. Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci Rep. 2019;9:10739. doi: 10.1038/s41598-019-47189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J, Sun Y, Ou Z, Yeh S, Huang CP, You B, Tsai YC, Sheu TJ, Zu X, Chang C. Androgen receptor-regulated circFNTA activates KRAS signaling to promote bladder cancer invasion. EMBO Rep. 2020;32:e48467. doi: 10.15252/embr.201948467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su Y, Yang W, Jiang N, Shi J, Chen L, Zhong G, Bi J, Dong W, Wang Q, Wang C, Lin T. Hypoxia-elevated circELP3 contributes to bladder cancer progression and cisplatin resistance. Int J Biol Sci. 2019;15:441–452. doi: 10.7150/ijbs.26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin C, Shi L, Li Z, Liu W, Zhao B, Qiu Y, Zhao Y, Li K, Li Y, Zhu Q. Circ_0039569 promotes renal cell carcinoma growth and metastasis by regulating miR-34a-5p/CCL22. Am J Transl Res. 2019;11:4935–4945. [PMC free article] [PubMed] [Google Scholar]
- 73.Wang C, Tao W, Ni S, Chen Q. Circular RNA circ-Foxo3 induced cell apoptosis in urothelial carcinoma via interaction with miR-191-5p. Onco Targets Ther. 2019;12:8085–8094. doi: 10.2147/OTT.S215823. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Yuan W, Zhou R, Wang J, Han J, Yang X, Yu H, Lu H, Zhang X, Li P, Tao J, et al. Circular RNA Cdr1as sensitizes bladder cancer to cisplatin by upregulating APAF1 expression through miR-1270 inhibition. Mol Oncol. 2019;13:1559–1576. doi: 10.1002/1878-0261.12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie F, Zhao N, Zhang H, Xie D. Circular RNA CircHIPK3 promotes gemcitabine sensitivity in bladder cancer. J Cancer. 2020;11:1907–1912. doi: 10.7150/jca.39722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen Z, Zhou L, Zhang C, Xu J. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020;468:88–101. doi: 10.1016/j.canlet.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 77.Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, Zhang H, Yang P, Cui X, Ren Y, et al. A noncoding regulatory RNAs network driven by Circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2020;71:130–147. doi: 10.1002/hep.30795. [DOI] [PubMed] [Google Scholar]
- 78.Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. 2019;10:4317. doi: 10.1038/s41467-019-11777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.