Abstract
Our previous study demonstrated the role of family with sequence similarity 83, member B (FAM83B) in endometrial cancer tumorigenesis and metastasis. FAM83B is involved in epithelial-to-mesenchymal transition (EMT). However, the regulatory network of EMT, which promotes endometrial cancer cell metastasis, involving microRNAs (miRNAs/miRs) and FAM83B, has not been elucidated. To investigate the potential mechanism underlying miR-199a/b-5p in endometrial cancer, the effect of miR-199a/b-5p and its targeted FAM83B gene on the biological behaviour of endometrial cancer cells was assessed. The Gene Expression Omnibus dataset analysis results revealed that the expression levels of 150 miRNAs in non-cancerous endometrial tissues were upregulated compared with those in endometrial cancer tissues. TargetScan predicted that the nucleotides 672–679 of FAM83B 3′-untranslated region (UTR) were the target sites of miR-199a/b-5p. The differentially expressed miRNAs were enriched in several Kyoto Encyclopedia of Genes and Genomes pathways. Reverse transcription-quantitative PCR analysis revealed that the expression levels of miR-199a/b-5p in the endometrial non-cancerous cell lines were significantly upregulated compared with those in the six endometrial cancer cell lines. miR-199a/b-5p inhibited the EMT signaling pathway by regulating the expression levels of E-cadherin, N-cadherin, Snail, α-smooth muscle actin, vimentin and Twist. This suggested that miR-199a/b-5p inhibited endometrial cancer cell proliferation and migration through the inhibition of the EMT signaling pathway. Furthermore, the nucleotides 672–679 of the FAM83B 3′-UTR were demonstrated to be the binding site of miR-199a/b-5p. These results suggested that miR-199a/b-5p inhibited endometrial cancer cell proliferation and metastasis by targeting the 3′-UTR of FAM83B, which is involved in the EMT signaling pathway.
Keywords: miR-199a/b-5p, endometrial cancer, FAM83B, EMT signaling, neoplasm metastasis
Introduction
Endometrial cancer is one of the most common gynecological cancers in China and is a major threat to women's health (1). The incidence and mortality rates of endometrial cancer in China are 8.56 and 1.94 per 100,000 individuals, respectively (2). Furthermore, patients diagnosed with late-stage endometrial cancer exhibit a poor prognosis (3). Thus, there is a need to elucidate the molecular mechanisms underlying endometrial cancer to develop reliable predictive biomarkers and effective therapeutic agents.
Epithelial-to-mesenchymal transition (EMT) is a phenotypic plasticity process that confers migratory and invasive properties to epithelial cells during the pathogenesis of endometrial cancer (4). EMT is mediated by cell adhesion molecules (E-cadherin and N-cadherin), transcription factors (Snail and Twist), α-smooth muscle actin (SMA) and vimentin (5). Additionally, various microRNAs (miRNAs/miRs) modulate the regulatory network of EMT. Drug resistance and prognosis in patients with various types of cancer, such as breast, colorectal, bladder, ovarian and endometrial cancer, are directly determined by EMT-associated factors (6,7). Previous studies have demonstrated that EMT-associated factors, which are upregulated in cancer, mediate cancer cell proliferation, migration and invasion. For example, Padmanaban et al (8) reported that E-cadherin promotes metastasis in multiple models of breast cancer, and Li et al (9) demonstrated that Snail-induced claudin-11 promotes tumor progression by increasing cancer cell migration. Thus, EMT-associated factors serve vital roles in the tumorigenesis of endometrial cancer.
Family with sequence similarity 83, member B (FAM83B) is reported to be a novel molecule involved in the development of various malignant tumors, including pancreatic ductal Adenocarcinoma (10) and gastric cancer (11). Previous studies have demonstrated that FAM83B can directly activate the PI3K/AKT/mTOR signaling pathway in tumors, including breast, lung, ovarian and cervical cancer, by binding to PI3K (12,13) or can indirectly activate the pathway by binding and hyper-activating EGFR in lung adenocarcinoma (14). Our previous study demonstrated that FAM83B knockdown inhibits endometrial cancer cell proliferation and metastasis through the inhibition of the PI3K/AKT/mTOR signaling pathway (15). Collectively, the aforementioned studies suggest that the pathogenesis of various types of cancer is mediated by FAM83B.
miR-199 is a highly conserved miRNA family that comprises miR-199a and miR-199b (16). A previous study demonstrated that miR-199a-5p inhibits the proliferation, migration and invasion of cancer cells (17). Additionally, miR-199a-5p activates EMT-associated signaling pathways (18). In endometrial carcinoma, the dysregulation of miR-199b in tissues and plasma results in the upregulation of the mTOR kinase (19). Previous studies have reported a one-to-one or one-to-two association of miR-199, FAM83B, EMT, cell metastasis and endometrial cancer. However, the role of miR-199-a/b-5p, which targets FAM83B, in endometrial cancer has not been elucidated (11,15,20–22).
In the present study, the target genes of miR-199-a/b-5p (including FAM83B) and the signaling pathways in which they are enriched were predicted through data mining. The expression levels of miR-199-a/b-5p, FAM83B, E-cadherin, N-cadherin, Snail, α-SMA, vimentin and Twist were examined in endometrial cancer cell lines. Additionally, the biological functions of miR-199-a/b-5p and FAM83B in the proliferation, migration, invasion, in vivo tumor formation and pulmonary metastasis of endometrial cancer cells were evaluated. Furthermore, the 3′-untranslated region (UTR) binding sites of miR-199-a/b-5p in FAM83B was investigated.
Materials and methods
Gene Expression Omnibus (GEO) dataset analysis
Wang et al (23) confirmed that GEO datasets can provide a more convenient way to screen gene expression between disease and non-disease cases, therefore GEO datasets were used to analyze the difference in miRNA expression between endometrial adenocarcinoma and non-cancerous tissues. Of the 48 samples in the dataset GSE25405_RAW.tar, the microarray data of 27 samples were downloaded from GEO of the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/geo). The datasets were divided into the following groups: Seven non-cancerous endometrial tissue datasets (GSM623853-GSM623855 and GSM623875-GSM623878) and 20 endometrioid adenocarcinoma datasets (GSM623856-GSM623874 and GSM623881). The Agilent-019118 Human miRNA Microarray 2.0 G4470B (GPL7731) data were analyzed using the Agilent GeneSpringGX Software (version 11.5; Agilent Technologies, Inc.). The differentially expressed miRNAs were identified based on the following criteria: P<0.01 and log2 fold change (FC) <-2. Heatmap and volcano plot were used to represent the differentially expressed miRNAs. To determine the expression in the microarray data, the differential gene expression profile data (GSE35794) of the top 250 miRNAs were analyzed using NCBI GEO2R (version R 3.2.3; http://www.ncbi.nlm.nih.gov/geo/geo2r/).
Target gene prediction
The putative miRNA targets were predicted using TargetScan 7.2 (www.targetscan.org/vert_72). Additionally, the poorly conserved 3′-UTR of FAM83B was predicted.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
The predicted target genes of miR-199-a/b-5p were annotated using Kobas (version 3.0; kobas.cbi.pku.edu.cn/kobas3). The KEGG pathway enrichment analysis was performed using R clusterProfiler (version 3.10.1; aipufu.com/index.html).
Cell culture and transfection
Human endometrial cancer cell lines (AN3 CA, HEC-1-B, RL95-2, HEC-1-A, Ishikawa) were purchased from the American Type Culture Collection, human endometrial epithelial cells (hEECs) were purchased from Procell Life Science & Technology Co., Ltd. (cat. no. CP-H058) and the JEC cell line was purchased from YRGene (cat. no. CC021). The cells were cultured in Dulbecco's modified Eagles medium (DMEM, HyClone; Cytiva) supplemented with 10% fetal bovine serum (FBS; HyClone; Cytiva), 100 µg/ml streptomycin and 100 U/ml penicillin (Thermo Fisher Scientific, Inc.) at 37°C and 5% CO2 conditions.
For transfection experiments, the six human endometrial cancer cell lines (2.5×105) were seeded in a 6-well plate and incubated overnight at 37°C with 5% CO2. Subsequently, cells were transfected with 100 nM miRNA mimics or inhibitors, 50 nM FAM83B small interfering (si)RNA or 50 nM FAM83B overexpression plasmid for 4 h. At 24 h post-transfection, cells were used for subsequent experiments. Sequences of siRNA and miRNA mimics/inhibitors, including the negative controls, were as follows: miR-199a-5p mimics, 5′-CCCAGUGUUCAGACUACCUGUUC-3′; miR-199b-5p mimics, 5′-CCCAGUGUUUAGACUAUCUGUUC-3′; miR-199a/b-5p mimics negative control, 5′-UCACAACCUCCUAGAGGAGAGA-3′; miR-199a-5p inhibitor, 5′-GAACAGGUAGUCUGAACACUGGG-3′; miR-199b-5p inhibitor, 5′-GAACAGGTAGTCTAAACACTGGG-3′; and miR-199a/b-5p inhibitor negative control, 5′-CAGUACUUUUGUGUAGUACAA-3′. The miR-199a/b-5p mimics/inhibitor, si-FAM83B constructs, FAM83B overexpression plasmid (pcDNA3.1 vector) and and their negative controls (scramble) were synthesized by Sangon Biotech Co., Ltd. To overexpress miR-199a/b-5p, the cells were transfected with miR-199a-5p and miR-199b-5p mimics mixed in a ratio of 1:1. All RNA transfections were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions.
Animal experiments
A total of 48 female BALB/c-nu/nu specific pathogen-free mice from Guangdong Medical Laboratory Animal Center (6-weeks old; 13–15 g) were used in the present study. Mice were housed in a temperature-controlled room (at 24±1°C) with 50±10% humidity, 12-h light/dark cycles, and standard rodent food and water ad libitum in polystyrene cages. All animal experiments were approved by the Animal Care and Use Committee of Guangzhou Medical University (Guangzhou, China; approval no. GD2019-036) and conducted according to the National Institutes of Health guidelines (24).
Subcutaneous xenograft animal model and in vivo pulmonary metastasis assay
In each subgroup, mice were lively, and daily observation revealed no abnormality in eating or any marked weight loss. Following terminal anesthesia by intraperitoneal injection with pentobarbital sodium (100 mg/kg body weight), mice were euthanized by cervical dislocation and the death of the mice was verified according to the following criteria: i) no breathing; ii) no nerve reflexes; iii) no heartbeat; and iv) relaxed muscles. The normal lung and tumor tissues were quickly dissected. The maximum tumor diameter observed for a single subcutaneous tumor during the xenograft assay was 19.7 mm.
The miR-199a/b-5p agomir/antagomir and miRNA agomir/antagomir control (scramble) constructs were purchased from Guangzhou RiboBio Co., Ltd. To overexpress miR-199a/b-5p, cells were transfected with miR-199a-5p agomir and miR-199b-5p agomir mixed in a ratio of 1:1. The sequences of the agomirs were as follows: miR-199a-5p agomir, 5′-CCCAGUGUUCAGACUACCUGUUC-3′; miR-199b-5p agomir, 5′-CCCAGUGUUUAGACUACCUGUUC-3′; miR-199a/b-5p agomir negative control, 5′-GUCUCCGUCUGUCCAAUCUAGCA-3′; miR-199a-5p antagomir, 5′-GAACAGGUAGUCUGAACACUGGG-3′; miR-199b-5p antagomir, 5′-GAACAGGTAGTCTAAACACTGGG-3′; and miR-199a/b-5p antagomir negative control, 5′-UAUUAGGCGCACAGAGGAUCGGA-3′. Ishikawa and JEC cells were transfected with 50 nM miR-199a/b-5p agomir or 100 nM antagomir at 37°C using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 4 h. At 24 h post-transfection, cells were used for subsequent experiments. Subequently, cells were cultured in complete medium at 37°C with 5% CO2. Transfected cells were harvested and resuspended in PBS. The resuspended cells (5×106) were subcutaneously injected into the right flank of 6-week-old female BALB/c nu/nu mice. Three weeks later, the mice were euthanized and the tumors were excised. The tumor volume was calculated using the following equation: Volume = (length × width2)/2. To establish an in vivo pulmonary metastasis model, 100 µl suspension of cancer cells (1×106) was injected into the tail vein of 6-week-old female BALB/c nu/nu mice. All mice were sacrificed at four weeks post-injection. Lung tissue was excised and fixed with 10% formalin for 48 h at 25°C, paraffin-embedded and sectioned into 4-µm-thick sections. At room temperature, sections were stained with hematoxylin for 4 min followed by staining with eosin for 90 sec. Stained sections were observed using a light microscope (Olympus Corporation; magnification, ×200).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from Ishikawa and JEC cell lines, as well as the lung tissues of laboratory mice using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The extracted RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.), following the manufacturer's instructions. The RT-qPCR analysis was performed using the SYBR Premix ExTaq II kit (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol, using a 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The primers used for RT-qPCR analysis were as follows: miR-199a-5p and miR-199b-5p forward, 5′-ACACTCCAGCTGGGCCCAGTGTTCAGACTAC-3′ and reverse, 5′-CTCAACTGGTGTCGTGGA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; FAM83B forward, 5′-AAAGCTCACCTCAGCATGGTT-3′ and reverse, 5′-AGCAAATGAACTAGGGACAC-3′; and GAPDH forward, 5′-GCTCATTTGCAGGGGGGAG-3′ and reverse, 5′-GTTGGTGGTGCAGGAGGCA-3′. The levels of target RNA were normalized to those of U6 or GAPDH. The relative miR-199a/b-5p and FAM83B expression levels were calculated using the 2−ΔΔCq method (25). All PCR experiments were performed in triplicate.
Western blotting
The harvested cells were lysed with ice-cold SDS lysis buffer (Beyotime Institute of Biotechnology). The protein concentration in the lysate was determined using a BCA protein assay kit (Nanjing KeyGen Biotech Co., Ltd.), following the manufacturer's instructions. Equal amounts of denatured proteins (20 µg/lane) were separated via 10% SDS-PAGE and transferred to a polyvinylidene fluoride membrane (EMD Millipore). The membrane was blocked with 5% BSA (Beijing Solarbio Science & Technology Co., Ltd.) and incubated with primary antibodies at 4°C overnight. Next, the membrane was incubated with HRP-conjugated goat anti-rabbit (1:10,000; cat. no. ab205718; Abcam) secondary antibodies for 2 h at 25°C. The protein bands were developed using ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.) and examined using the BIS 303 PC imaging system (DNR Bio-Imaging Systems Ltd.). GAPDH was used as a loading control. Protein expression levels were semi-quantified using ImageJ software (version 1.52n; National Institutes of Health). Primary antibodies against FAM83B (1:500; cat. no. PA5-28548; Invitrogen; Thermo Fisher Scientific, Inc.), E-Cadherin (1:5,000; cat. no. ab40772; Abcam), N-Cadherin (1 µg/ml; cat. no. ab18203; Abcam), Snail (0.3 µg/ml; cat. no. ab53519; Abcam), α-SMA (0.5 µg/ml; cat. no. ab5694; Abcam), vimentin (1:1,000; cat. no. ab92547; Abcam), Twist (2.5 µg/ml; cat. no. ab49254; Abcam) and GAPDH (1:10,000; cat. no. ab181602; Abcam) were used.
Transwell migration and invasion assay
Cell migration and invasion assays were performed using 24-well Transwell chambers (BD Biosciences). For the invasion assay, the Transwell chambers were precoated with Matrigel® (BD Biosciences) for 6 h at 37°C. The harvested cells (1×105) were placed in the top chamber containing serum-free medium at 37°C with 5% CO2. The bottom chamber contained complete medium supplemented with 10% FBS. The cells were incubated at 37°C and 5% CO2 for 48 h. The migrated and invaded cells on the reverse side of the chamber inserts were fixed with methanol for 30 min and stained with 0.1% crystal violet for 15 min at room temperature. The number of cells was measured in three randomly selected high-power fields across the center and the periphery of the membrane using a light microscope (Olympus Corporation; magnification, ×200). All experiments were performed in triplicates.
Dual-Luciferase reporter assay
The amplified 3′-UTR fragments of FAM83B were cloned into the psi-CHECK-2 luciferase miRNA expression reporter vector (Promega Corporation). The 293T cells (2×104; cat. no. CRL-11268; ATCC) were cultured in 24-well plates with DMEM (HyClone; Cytiva) supplemented with 10% FBS. Subsequently, cells were transfected with 50 nM miR-199a/b-5p mimics, 100 nM miR-199a/b-5p inhibitor or mimic negative control (NC), 0.5 µg of psi-CHECK-2 luciferase reporter vector containing the wild-type (WT) or mutant (Mut) 3′-UTR sequences of FAM83B, or empty vector. Transfections were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. At 48 h post-transfection, the luciferase activity was assessed using the Dual-Luciferase Reporter Assay System (GloMax; Promega). Firefly luciferase activities were normalized to Renilla luciferase activities. Three independent experiments were performed in duplicates.
Statistical analysis
Data are presented as the mean ± standard deviation. Multiple comparisons were performed using one-way ANOVA followed by Dunnett's post-hoc test. All statistical analyses were performed using SPSS (version 19.0; IBM Corp.). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-199a/b-5p expression is downregulated in patients with endometrial cancer
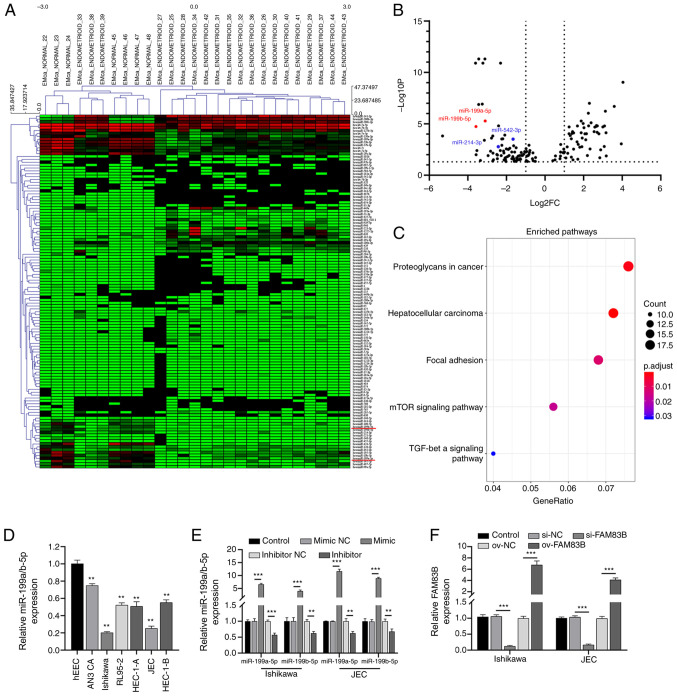
The cluster analysis of the GSE25405 dataset revealed 150 differentially expressed miRNAs between the non-cancerous endometrial tissues and human endometrial cancer tissues (Fig. 1A and B). Based on the criteria of P<0.01 and log2 FC >2, 24 miRNAs were screened out with significantly different expression. Our previous study confirmed that upregulated FAM38B expression in endometrial cancer can affect the proliferation and metastasis of cancer cells (15). Therefore, FAM38 was chosen for further analysis. According to TargetScan 7.2 analysis, 4/24 significantly differentially expressed miRNAs (Fig. 1B) had binding sites with FAM38B. Therefore, miR-199a-5p and miR-199b-5p were chosen for further analysis. The KEGG pathway enrichment analysis revealed that the target genes of miR-199a/b-5p were enriched in several EMT-associated pathways, including TGF-β and mTOR (Fig. 1C).
Figure 1.
miR-199a/b-5p expression is downregulated in endometrial cancer. (A) Heatmap revealing 150 differentially expressed miRNAs. (B) Volcano plot showing the downregulated expression of miR-199a/b-5p in endometrial tissues (P<0.01; log2FC <-2. (C) The signaling pathways identified via KEGG enrichment analysis. RT-qPCR analysis of miR-199a/b-5p expression in (D) six cancer cell lines and hEECs and (E) Ishikawa and JEC cells after transfection with miR-199a-5p and miR-199b-5p mimics and inhibitors. (F) RT-qPCR analysis of FAM83B expression in Ishikawa and JEC cells after FAM83B knockdown or overexpression. **P<0.01 and ***P<0.001 vs. hEEC or NC. miR/miRNA, microRNA; si, small interfering; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; hEEC, human endometrial epithelial cell; FAM83B, family with sequence similarity 83, member B; ov, overexpression.
miR-199a/b-5p expression is downregulated in human endometrial cancer cell lines
The expression levels of miR-199a/b-5p were examined in different endometrial cancer cells. RT-qPCR analysis revealed that the expression levels of miR-199a/b-5p were significantly downregulated in all six endometrial cancer cell lines (AN3 CA, HEC-1-B, RL95-2, HEC-1-A, Ishikawa and JEC) compared with those in the hEECs (Fig. 1D). The JEC and Ishikawa cell lines were chosen for further analysis as they exhibited the lowest expression levels of miR-199a/b-5p. Additionally, the RT-qPCR analysis revealed that compared with the respective negative control, the expression levels of miR-199a/b-5p in Ishikawa and JEC cells were significantly upregulated after transfection with miR-199a-5p and miR-199b-5p mimics, and significantly downregulated after transfection with miR-199a-5p and miR-199b-5p inhibitors (Fig. 1E), which indicated that the miR-199a-5p and miR-199b-5p mimics and inhibitors were effective. Transfection with si-FAM83B knocked down FAM83B with an efficiency of >80% in both Ishikawa and JEC cells (Fig. 1F). Compared with the mimic NC-transfected group, the FAM83B mimic-transfected group exhibited significantly increased FAM83B expression (Fig. 1F), which indicated that the overexpression of FAM83B was effective.
miR-199a/b-5p inhibits the migration and invasion of JEC and Ishikawa cells
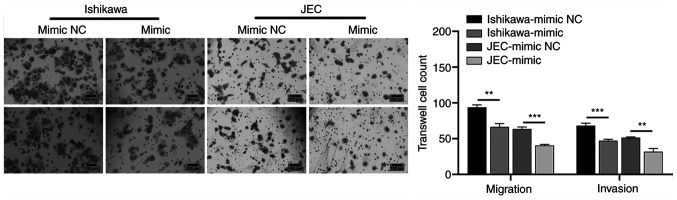
The Ishikawa and JEC cells were transfected with miR-199a/b-5p mimics to examine the effect of miR-199a/b-5p on the migration and invasion of cancer cells. The results of the Transwell assay revealed that the migratory and invasive abilities of the miR-199a/b-5p mimic-transfected cells at 48 h post-transfection were significantly lower compared with those of the mimic NC-transfected cells (Fig. 2).
Figure 2.
miR-199a/b-5p decreases the migratory and invasive abilities of Ishikawa and JEC cells. Transwell results demonstrated that, compared with those in the NC group, the migration and invasion of Ishikawa and JEC cells were significantly lower in the miR-199a/b-5p mimic-transfected group (magnification, ×200). **P<0.01; ***P<0.001. NC, negative control; miR, microRNA.
miR-199a/b-5p inhibits the EMT signaling pathway in Ishikawa and JEC cells
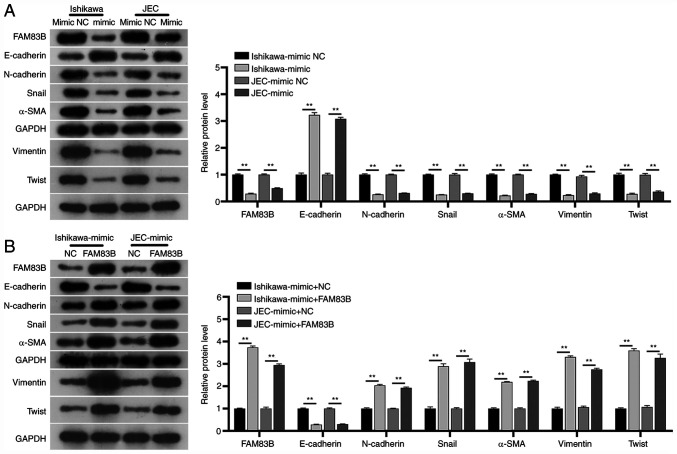
The signaling pathway through which miR-199a/b-5p mediates cell migration and invasion in the Ishikawa and JEC cells was investigated. Western blot analysis revealed that the protein expression levels of FAM83B, N-cadherin, Snail, α-SMA, vimentin and Twist in the miR-199a/b-5p mimic-transfected Ishikawa and JEC cells were significantly lower compared with those in the mimic NC-transfected Ishikawa and JEC cells at 48 h post-transfection. By contrast, transfection with the miR-199a/b-5p mimics significantly upregulated the expression levels of E-cadherin compared with those of the mimic NC-transfected Ishikawa and JEC cells (Fig. 3A). These findings suggested that miR-199a/b-5p inhibited the EMT signaling pathway in Ishikawa and JEC cells.
Figure 3.
Effect of miR-199a/b-5p/FAM83B network on the EMT signaling pathway. (A) Western blot analysis indicated that transfection with miR-199a/b-5p mimics inhibited EMT signaling in the Ishikawa and JEC cells. The protein expression levels of FAM83B, N-cadherin, Snail, α-SMA, vimentin and Twist were downregulated, whereas those of E-cadherin were upregulated in the miR-199a/b-5p mimic-transfected cells. (B) Western blot analysis revealed that transfection with miR-199a/b-5p and FAM83B mimics activated the EMT signaling pathway in Ishikawa and JEC cells. The protein expression levels of FAM83B, N-cadherin, Snail, α-SMA, vimentin and Twist were upregulated, whereas those of E-cadherin were downregulated in the FAM83B mimic-transfected cells. **P<0.01. EMT, epithelial-to-mesenchymal transition; miR, microRNA; NC, negative control; α-SMA, α-smooth muscle actin; FAM83B, family with sequence similarity 83, member B.
FAM83B attenuates the miR-199a/b-5p-induced EMT signaling pathway inhibition in Ishikawa and JEC cells
The expression levels of N-cadherin, Snail, α-SMA, vimentin and Twist were evaluated in the FAM83B mimic-transfected Ishikawa and JEC cells at 48 h post-transfection. Transfection with the FAM83B mimic significantly attenuated the miR-199a/b-5p mimic-induced downregulation of the expression levels of N-cadherin, Snail, α-SMA, vimentin and Twist in the Ishikawa and JEC cells (Fig. 3B). Additionally, transfection with FAM83B mimics significantly downregulated the expression levels of E-cadherin compared with those of mimic NC-transfected Ishikawa and JEC cells (Fig. 3B). These findings suggested that FAM83B may activate the EMT signaling pathway in Ishikawa and JEC cells.
FAM83B attenuates the miR-199a/b-5p-induced inhibition of the migratory and invasive abilities of Ishikawa and JEC cells
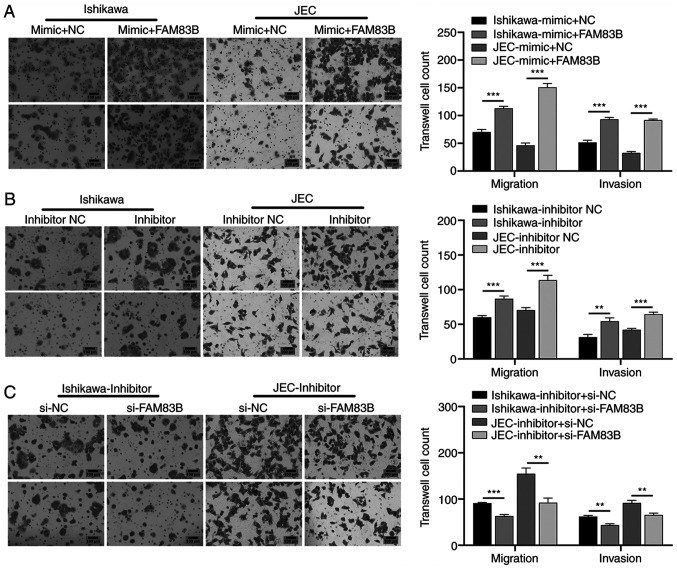
To evaluate the effects of activating the EMT signaling pathway on cancer cell migration and invasion, Ishikawa and JEC cells were further transfected with FAM83B mimics for 48 h. Compared with the miR-199a/b-5p mimic-transfected Ishikawa and JEC cells, the FAM83B mimic-transfected Ishikawa and JEC cells exhibited significantly higher migratory and invasive abilities (Fig. 4A). Additionally, the effects of miR-199a/b-5p inhibitors and FM83B knockdown on the migration and invasion of Ishikawa and JEC cells were examined. Transfection with miR-199a/b-5p inhibitors significantly enhanced the migration and invasion of Ishikawa and JEC cells compared with those of inhibitor NC-transfected Ishikawa and JEC cells (Fig. 4B). By contrast, transfection with si-FAM83B significantly attenuated the miR-199a/b-5p inhibitor-induced migration and invasion of Ishikawa and JEC cells (Fig. 4C). These findings indicated that FAM83B attenuated the miR-199a/b-5p-induced inhibition of the migration and invasion of Ishikawa and JEC cells.
Figure 4.
Effect of miR-199a/b-5p/FAM83B network on migration and invasion of Ishikawa and JEC cells. (A) Migration and invasion of the miR-199a/b-5p mimic-transfected Ishikawa and JEC cells were significantly lower than those in the FAM83B mimic-transfected Ishikawa and JEC cells (magnification, ×200). (B) Transfection with miR-199a/b-5p inhibitors significantly enhanced the migration and invasion of Ishikawa and JEC cells (magnification, ×200). (C) Migration and invasion of the miR-199a/b-5p inhibitor-transfected Ishikawa and JEC cells were significantly higher than those of the si-FAM83B-transfected Ishikawa and JEC cells (magnification, ×200). **P<0.01; ***P<0.001. NC, negative control; miR, microRNA; si, small interfering; FAM83B, family with sequence similarity 83, member B.
miR-199a/b-5p regulates metastasis
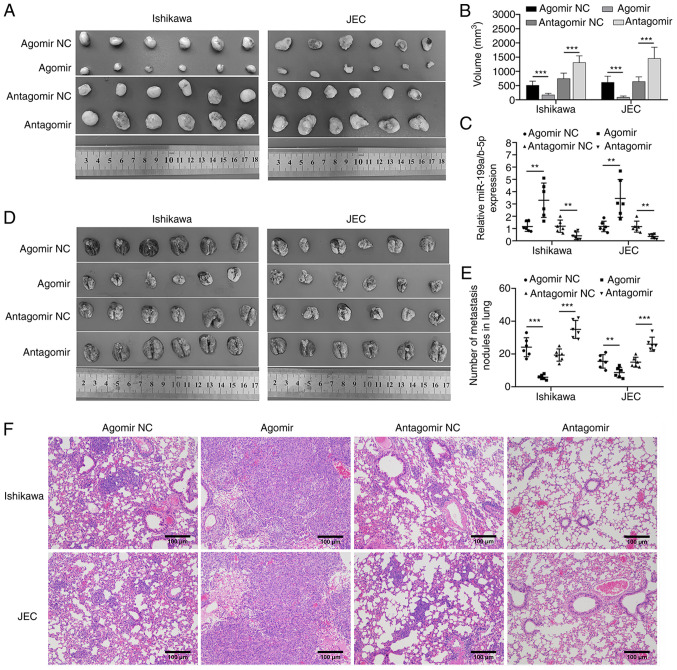
The role of miR-199a/b-5p in regulating metastasis of endometrial cancer cells was examined using the BALB/c nu/nu mouse subcutaneous tumor and pulmonary metastasis models. The administration of miR-199a/b-5p agomir-transfected cancer cells significantly inhibited the growth of subcutaneous tumors in the nude mice compared with that of the agomir NC-transfected group (Fig. 5A and B). By contrast, the administration of miR-199a/b-5p antagomir-transfected cancer cells significantly enhanced the growth of subcutaneous tumors compared with that of the antagomir NC-transfected group (Fig. 5A and B). These findings suggested that miR-199a/b-5p inhibited the growth of endometrial cancer cells in vivo. Meanwhile, miR-199a/b-5p expression levels in tumors were also analyzed via RT-qPCR. The results confirmed successful transfection of miR-199a/b-5p agomir and antagomir in subcutaneous tumors. (Fig. 5C). Additionally, a pulmonary metastasis model was established by injecting the cancer cells through the tail vein. Compared with the respective control groups, miR-199a/b-5p agomir significantly inhibited the lung metastasis and tumor nodules, whereas miR-199a/b-5p antagomir significantly enhanced lung metastasis and tumor nodules. (Fig. 5D-F).
Figure 5.
miR-199a/b-5p inhibited the growth of subcutaneous tumors and lung metastasis of cancer cells. (A) Injecting nude mice with miR-199a/b-5p agomir-transfected Ishikawa and JEC cells significantly inhibited the growth of subcutaneous tumors. By contrast, injecting nude mice with miR-199a/b-5p antagomir-transfected Ishikawa and JEC cells significantly promoted the growth of subcutaneous tumors. (B) Tumor volume of each subgroup of tumor tissues. (C) Reverse transcription-quantitative PCR analysis of the expression levels of miR-199a/b-5p in each subgroup of tumor tissues. (D) Lung tissues were dissected and the diffusion effects of Ishikawa and JEC cells transfected with agomir NC, agomir, antagomir NC or antagomir were analyzed. (E) Number of tumor nodules on the surface of lung tissues. (F) Hematoxylin and eosin staining analysis revealed that compared with that in the control groups, the lung metastasis of cancer cells was lower in mice injected with agomir-transfected Ishikawa and JEC cells and higher in mice injected with antagomir-transfected Ishikawa and JEC cells (magnification, ×200). **P<0.01; ***P<0.001. NC, negative control; miR, microRNA.
FAM83B is a direct target of miR-199a/b-5p
The functions of miR-199a/b-5p in endometrial cancer cell was validated using FAM83B as a target. Single putative miR-199a/b-5p recognition site was predicted within the FAM83B 3′-UTR sequences. FAM83B contained a 7-nucleotide site within its 3′-UTR, which matched with the seed region of miR-199a/b-5p (Fig. 6A).
Figure 6.
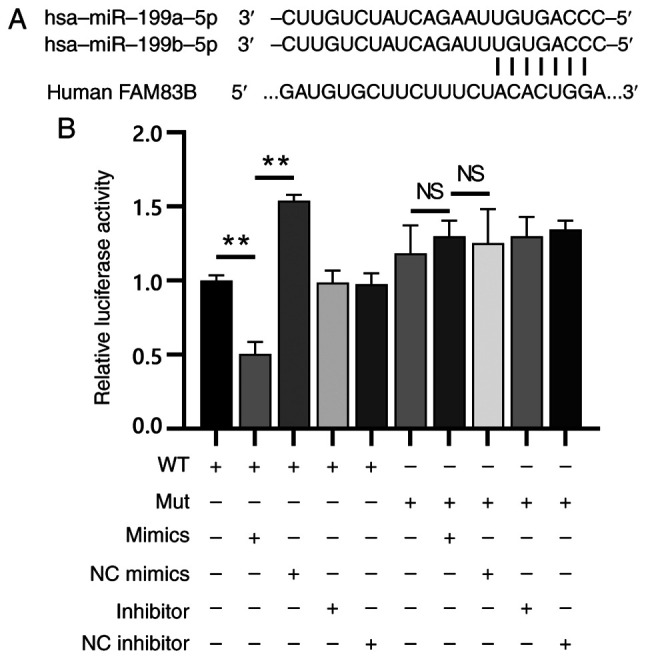
FAM83B is a direct target of miR-199a/b-5p. (A) TargetScan predicted that the nucleotides 672–679 of the FAM83B 3′-UTR were the target site of miR-199a/b-5p. (B) Dual-luciferase reporter assay revealed that the luciferase activity of the WT FAM83B 3′-UTR construct in the miR-199a/b-5p mimic-transfected group was significantly lower than that in the mimic NC-transfected and WT (blank) groups. The luciferase activity of the mutant FAM83B 3′-UTR construct in the miR-199a/b-5p mimic-transfected group was similar to that in the mimic NC-transfected and Mut (blank) groups. **P<0.01; NS, not significant (P>0.05). NC, negative control; miR, microRNA; UTR, untranslated region; WT, wild-type; Mut, mutant; FAM83B, family with sequence similarity 83, member B.
The mechanism through which miR-199a/b-5p modulates FAM83B expression was examined by introducing the 3′-UTR of FAM83B containing one miR-199a/b-5p binding site downstream of a luciferase reporter. The luciferase activity of WT FAM83B 3′-UTR construct in cells transfected with miR-199a/b-5p mimics was significantly lower compared with that in cells transfected with the mimic NC (Fig. 6B). A luciferase reporter containing the Mut FAM83B 3′-UTR binding site of miR-199a/b-5p was also constructed. The luciferase activity of the Mut FAM83B 3′-UTR construct was not suppressed upon transfection with miR-199a/b-5p mimics or inhibitor (Fig. 6B). This indicated that miR-199a/b-5p may bind selectively to mRNAs and that the single recognition element identified in the 3′-UTR of the FAM83B mRNA may be sufficient for miR-199a/b-5p activity. These results indicated that miR-199a/b-5p directly targeted FAM83B.
Discussion
Globally, the incidence and mortality rates of endometrial cancer, which is one of the most common malignancies among women, have increased (26). Previous studies have suggested that various abnormally expressed miRNAs are potential predictive biomarkers for the diagnosis or prognosis of endometrial cancer (27,28). However, the molecular mechanisms underlying the pathogenesis of endometrial cancer have not been elucidated. In the present study, miR-199a/b-5p expression was downregulated in human endometrial cancer cells compared with in hEECs. Additionally, miR-199a/b-5p inhibited the migration and invasion of the endometrial cancer cells through the suppression of EMT-associated factors and EMT signaling pathway. However, FAM83B attenuated the miR-199a/b-5p-induced inhibition of cancer cell migration and invasion through the activation of the EMT signaling pathway. Additionally, miR-199a/b-5p was bound to the nucleotides 672–679 of the FAM83B 3′-UTR.
Our previous study demonstrated that the upregulated FAM83B expression is associated with decreased survival in patients with endometrial cancer (15). Grant (29) demonstrated that FAM83B is a candidate oncogene that mediates tyrosine kinase inhibitor resistance. In contrast to FAM83B expression, the expression levels of miR-199a/b-5p in endometrial cancer cells were significantly downregulated compared with those in hEECs. Previous studies on cancer, including gastric (11), lung (30) and colon (31) cancer, have focused on either FAM83B or miR-199a/b-5p, but have not evaluated these two markers simultaneously. Hwang et al (32) demonstrated that miR-199b-5p may be a novel biomarker for sporadic and hereditary parathyroid tumors and that both miR-199b-5p and FAM83B may be potential diagnosis or prognosis markers for parathyroid tumors. Compared with healthy individuals, patients with endometrial cancer exhibited downregulated miR-199a/b-5p expression and upregulated FAM83B expression (32). Therefore, the simultaneous analysis of miR-199a/b-5p and FAM83B may improve the accuracy and precision of cancer diagnosis.
EMT, which is reported in various types of tumor, including breast and lung cancer (33), can promote tumor metastasis and invasion (34). The inhibition of EMT is a novel therapeutic strategy for cancer (33,35,36). A previous study demonstrated that miR-199b-5p attenuates TGF-β1-induced EMT by binding to the 3′-UTR of N-cadherin mRNA (37). In the present study, miR-199a/b-5p inhibited EMT by binding to the 672–679 nucleotides of the FAM83B 3′-UTR. Mechanistically, FAM83B attenuates the miR-199a/b-5p-induced inhibition of cancer cell migration and invasion through the activation of the EMT signaling pathway. The current findings indicated that the regulatory network of EMT, which promotes endometrial cancer metastasis, may involve FAM83B and miRNAs, such as miR-199a/b-5p.
The present study had a number of limitations. First, the in vitro regulatory mechanism underlying miR-199a/b-5p-mediated FAM83B expression was not investigated in the present study, although KEGG enrichment studies were performed. Additionally, the effects of other hypothesized target genes that were predicted to bind to miR-199a/b-5p in endometrial cancer cells were not investigated. Therefore, further investigations are acquired.
In conclusion, the present study demonstrated that miR-199a/b-5p was downregulated in endometrial cancer and that FAM83B promoted cancer cell metastasis and invasion by activating the EMT signaling pathway. Additionally, miR-199a/b-5p was demonstrated to bind to the nucleotides 672–679 of the FAM83B 3′-UTR. The current findings may improve the understanding of the role of FAM83B in endometrial cancer. Furthermore, the present study indicated that FAM83B may be a potential therapeutic target for endometrial cancer. The elucidation of the role of miR-199a/b-5p and FAM83B in EMT will aid in improving diagnosis and developing therapeutic agents for endometrial cancer, as well as evaluating drug efficacy and prognosis in patients with endometrial cancer. The role of miRNAs and FAM83B in endometrial cancer should be evaluated using an independent large population in future studies.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- EMT
epithelial-to-mesenchymal transition
- UTR
untranslated region
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- α-SMA
α-smooth muscle actin
- RT-qPCR
reverse transcription-quantitative PCR
- FC
fold change
- miRNA/miR
microRNA
- siRNA
small interfering RNA
- FAM83B
family with sequence similarity 83, member B
- hEECs
human endometrial epithelial cells
Funding Statement
The present study was supported by the Guangzhou Medical University PhD Start Fund (grant no. 2016C22), Guangzhou Medical University PhD Start Fund (grant no. 2016C24), Guangzhou Medical University Affiliated Third Hospital Elite Talent Program Initiation Fund.
Funding
The present study was supported by the Guangzhou Medical University PhD Start Fund (grant no. 2016C22), Guangzhou Medical University PhD Start Fund (grant no. 2016C24), Guangzhou Medical University Affiliated Third Hospital Elite Talent Program Initiation Fund.
Availability of data and materials
The datasets generated and/or analyzed during the current study (GSE25405 and GSE35794) are available in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) repository. All other data are available from the corresponding author on reasonable request.
Authors' contributions
HX, NW and QL conceived and designed the study and developed the methodology. HX, NW, HC, MZ and QL performed the experiments and collected the data. HX, NW and QL analyzed and interpreted the data. HX and NW drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures involving mice and the corresponding experimental protocols were approved by the Animal Care and Use Committee of Guangzhou Medical University (Guangzhou, China; approval no. GD2019-036).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387:1094–1108. doi: 10.1016/S0140-6736(15)00130-0. [DOI] [PubMed] [Google Scholar]
- 2.Geng YH, Wang ZF, Jia YM, Zheng LY, Chen L, Liu DG, Li XH, Tian XX, Fang WG. Genetic polymorphisms in CDH1 are associated with endometrial carcinoma susceptibility among Chinese Han women. Oncol Lett. 2018;16:6868–6878. doi: 10.3892/ol.2018.9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez AM, Schmeler KM, Kuo YF. Lack of improvement in survival rates for women under 50 with endometrial cancer, 2000–2011. J Cancer Res Clin Oncol. 2016;142:783–793. doi: 10.1007/s00432-015-2092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang HH, Li R, Li YJ, Yu XX, Sun QN, Li AY, Kong Y. eIF4Erelated miR320a and miR3405p inhibit endometrial carcinoma cell metastatic capability by preventing TGF-β-1induced epithelial-mesenchymal transition. Oncol Rep. 2020;43:447–460. doi: 10.3892/or.2019.7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su J, Morgani SM, David CJ, Wang Q, Er EE, Huang YH, Basnet H, Zou Y, Shu W, Soni RK, et al. TGF-beta orchestrates fibrogenic and developmental EMTs via the RAS effector RREB1. Nature. 2020;577:566–571. doi: 10.1038/s41586-019-1897-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qureshi R, Arora H, Rizvi MA. EMT in cervical cancer: Its role in tumour progression and response to therapy. Cancer Lett. 2015;356:321–331. doi: 10.1016/j.canlet.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 7.Mutlu M, Raza U, Saatci O, Eyupoglu E, Yurdusev E, Sahin O. miR-200c: A versatile watchdog in cancer progression, EMT, and drug resistance. J Mol Med (Berl) 2016;94:629–644. doi: 10.1007/s00109-016-1420-5. [DOI] [PubMed] [Google Scholar]
- 8.Padmanaban V, Krol I, Suhail Y, Szczerba BM, Aceto N, Bader JS, Ewald AJ. E-cadherin is required for metastasis in multiple models of breast cancer. Nature. 2019;573:439–444. doi: 10.1038/s41586-019-1526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li CF, Chen JY, Ho YH, Hsu WH, Wu LC, Lan HY, Hsu DS, Tai SK, Chang YC, Yang MH. Snail-induced claudin-11 prompts collective migration for tumour progression. Nat Cell Biol. 2019;21:251–262. doi: 10.1038/s41556-018-0268-z. [DOI] [PubMed] [Google Scholar]
- 10.Shen CQ, Yan TT, Liu W, Zhu XQ, Tian XL, Fu XL, Hua R, Zhang JF, Huo YM, Liu DJ, et al. High expression of FAM83B predicts poor prognosis in patients with pancreatic ductal adenocarcinoma and correlates with cell cycle and cell proliferation. J Cancer. 2017;8:3154–3165. doi: 10.7150/jca.20086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou Z, Ma T, He X, Zhou J, Ma H, Xie M, Liu Y, Lu D, Di S, Zhang Z. Long intergenic non-coding RNA 00324 promotes gastric cancer cell proliferation via binding with HuR and stabilizing FAM83B expression. Cell Death Dis. 2018;9:717. doi: 10.1038/s41419-018-0758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cipriano R, Graham J, Miskimen KL, Bryson BL, Bruntz RC, Scott SA, Brown HA, Stark GR, Jackson MW. FAM83B mediates EGFR- and RAS-driven oncogenic transformation. J Clin Invest. 2012;122:3197–3210. doi: 10.1172/JCI60517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. FAM83B-mediated activation of PI3K/AKT and MAPK signaling cooperates to promote epithelial cell transformation and resistance to targeted therapies. Oncotarget. 2013;4:729–738. doi: 10.18632/oncotarget.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamaura T, Ezaki J, Okabe N, Takagi H, Ozaki Y, Inoue T, Watanabe Y, Fukuhara M, Muto S, Matsumura Y, et al. Family with sequence similarity 83, member B is a predictor of poor prognosis and a potential therapeutic target for lung adenocarcinoma expressing wild-type epidermal growth factor receptor. Oncol Lett. 2018;15:1549–1558. doi: 10.3892/ol.2017.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Q, Chen H, Zhang M, Xiong H, Jiang Q. Knocking down FAM83B inhibits endometrial cancer cell proliferation and metastasis by silencing the PI3K/AKT/mTOR pathway. Biomed Pharmacother. 2019;115:108939. doi: 10.1016/j.biopha.2019.108939. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Ye B, Wang P, Yao F, Zhang C, Yu G. Overview of microRNA-199a Regulation in Cancer. Cancer Manag Res. 2019;11:10327–10335. doi: 10.2147/CMAR.S231971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye H, Pang L, Wu Q, Zhu Y, Guo C, Deng Y, Zheng X. A critical role of mir-199a in the cell biological behaviors of colorectal cancer. Diagn Pathol. 2015;10:65. doi: 10.1186/s13000-015-0260-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y, Liu J, Jiang B, Chen J, Fu Z, Bai F, Jiang J, Tang Z. MiR-199a-5p loss up-regulated DDR1 aggravated colorectal cancer by activating epithelial-to-mesenchymal transition related signaling. Dig Dis Sci. 2014;59:2163–2172. doi: 10.1007/s10620-014-3136-0. [DOI] [PubMed] [Google Scholar]
- 19.Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T, Cassandrini P, Zamboni G, Maciejewski R. Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer. 2012;12:369. doi: 10.1186/1471-2407-12-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang HY, Li CH, Wang XC, Luo YQ, Cao XD, Chen JJ. MiR-199 inhibits EMT and invasion of hepatoma cells through inhibition of Snail expression. Eur Rev Med Pharmacol Sci. 2019;23:7884–7891. doi: 10.26355/eurrev_201909_18998. [DOI] [PubMed] [Google Scholar]
- 21.Chu Y, Wang Y, Peng W, Xu L, Liu M, Li J, Hu X, Li Y, Zuo J, Ye Y. STAT3 activation by IL-6 from adipose-derived stem cells promotes endometrial carcinoma proliferation and metastasis. Biochem Biophys Res Commun. 2018;500:626–631. doi: 10.1016/j.bbrc.2018.04.121. [DOI] [PubMed] [Google Scholar]
- 22.Cai J, Zhang Y, Huang S, Yan M, Li J, Jin T, Bao S. MiR-100-5p, miR-199a-3p and miR-199b-5p induce autophagic death of endometrial carcinoma cell through targeting mTOR. Int J Clin Exp Pathol. 2017;10:9262–9272. [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Monteiro CD, Jagodnik KM, Fernandez NF, Gundersen GW, Rouillard AD, Jenkins SL, Feldmann AS, Hu KS, McDermott MG, et al. Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd. Nat Commun. 2016;7:12846. doi: 10.1038/ncomms12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Yu D, Liu Z, Zhou F, Dai J, Wu B, Zhou J, Heng BC, Zou XH, Ouyang H, et al. Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res Ther. 2017;8:189. doi: 10.1186/s13287-017-0632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Sorosky JI. Endometrial cancer. Obstet Gynecol. 2012;120:383–397. doi: 10.1097/AOG.0b013e3182605bf1. [DOI] [PubMed] [Google Scholar]
- 27.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, Yang X, Wang C, Zhang S, Wang Z, Li M, Wang Y, Wang X, Yang X. HDAC6, modulated by miR-206, promotes endometrial cancer progression through the PTEN/AKT/mTOR pathway. Sci Rep. 2020;10:3576. doi: 10.1038/s41598-020-60271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant S. FAM83A and FAM83B: Candidate oncogenes and TKI resistance mediators. J Clin Invest. 2012;122:3048–3051. doi: 10.1172/JCI64412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richtmann S, Wilkens D, Warth A, Lasitschka F, Winter H, Christopoulos P, Herth FJF, Muley T, Meister M, Schneider MA. FAM83A and FAM83B as prognostic biomarkers and potential new therapeutic targets in NSCLC. Cancers (Basel) 2019;11:652. doi: 10.3390/cancers11050652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao CC, Wu PH, Huang HC, Chung HY, Chou YC, Cai BH, Kannagi R. Downregulation of miR-199a/b-5p is associated with GCNT2 induction upon epithelial-mesenchymal transition in colon cancer. FEBS Lett. 2017;591:1902–1917. doi: 10.1002/1873-3468.12685. [DOI] [PubMed] [Google Scholar]
- 32.Hwang S, Jeong JJ, Kim SH, Chung YJ, Song SY, Lee YJ, Rhee Y. Differential expression of miRNA199b-5p as a novel biomarker for sporadic and hereditary parathyroid tumors. Sci Rep. 2018;8:12016. doi: 10.1038/s41598-018-30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pattabiraman DR, Bierie B, Kober KI, Thiru P, Krall JA, Zill C, Reinhardt F, Tam WL, Weinberg RA. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science. 2016;351:aad3680. doi: 10.1126/science.aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamilton G, Rath B. Mesenchymal-epithelial transition and circulating tumor cells in small cell lung cancer. Adv Exp Med Biol. 2017;994:229–245. doi: 10.1007/978-3-319-55947-6_12. [DOI] [PubMed] [Google Scholar]
- 35.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 36.Wang ZC, Gao Q, Shi JY, Guo WJ, Yang LX, Liu XY, Liu LZ, Ma LJ, Duan M, Zhao YJ, et al. Protein tyrosine phosphatase receptor S acts as a metastatic suppressor in hepatocellular carcinoma by control of epithermal growth factor receptor-induced epithelial-mesenchymal transition. Hepatology. 2015;62:1201–1214. doi: 10.1002/hep.27911. [DOI] [PubMed] [Google Scholar]
- 37.Zhou SJ, Liu FY, Zhang AH, Liang HF, Wang Y, Ma R, Jiang YH, Sun NF. MicroRNA-199b-5p attenuates TGF-beta1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Br J Cancer. 2017;117:233–244. doi: 10.1038/bjc.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study (GSE25405 and GSE35794) are available in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) repository. All other data are available from the corresponding author on reasonable request.